Abstract
Free full text

Ligand-dependent interaction of the glucocorticoid receptor with p53 enhances their degradation by Hdm2
Abstract
The glucocorticoid receptor (GR) and the tumor supressor p53 mediate different stress responses. We have studied the mechanism of their mutual inhibition in normal endothelial cells (HUVEC) in response to hypoxia, a physiological stress, and mitomycin C, which damages DNA. Dexamethasone (Dex) stimulates the degradation of endogenous GR and p53 by the proteasome pathway in HUVEC under hypoxia and mitomycin C treatments, and also in hepatoma cells (HepG2) under normoxia. Dex inhibits the functions of p53 (apoptosis, Bax, and p21WAF1/CIP1 expression) and GR (PEPCK and G-6-Pase expression). Endogenous p53 and GR form a ligand-dependent trimeric complex with Hdm2 in the cytoplasm. Disruption of the p53–HDM2 interaction prevents Dex-induced ubiquitylation of GR and p53. The ubiquitylation of GR requires p53, the interaction of p53 with Hdm2, and E3 ligase activity of Hdm2. These results provide a mechanistic basis for GR and p53 acting as opposing forces in the decision between cell death and survival.
The development of most tumors is associated with loss of function of the tumor suppressor p53. Different physiological stress, including DNA damage and hypoxia, activate p53. p53 is a transcription factor that regulates genes involved in growth arrest and apoptosis. p53 is down-regulated by its target gene product Mdm2 (human homolog Hdm2). Mdm2 forms an autoregulatory loop with p53 by binding to its N-terminal domain, inhibiting its transcriptional activity and increasing its degradation by the ubiquitin proteasome pathway (for reviews, see Jimenez et al. 1999; Lakin and Jackson 1999; Sionov and Haupt 1999). Mdm2 is a RING finger-dependent ubiquitin protein ligase for p53 and itself (Argentini et al. 2000; Fang et al. 2000; Honda and Yasuda 2000). Mdm2 also inhibits p53 by nuclear export through a mechanism involving either the nuclear export signal (NES) of Mdm2 (Tao and Levine 1999) or the RING finger of Mdm2 and the NES of p53 (Boyd et al. 2000; Geyer et al. 2000). The NES of p53 is masked in the transcriptionally active heterodimer, but is exposed in the monomeric form of p53 (Stommel et al. 1999). MdmX, a Mdm2 homolog that lacks a NES, stabilizes p53 by retention in the nucleus (Jackson and Berberich 2000; Stad et al. 2000). Nuclear p53 levels can also be maintained by ARF, which blocks nucleo-cytoplasmic shuttling of Mdm2 (Sherr and Weber 2000). Most studies of p53 involve DNA damaging drugs and radiation. Much less is known about the physiological stress, hypoxia. Under hypoxic conditions, p53 is stabilized by mitochondria through a redox-dependent mechanism (Chandel et al. 2000) and by HIF-1α (An et al. 1998), whereas p53 induces degradation of HIF-1α (Ravi et al. 2000). The response to hypoxia in vivo also involves the glucocorticoid receptor (GR) (Bauer et al. 1999).
GR is a member of the steroid receptor superfamily that mediates physiological processes controlled by glucocorticoids. In the unbound state GR is located in the cytoplasm bound to chaperones. Upon ligand binding the chaperones dissociate, exposing the NLS that enables GR to enter the nucleus and regulate transcription (Cheung and Smith 2000). GR activates transcription by binding to glucocorticoid response elements (GREs), and represses by binding to negative and composite elements (Kellendonk et al. 1999). GR mediates stress responses, and animals that overexpress GR are more resistant to stress and endotoxic shock (Reichardt et al. 2000).
Recently, evidence has been growing for cross talk between the p53 and GR-mediated responses to stress. p53 physically interacts with and represses the activities of GR (Sengupta et al. 2000b). Modulation of glucocorticoid-induced apoptosis has been linked to p53 gene dosage in mice (Mori et al. 1999). Glucocorticoids prevent p53-induced apoptosis in immortalized human granulosa cells (Sasson et al. 2001). p53 and GR regulate in opposite manners the type II hexokinase gene, whose overexpression is associated with increased glucose catabolism in cancer cells (Mathupala et al. 1997). Hypoxia leads to p53 activation and decreased glucose metabolism (Riva et al. 1998). Because little is known about the functional interactions between the signalling pathways mediated by p53 and GR under physiological conditions, we have investigated how p53 and GR regulate each other under hypoxia. We show that they mutually inhibit each other's activity by cytoplasmic sequestration in a ligand-dependent manner, which leads to increased degradation through recruitment of the E3 ubiquitin ligase Hdm2 and the proteasome pathway.
Results
Dexamethasone enhances the interaction of GR with the core domain and NLS of p53
p53
We have reported previously that p53 and GR interact in vivo (Sengupta et al. 2000b). We used in vitro GST pull-down assays to determine which region of p53 interacts with GR (Fig. (Fig.1A,1A, left). The GST fusion proteins were produced in Escherichia coli, purified on Glutathione Sepharose beads, and equivalent amounts of protein (judged by Coomassie blue staining, Fig. Fig.1B),1B), were used. Full-length p53 interacted with GR weakly but specifically in the absence of ligand (Fig. (Fig.1A,C,1A,C, lanes 3 and 6). This interaction was enhanced by the agonists Dexamethasone (Dex) or Cortisol (Cort), and inhibited by the antagonist RU486 (Fig. (Fig.1C,1C, lanes 2–6). These results show that the binding of GR to p53 is enhanced by agonists in vitro. In the presence of ligand, GR interacted with both the core domain [GST–p53(76–304)] and the NLS [GST–p53(305–320)] of p53, and more strongly with a mutant containing both regions [GST–p53(76–320)]. No interaction was detected between GR and either the N-terminal [GST–p53(1–75)] or C-terminal [GST–p53(321–362) and GST–p53(363–393)] domains. The mutant containing both interaction domains [GST–p53(76–320)] interacted more strongly than the full-length protein, raising the possibility that it lacks sequences that inhibit the interaction with GR. The interactions between GR and deletion mutants containing the NLS [GST–p53(76–320) and GST–p53(305–320)] were inhibited by a peptide containing the NLS (amino acids 305–320, Pep, Fig. Fig.1C,1C, lanes 7–11), confirming that the GR–p53 NLS interaction is specific.
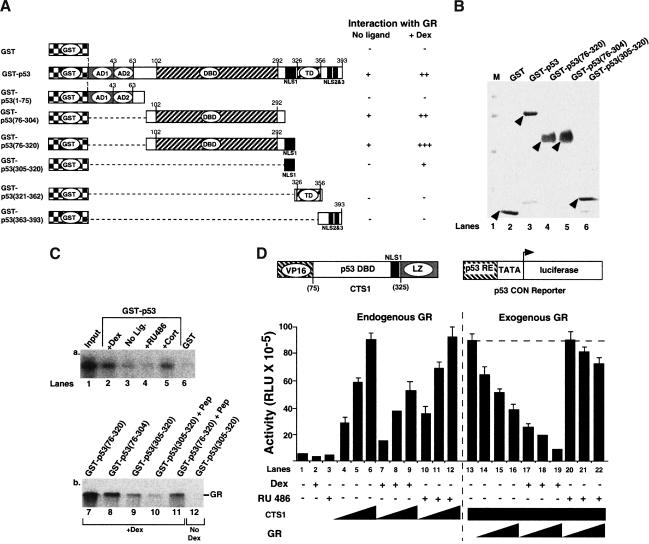
The core domain and the NLS of p53 interact with GR and attenuate transactivation by GR. (A) Schemes. The GST constructs are shown on the left, and the extent of their interaction with full-length GR is summarized on the right. AD1, activation domain 1; AD2, activation domain 2; DBD, DNA binding domain; NLS, nuclear localization signal; TD, tetramerization domain; +, detectable interaction; −, undetectable interaction. The numbers indicate the amino acids of human p53. (B) Coomassie stained gel. The proteins of interest are indicated by arrows. (Lane 1) molecular weight marker (M); (lane 2) GST; (lane 3) GST–p53; (lane 4) GST–p53(76–320); (lane 5) GST–p53(76–304); (lane 6) GST–p53(305–320). (C) Equal amounts of in vitro translated GR (lanes 2–12) were incubated with wild-type or mutant p53 fusion proteins, vehicle (ethanol), 10−7 M ligands or 50 μM peptide (Pep), as indicated. (Lane 1) Input, 5%; (lane 2) GST–p53 +
+ Dex; (lane 3) GST–p53; (lane 4) GST–p53
Dex; (lane 3) GST–p53; (lane 4) GST–p53 +
+ RU486; (lane 5) GST–p53
RU486; (lane 5) GST–p53 +
+ Cortisol; (lane 6) GST; (lane 7) GST–p53(76–320)
Cortisol; (lane 6) GST; (lane 7) GST–p53(76–320) +
+ Dex; (lane 8) GST–p53(76–304)
Dex; (lane 8) GST–p53(76–304) +
+ Dex; (lane 9) GST–p53(305–320)
Dex; (lane 9) GST–p53(305–320) +
+ Dex; (lane 10) GST–p53(305–320)
Dex; (lane 10) GST–p53(305–320) +
+ Pep
Pep +
+ Dex; (lane 11) GST–p53(76–320)
Dex; (lane 11) GST–p53(76–320) +
+ Pep
Pep +
+ Dex; (lane 12) GST–p53(305–320). (D) p53 null HSC-2 cells were transfected with CTS1 (top left), and p53 activity was measured with the p53 CON reporter (top right, all lanes). Vehicle (ethanol, −), 10−7 M Dex or 10−7 M RU486 were added as indicated. (Lane 1) No ligand; (lane 2) +Dex; (lane 3) +RU486; (lanes 4–6) CTS1 (125, 250, 500 ng/mL); (lanes 7–9) CTS1 (125, 250, 500 ng/mL) +Dex; (lanes 10–12) CTS1 (125, 250, 500 ng/mL)
Dex; (lane 12) GST–p53(305–320). (D) p53 null HSC-2 cells were transfected with CTS1 (top left), and p53 activity was measured with the p53 CON reporter (top right, all lanes). Vehicle (ethanol, −), 10−7 M Dex or 10−7 M RU486 were added as indicated. (Lane 1) No ligand; (lane 2) +Dex; (lane 3) +RU486; (lanes 4–6) CTS1 (125, 250, 500 ng/mL); (lanes 7–9) CTS1 (125, 250, 500 ng/mL) +Dex; (lanes 10–12) CTS1 (125, 250, 500 ng/mL) +
+ RU486; (lane 13) CTS1 (500 ng/mL); (lanes 14–16) CTS1 (500 ng/mL)
RU486; (lane 13) CTS1 (500 ng/mL); (lanes 14–16) CTS1 (500 ng/mL) +
+ GR (250, 500, 1250 ng/mL); (lanes 17–19) CTS1 (500 ng/mL)
GR (250, 500, 1250 ng/mL); (lanes 17–19) CTS1 (500 ng/mL) +
+ GR (250, 500, 1250 ng/mL) +Dex; (lanes 20–22) p53 (500 ng/mL)
GR (250, 500, 1250 ng/mL) +Dex; (lanes 20–22) p53 (500 ng/mL) +
+ GR (250, 500, 1250 ng/mL)
GR (250, 500, 1250 ng/mL) +
+ RU486. Luciferase activity was measured 24 h post-transfection and corrected for transfection efficiency. The values shown are from four independent experiments. VP16, HSV1 VP16 transcription activation domain; DBD, DNA binding domain; NLS, nuclear localization signal; LZ, leucine zipper; p53 RE, p53 responsive element; TATA, minimal promoter element.
RU486. Luciferase activity was measured 24 h post-transfection and corrected for transfection efficiency. The values shown are from four independent experiments. VP16, HSV1 VP16 transcription activation domain; DBD, DNA binding domain; NLS, nuclear localization signal; LZ, leucine zipper; p53 RE, p53 responsive element; TATA, minimal promoter element.
GR interacts with the central region of p53, suggesting that it should inhibit the Chimeric Tumor Supressor, CTS1, that harbors this domain of p53 linked to heterologous activation and dimerization domains (Conseiller et al. 1998) (Fig. (Fig.1D,1D, top left). The transcriptional activity of CTS1 was measured in p53 null HSC-2 cells with the p53CON reporter (Fig. (Fig.1D,1D, top right). Vehicle alone (ethanol), Dex, and RU486 did not affect the basal activity of the reporter (Fig. (Fig.1D,1D, lanes 1–3). Increasing amounts of CTS1 stimulated reporter activity in a dose-dependent manner (Fig. (Fig.1D,1D, lanes 4–6). CTS1 was inhibited around threefold by endogenous GR activated with Dex, but was not affected by RU486 (Fig. (Fig.1D,1D, lanes 7–12). With a constant level of CTS1, exogenous GR inhibited CTS1 activity to a small extent (Fig. (Fig.1D,1D, lanes 13–16), and the addition of Dex increased this inhibition (Fig. (Fig.1D,1D, lanes 17–19). In contrast, GR, in the presence of RU486, had little effect (Fig. (Fig.1D,1D, lanes 20–22). Similar results were obtained in Saos-2 cells (data not shown). These results indicate that the region of p53 that interacts with GR in vitro is sufficient for efficient repression by GR in vivo.
Dex decreases p53 and GR levels and inhibits expression of their target genes in HepG2 cells
cells
Cytoplasmic p53 in neuroblastoma cells is released from its complex with GR and translocates to the nucleus following treatment with GR antagonists (Sengupta et al. 2000b). We investigated whether cytoplasmic p53 in other tumor cell types, such as HepG2 (hepatoma) and SA1 (osteosarcoma) (Wasylyk et al. 1999; Lu et al. 2000), behaved similarly. Under normal growth conditions, Dex increased the amount of p53 and GR in the cytoplasm of both cell lines, although some p53 always remained nuclear. In contrast, RU486 significantly increased the amount of nuclear p53 (data not shown). Interestingly, in HepG2 cells, Dex caused a drastic decrease in the protein levels of both p53 and GR (Fig. (Fig.2A,2A, lanes 1,2). This effect was blocked by the proteasome inhibitor LLnL (Fig. (Fig.2A,2A, lanes 3,4), suggesting that the decrease is due to enhanced degradation. We investigated whether Dex can inhibit the expression of genes downstream from both p53 (Bax, Hdm2, and p21WAF1/CIP1) and GR (phospho enol pyruvate carboxy kinase, PEPCK; Imai et al. 1990). Dex decreased Bax and p21WAF1/CIP1 protein levels (Fig. (Fig.2B)2B) and promoter activity (reporter assays; data not shown). Dex did not affect either the level of Hdm2 protein (Fig. (Fig.2B),2B), or the activity of the promoter (reporter assay, data not shown), showing that Hdm2 expression is less sensitive than Bax or p21WAF1/CIP1 to changes in p53 levels. Dex decreased PEPCK protein levels (Fig. (Fig.2B).2B). These results show that Dex down-regulates both p53 and GR protein levels and the expression of their downstream targets in HepG2 cells.
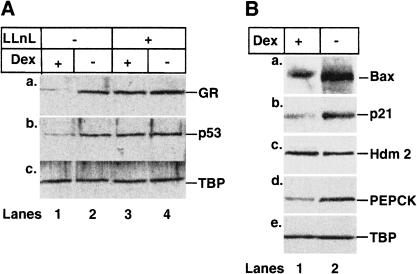
p53 and GR undergo ligand and proteasome-dependent degradation in HepG2 cells. (A) HepG2 cells were grown for 12 h with vehicle (ethanol), 10−7 M Dex, and 50 μM LLnL, as indicated. Proteins (50 μg) were analyzed by Western blotting with antibodies to: (a) GR (E-20 and P-20); (b) p53 (DO-1); (c) TBP (3G3). (Lane 1) +Dex; (lane 2) (−, vehicle); (lane 3) +Dex +
+ LLnL; (lane 4) (−, vehicle)
LLnL; (lane 4) (−, vehicle) +
+ LLnL. (B) Lysates from cells incubated in the absence of LLnL were Western blotted with antibodies to: (a) Bax (N-20); (b) p21WAF1/CIP1 (1WA-IC581); (c) Hdm2 (2A10); (d) PEPCK; (e) TBP (3G3). (Lane 1) +Dex; (lane 2) (−, vehicle).
LLnL. (B) Lysates from cells incubated in the absence of LLnL were Western blotted with antibodies to: (a) Bax (N-20); (b) p21WAF1/CIP1 (1WA-IC581); (c) Hdm2 (2A10); (d) PEPCK; (e) TBP (3G3). (Lane 1) +Dex; (lane 2) (−, vehicle).
Dex activation of endogenous GR inhibits p53 induction by hypoxia in normal endothelial cells (HUVEC)
(HUVEC)
HepG2 is a tumor cell line with undefined genetic alterations. Hence, we investigated in normal human cells (HUVEC) the response to hypoxia, a natural inducer of p53. Normally growing HUVEC express very little or no p53 protein (Fig. (Fig.3A,3A, lane 1). p53 was strongly induced after 12 h of hypoxia, and increased further after 18 h (Fig. (Fig.3A,3A, lanes 3,5). Dex inhibited the induction of p53 up to 36 h (Fig. (Fig.3A,3A, lanes 2–5; data not shown). The proteasome inhibitor, LLnL, decreased Dex-dependent inhibition of p53 induction, without substantially affecting the level of p53 in the absence of ligand (Fig. (Fig.3A,3A, lanes 6–9). p53 RNA levels detected by Northern blotting were not affected by Dex (Fig. (Fig.3B),3B), showing that the inhibitory effect of Dex on p53 is not due to changes in transcription or RNA stability. In conclusion, Dex decreases p53 stability by enhanced proteasomal degradation.
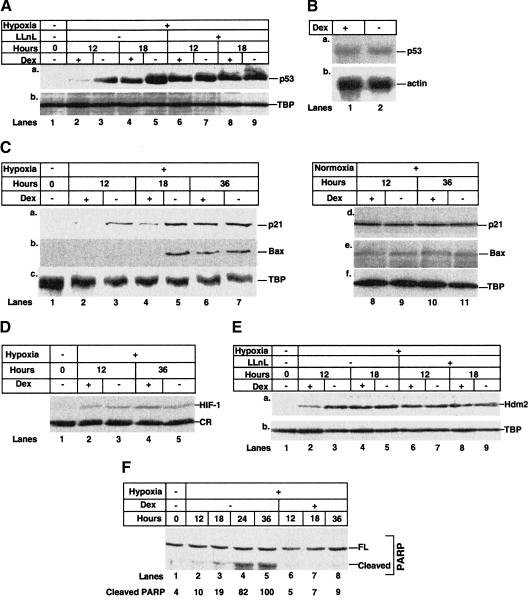
Dex inhibits induction by hypoxia of p53 protein levels, expression of p53 target genes, and apoptosis. (A) HUVEC cells were grown for different times with, as indicated, vehicle (ethanol), 10−7 M Dex, and 50 μM LLnL, under normoxic (−, lane 1) or hypoxic (lanes 2–9) conditions. Lysates (50 μg) were analyzed by SDS-PAGE and Western blotting with antibodies against (a) p53 (DO-1); (b) TBP (3G3). (Lanes 1,3,5) (−, vehicle); (lanes 2,4) +Dex; (lanes 6,8) +Dex +
+ LLnL; (lanes 7,9) (−, vehicle)
LLnL; (lanes 7,9) (−, vehicle) +
+ LLnL. (B) Total RNA was extracted from HUVEC cells grown with vehicle (ethanol) or 10−7 M Dex and hypoxia for 18 h. The RNAs (40 μg) were run on formaldehyde–agarose gels and northerns were hybridized with probes against (a) p53, (b) β-actin. (Lane 1) +Dex; (lane 2) (−, vehicle). (C) HUVEC were grown under normoxia (lanes 1,8–11) or hypoxia (lanes 2–7) with vehicle (ethanol) or Dex. Lysates (50 μg, lanes 1–7; 200 μg, lanes 8–11) were Western blotted with antibodies against (a,d) p21WAF1/CIP1 (1WA-IC581); (b,e) Bax (N-20); (c,f) TBP (3G3). (Lanes 1, 3,5,7,9,11) (−, vehicle); (lanes 2,4,6,8,10) +Dex. (D) HUVEC were grown under normoxia (lane 1) or hypoxia (lanes 2–5) with vehicle (ethanol) or Dex. Lysates (50 μg) were Western blotted with antibodies against HIF-1α (C-19). (Lanes 1,3,5) (−, vehicle); (lanes 2,4) +Dex. CR, cross-reacting band. (E) HUVEC were grown under normoxia (lane 1) or hypoxia (lanes 2–9) with vehicle (ethanol), Dex, and 50 μM LLnL, as indicated. Lysates (50 μg) were Western blotted with antibodies to (a) Hdm2 (2A10), (b) TBP (3G3). (Lanes 1,3,5,7,9) (−, vehicle); (lanes 2,4,6,8) +Dex. (F) HUVEC were grown under normoxia (lane 1) or hypoxia (lanes 2–8). Lysates (50 μg) were Western blotted with polyclonal antibodies against PARP. (Lanes 1–5) (−, vehicle); (lanes 6–8) +Dex. FL, full length. The cleavage products were scanned and the values are the amount of the cleaved product relative to lane 5.
LLnL. (B) Total RNA was extracted from HUVEC cells grown with vehicle (ethanol) or 10−7 M Dex and hypoxia for 18 h. The RNAs (40 μg) were run on formaldehyde–agarose gels and northerns were hybridized with probes against (a) p53, (b) β-actin. (Lane 1) +Dex; (lane 2) (−, vehicle). (C) HUVEC were grown under normoxia (lanes 1,8–11) or hypoxia (lanes 2–7) with vehicle (ethanol) or Dex. Lysates (50 μg, lanes 1–7; 200 μg, lanes 8–11) were Western blotted with antibodies against (a,d) p21WAF1/CIP1 (1WA-IC581); (b,e) Bax (N-20); (c,f) TBP (3G3). (Lanes 1, 3,5,7,9,11) (−, vehicle); (lanes 2,4,6,8,10) +Dex. (D) HUVEC were grown under normoxia (lane 1) or hypoxia (lanes 2–5) with vehicle (ethanol) or Dex. Lysates (50 μg) were Western blotted with antibodies against HIF-1α (C-19). (Lanes 1,3,5) (−, vehicle); (lanes 2,4) +Dex. CR, cross-reacting band. (E) HUVEC were grown under normoxia (lane 1) or hypoxia (lanes 2–9) with vehicle (ethanol), Dex, and 50 μM LLnL, as indicated. Lysates (50 μg) were Western blotted with antibodies to (a) Hdm2 (2A10), (b) TBP (3G3). (Lanes 1,3,5,7,9) (−, vehicle); (lanes 2,4,6,8) +Dex. (F) HUVEC were grown under normoxia (lane 1) or hypoxia (lanes 2–8). Lysates (50 μg) were Western blotted with polyclonal antibodies against PARP. (Lanes 1–5) (−, vehicle); (lanes 6–8) +Dex. FL, full length. The cleavage products were scanned and the values are the amount of the cleaved product relative to lane 5.
The products of the p53 downstream genes Bax and p21WAF1/CIP1 were induced by hypoxia (Fig. (Fig.3C,3C, lanes 1,3,5,7). p21WAF1/CIP1 has also been shown to be induced by hypoxia in normal fibroblasts but not in several transformed cell lines (Ashcroft et al. 2000; Koumenis et al. 2001). Bax and p21WAF1/CIP1 induction was inhibited by Dex (Fig. (Fig.3C,3C, lanes 2,4,6). Under normoxia, in which p53 levels are very low, Dex had no effect, indicating that Dex inhibition of p53 downstream genes is p53 dependent (Fig. (Fig.3C,3C, lanes 8–11). HIF-1α stabilizes p53 in response to hypoxia (An et al. 1998), which raised the possibility that it, too, is affected by Dex. However, HIF-1α induction by hypoxia in HUVEC was not significantly affected by Dex (Fig. (Fig.3D).3D). The apparently weak induction of HIF-1α is consistent with the low level of HIF-1α RNA in endothelial cells (Tian et al. 1997). Dex did, however, inhibit Hdm2 induction until 12 h under hypoxia (Fig. (Fig.3E,3E, lanes 2,3). In the presence of LLnL the difference in the level of induction was no longer observed, indicating that the difference is due to Dex-induced proteasomal degradation (Fig. (Fig.3E,3E, lanes 6,7). After 18 and up to 36 h the levels of Hdm2 were similar, irrespective of the presence of Dex and LLnL (Fig. (Fig.3E,3E, lanes 4,5,8,9; data not shown). Hypoxia induces apoptosis of HUVEC in a p53-dependent manner (Stempien-Otero et al. 1999). We, therefore, investigated whether Dex affected apoptosis by following PARP cleavage and DNA fragmentation. PARP cleavage, an early marker of apoptosis, was detected at 18 h and increased up to 36 h (Fig. (Fig.3F,3F, lanes 1–5). Dex inhibited PARP cleavage up to 36 h under hypoxia (Fig. (Fig.3F,3F, lanes 6–8). Similar results were obtained using DNA fragmentation (data not shown). These results show that the activation of endogenous GR with Dex in HUVEC, as well as HepG2, dramatically decreases p53 downstream functions.
Dex-dependent GR degradation by a proteasome pathway and inhibition of its target genes under hypoxia
hypoxia
Hypoxia has been shown to affect GR functions and levels of expression in several systems (Jenq et al. 1996; Bauer et al. 1999). Therefore, we investigated whether hypoxia affects GR in HUVEC. Hypoxia alone caused a reproducible increase in GR protein levels (Fig. (Fig.4A,4A, lanes 1,3,7), which was down-regulated by Dex (Fig. (Fig.4A,4A, lanes 2,3,6,7). This down-regulation was blocked by the proteasome inhibitor LLnL (Fig. (Fig.4A,4A, lanes 4,5,8,9). As Dex had no effect on GR at the RNA level under hypoxia (data not shown), we conclude that GR is decreased at the protein level by a proteasome-mediated process in response to Dex under hypoxic conditions, similar to p53 (see above).
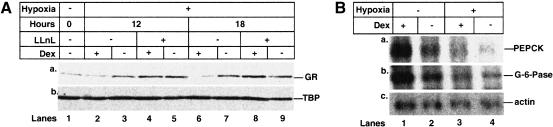
Dex enhances proteasome-mediated degradation of GR and prevents up-regulation of its target genes under hypoxia. (A) HUVEC were grown with vehicle (ethanol), 10−7 M Dex, and 50 μM LLnL, as indicated under normal (lane 1) or hypoxic (lanes 2–9) conditions. Lysates (50 μg) were Western blotted with antibodies to (a) GR (E-20 and P-20); (b) TBP (3G3). (Lanes 1,3,5) (−, vehicle); (lanes 2,4) +Dex; (lanes 6,8) +Dex +
+ LLnL; (lanes 7,9) (−, vehicle)
LLnL; (lanes 7,9) (−, vehicle) +
+ LLnL. (B) Total RNA from HUVEC grown in normoxia (lanes 1,2) or hypoxia (lanes 3,4) for 18 h with vehicle (ethanol) or 10−7 M Dex. Northerns were hybridized with probes for (a) PEPCK; (b) G-6-Pase; (c) β-actin. (Lanes 1,3) +Dex; (lanes 2,4) (−, vehicle).
LLnL. (B) Total RNA from HUVEC grown in normoxia (lanes 1,2) or hypoxia (lanes 3,4) for 18 h with vehicle (ethanol) or 10−7 M Dex. Northerns were hybridized with probes for (a) PEPCK; (b) G-6-Pase; (c) β-actin. (Lanes 1,3) +Dex; (lanes 2,4) (−, vehicle).
We next investigated whether hypoxia affects Dex induction of the GR downstream genes PEPCK (Imai et al. 1990) and Glucose-6-Phosphatase (G-6-Pase) (Argaud et al. 1996). Whereas Dex induced RNA expression from both of these genes under normoxia (Fig. (Fig.4B,4B, lanes 1,2), hypoxia blocked this induction (Fig. (Fig.4B,4B, lanes 3,4). Similarly, hypoxia blocked PEPCK protein induction (data not shown). These results concur with studies in rat hepatocytes, where in vivo hypoxic exposure inhibits gluconeogenesis at the PEPCK level due to decreased PEPCK gene transcription (Pison et al. 1995).
p53, GR, and Hdm2 colocalize in the cytoplasm in the presence of ligand
ligand
Our experiments show that p53 and GR are degraded by proteasomes in HepG2 under normal growth conditions and in HUVEC under hypoxia. Because subcellular localization affects p53 degradation, we studied the localization of p53 and GR in HUVEC cells (Fig. (Fig.5).5). As expected, under normal conditions p53 was not detectable (Fig. (Fig.5A),5A), because of its very low level of expression (see above). GR was located in the cytoplasm in the absence of ligand, but accumulated in the nucleus in the presence of Dex. Under hypoxic conditions, in the absence of Dex, p53 was detectable and was localized to both the nucleus and the cytoplasm in the majority of cells (65%), and exclusively in the nucleus in about 25%, whereas GR remained in the cytoplasm (Fig. (Fig.5B).5B). In the presence of Dex, p53 was completely cytoplasmic in around 60% of the cells, and was colocalized extensively with GR. The immunofluorecence of both GR and p53 was, however, considerably lower, as expected from their degradation (see above). To confirm these data, LLnL was used to stabilize the proteins and increase the signal. Typical cells are shown in Figure Figure5C–E,5C–E, and the quantification in Figure Figure5F.5F. The presence of LLnL did not change the results overall, but the effects were more pronounced. p53 was detected in the nucleus in almost 90% of the cells in the absence of Dex. However, in the presence of ligand, p53 and GR colocalized extensively in the cytoplasm (Fig. (Fig.5C).5C).
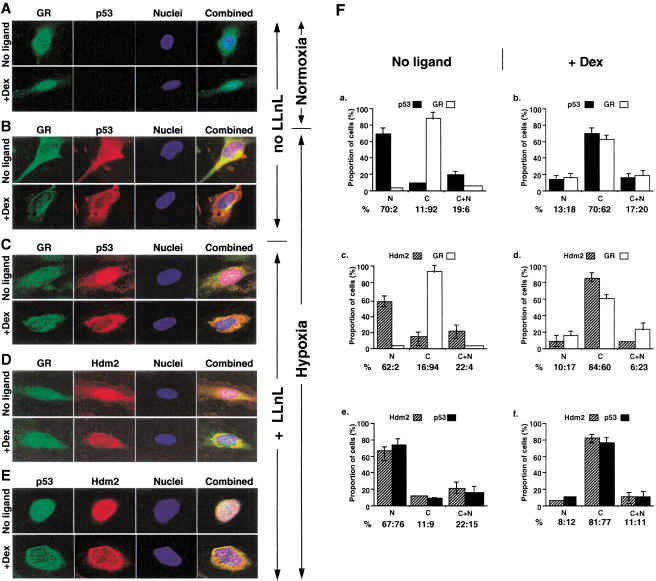
Dexamethasone induces cytoplasmic colocalization of p53, GR, and Hdm2 under hypoxia. (A–E) HUVEC were grown with vehicle (no ligand) or 10−7 M Dex for 18 h in normoxia (A) or hypoxia (B–E), without (A,B) or with (C–E) 50 μM LLnL. Fixed cells were incubated with antibodies against (A–C) GR (E-20 and P-200) and p53 (DO-1); (D) GR (E-20 and P-20) and Hdm2 (2A10); (E) p53 (588) and Hdm2 (2A10), secondary antibodies, and analyzed by confocal microscopy. (F) Quantitation of panels (C–E). The cells were grown without (a,c,e) or with (b,d,f) 10−7 M Dex. At least 500 cells were counted for each graph. The percentage of cells with protein in the nucleus (N), cytoplasm (C), and both (N +
+ C) are shown. (a,b) p53
C) are shown. (a,b) p53 +
+ GR; (c,d) Hdm2
GR; (c,d) Hdm2 +
+ GR; (e,f) Hdm2
GR; (e,f) Hdm2 +
+ p53.
p53.
Hdm2 is involved in p53 degradation and export from the nucleus, raising the possibility that it may also be implicated in GR turnover and localization. As expected, Hdm2 was found to be predominantly nuclear under hypoxia and in the absence of ligand. Interestingly, Hdm2 was cytoplasmic and extensively colocalized with GR in the presence of ligand (Fig. (Fig.5D).5D). Similarly, Hdm2 was found to colocalize with p53 in the nucleus in the absence of ligand and in the cytoplasm in the presence of Dex (Fig. (Fig.55E).
The ligand-dependent trimeric complex between p53, GR, and Hdm2 enhances proteasomal degradation
degradation
The colocalization of p53, GR, and Hdm2 in the cytoplasm led us to investigate whether they form a trimeric complex. Coimmunoprecipitation assays were performed on extracts from HUVEC that had been exposed to hypoxia in the presence of LLnL and in the presence or absence of Dex. Equivalent amounts of p53 were immunoprecipitated with the anti-p53 antibody, irrespective of the presence of ligand, as expected from the inhibition of p53 degradation by LLnL (Fig. (Fig.6A,6A, c). The p53-antibody immunoprecipitate contained both GR and Hdm2 in significantly higher amounts in the presence of Dex than in its absence (Fig. (Fig.6A,6A, a,b, lanes 3,4). In the reciprocal experiment, the GR-antibody immunoprecipitates contained equivalent amounts of GR irrespective of Dex treatment (Fig. (Fig.6A,6A, f). They also contained both p53 and Hdm2 in significantly higher amounts in the presence of Dex (Fig. (Fig.6A,6A, d,e, lanes 7,8). Similar results were obtained in HepG2 cells, where p53, GR and Hdm2 formed a trimeric complex more efficiently following Dex treatment of the cells (Fig. (Fig.6B).6B).
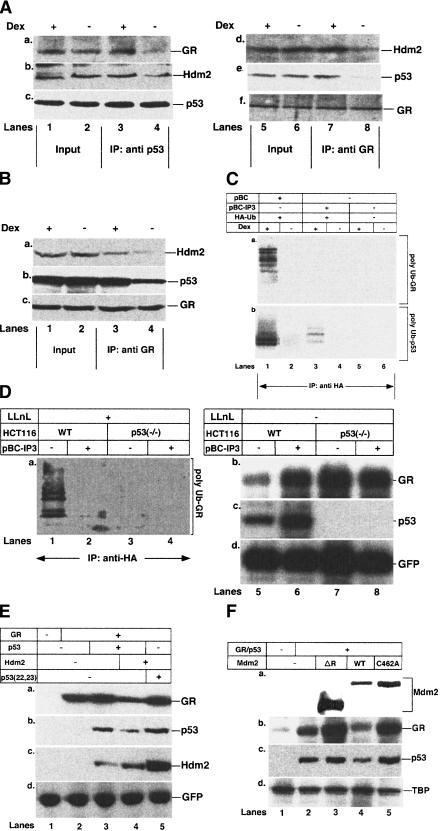
Dex stimulates the formation of a trimeric complex between p53, GR, and Hdm2 and ubiquitylation of GR and p53 by the E3 ligase activity of Hdm2. (A) HUVEC were grown in the presence of 50 μM LLnL and vehicle (−, ethanol) or 10−7 M Dex for 18 h under hypoxic conditions. Lysates (500 μg) were immunoprecipitated with antibodies against p53 (421) or GR (E-20 and P-20). Immunoprecipitates (IP) were Western blotted with antibodies against GR and Hdm2 (for p53 IP; a,b) or Hdm2 and p53 (for GR IP; d,e). The efficiency of IP was verified with self-antibodies (c,f). DO-1 was used to reveal p53 on the Western blots. (Lanes 1,3,5,7) +Dex; (lanes 2,4,6,8) (−, vehicle). Input, 10% lysate used for IP. (B) HepG2 cells were grown in the presence of 50 μM LLnL and vehicle (−, ethanol) or 10−7 M Dex for 12 h. Lysates (500 μg) were immunoprecipitated with antibodies against GR (E-20 and P-20). The IPs were Western blotted with antibodies against Hdm2 and p53 (a,b). The efficiency of IP was checked with self-antibodies (c). (Lanes 1,3) +Dex; (lanes 2,4) (−, vehicle). Input, 10% lysate used for IP. (C) HUVEC cells were transfected (lanes 1–4) with HA-Ub, pBC–IP3.2, or pBC, and 24 h posttransfection subjected to hypoxia for 18 h with LLnL (50 μM) and either vehicle (−, ethanol) or 10−7 M Dex. Lysates (500 μg) were immunoprecipitated with antibodies against HA (12CA5). The IPs were Western blotted with antibodies against with GR (E-20 and p-20) (a) and p53 (DO-1) (b). (D) HCT116 p53 (WT) (lanes 1,2,5,6) or HCT116 p53−/− (lanes 3,4,7,8) cells were transfected with pHA–Ub, pBC–IP3 (where indicated), and pEGGFP-C1 (lanes 5–8, to measure transfection efficiency). Twelve hours posttransfection the cells were incubated with 10−7 M Dex under hypoxia for 12 h, with (lanes 1–4) or without (lanes 5–8) 50 μM LLnL. Lysates (500 μg) from cells grown in the presence of LLnL were immunoprecipitated with antibodies against HA (12CA5). The IPs were Western blotted with antibodies against GR (E-20 and p-20; a). Lysates (50 μg) from cells with LLnL were Western blotted with antibodies to (b) GR (E-20 and P-20); (c) p53 (DO-1); (d) GFP (monoclonal). (Lanes 1,3,5,7) −IP3; (lanes 2,4,6,8) +IP3. (E) HCT116 p53−/− cells were transfected with pcDNA3–GR, pRC p53 (WT) pRC p53(22,23), pxMDM2, and pEGGFP-C1 (to measure transfection efficiency). Twelve hours posttransfection the cells were subjected to hypoxia for 12 h with 10−7 M Dex. Lysates (50 μg) were Western blotted with antibodies against (a) GR (E-20 and P-20); (b) p53 (1801); c) Hdm2 (2A10); (d) GFP (monoclonal). (Lane 1) Mock transfection; (lane 2) GR; (lane 3) p53 +
+ GR; (lane 4) p53
GR; (lane 4) p53 +
+ GR
GR +
+ Hdm2; (lane 5) p53(22,23)
Hdm2; (lane 5) p53(22,23) +
+ GR
GR +
+ Hdm2. (F) HCT116 p53−/− cells were transfected with pcDNA3–GR, pRC p53 (WT), pxMDM2, px ΔR, and pxC462A. Twelve hours posttransfection the cells were subjected to hypoxia for 12 h with 10−7 M Dex. Lysates (50 μg) were Western blotted with antibodies against (a) Hdm2 (2A10); (b) GR (E-20 and P-20); (c) p53 (DO-1); (d) TBP (3G3). (Lane 1) Mock transfection; (lane 2) p53
Hdm2. (F) HCT116 p53−/− cells were transfected with pcDNA3–GR, pRC p53 (WT), pxMDM2, px ΔR, and pxC462A. Twelve hours posttransfection the cells were subjected to hypoxia for 12 h with 10−7 M Dex. Lysates (50 μg) were Western blotted with antibodies against (a) Hdm2 (2A10); (b) GR (E-20 and P-20); (c) p53 (DO-1); (d) TBP (3G3). (Lane 1) Mock transfection; (lane 2) p53 +
+ GR; (lane 3) p53
GR; (lane 3) p53 +
+ GR
GR +
+ ΔR; (lane 4) p53
ΔR; (lane 4) p53 +
+ GR
GR +
+ Hdm2; (lane 5) p53
Hdm2; (lane 5) p53 +
+ GR
GR +
+ C462A.
C462A.
GR has been postulated to be degraded by the ubiquitin–proteasome pathway (Whitesell and Cook 1996; Segnitz and Gehring 1997), but there was no direct evidence. Our results do indicate, however, that during cross talk between the GR and p53 pathways, GR is degraded concurrently with p53 by a pathway involving the E3 ubiquitin ligase activity of MDM2. To test this hypothesis HUVEC cells were transfected with HA-tagged ubiquitin (HA-Ub) in the presence or absence of IP3, a small peptide that specifically disrupts the p53–Hdm2 interaction (Wasylyk et al. 1999). Twenty-four hours post-transfection, the cells were placed under hypoxic conditions in the presence of the proteasome inhibitor, LLnL. Western blotting of HA-immunoprecipitates with GR antibodies showed that GR was poly-ubiquitinated, but only when the cells were incubated in the presence of Dex (Fig. (Fig.6C,6C, a, lanes 1,2). IP3 expression inhibited GR poly-ubiquitylation, showing that it requires the p53–Hdm2 interaction (Fig. (Fig.6C,6C, a, lanes 1,3). The poly-ubiquitinated forms are specific, because they were not detected when HA-Ub was not transfected (Fig. (Fig.6C,6C, a, lanes 5,6). Interestingly, polyubiquitylation of p53 is also enhanced in the presence of Dex (Fig. (Fig.6C,6C, b, lanes 1,2). Polyubiquitylation of p53 was inhibited by the expression of IP3, as expected from the disruption of the p53–Hdm2 interaction (Fig. (Fig.6C,6C, b, lanes 1,3). These results show that Dex stimulates GR and p53 polyubiquitylation through a pathway involving Hdm2 and its interaction with p53.
The E3 ligase activity of Hdm2 is responsible for ligand-dependent degradation of GR
GR
To study GR degradation in transfection assays, in which exogenous proteins are expressed in cells, we used HCT116 isogenic cell lines in which p53 is either wild type or disrupted by homologous recombination (Bunz et al. 1998). The isogenic cell lines were transfected with recombinants that express HA-Ub and IP3. Posttransfection, the cells were subjected to hypoxia in the presence of Dex and LLnL. HA-antibody immunoprecipitates contained lower mobility HA-tagged polyubiquitylated forms of GR (Fig. (Fig.6D,6D, a, lane 1). IP3 expression inhibited ubiquitylation of GR (Fig. (Fig.6D,6D, a, lane 2). Ubiquitylated GR was not detected in cells lacking p53, whether or not IP3 was expressed (Fig. (Fig.6D,6D, a, lanes 3,4). These results indicate that GR ubiquitylation requires p53 and its interaction with Hdm2. In cells that were not treated with LLnL, IP3 expression increased the levels of both GR and p53 (Fig. (Fig.6D,6D, a, lanes 5,6), as expected from decreased degradation of both proteins by a common mechanism involving Hdm2. Irrespective of the presence or absence of IP3, GR levels were higher in the isogenic p53−/− cells (Fig. (Fig.6D,6D, a, lanes 7,8), as expected from the lack of p53 that is required to recruit Hdm2.
To confirm that p53–Hdm2 interactions are required for GR degradation, we used p53(22,23), which cannot interact with Hdm2 (Lin et al. 1994). Expression of wild-type p53, Hdm2, and exogenous GR in HCT116 p53−/− cells in the presence of Dex resulted in a significant decrease in GR levels (Fig. (Fig.6E,6E, lanes 1–4). In contrast, the GR levels remained unchanged when the p53(22,23) mutant was expressed (Fig. (Fig.6E,6E, lane 5). Interestingly, similar to GR, the levels of p53 and Hdm2 were also higher with p53(22,23) compared to p53 wild type (Fig. (Fig.6E,6E, lanes 5,6). These results suggest that Dex-dependent ternary complex formation induces degradation of all three proteins.
To determine whether the E3 ligase activity of Hdm2 is responsible for GR ubiquitylation and degradation, HCT116 p53−/− cells were transfected with GR, p53, and the Mdm2 mutants ΔR and C462A, which abolish the E3 ligase activity of Mdm2 (Argentini et al. 2000). Mdm2 promoted the degradation of both p53 and GR, whereas the mutants protected both proteins from degradation (Fig. (Fig.6F).6F). These results show that Mdm2 E3 ligase activity is required for GR degradation.
Dex-dependent ternary complexes involving p53, GR, and Hdm2 form during mitomycin C treatment
treatment
To determine whether DNA damage could stimulate Dex-dependent degradation of GR, we treated HUVEC cells with mitomycin C (Fritsche et al. 1993). Similar to hypoxia, mitomycin C stabilized p53 and GR in HUVEC cells (Fig. (Fig.7A,7A, lanes 1,3). Dex reduced induction of both GR and p53 (Fig. (Fig.7A,7A, lanes 2,3), and inhibited a downstream function of p53, apoptosis (data not shown). Inhibition of proteasome-mediated degradation with LLnL stabilized both proteins (Fig. (Fig.7A,7A, lanes 4,5). These results show that both p53 and GR undergo enhanced degradation in the presence of Dex during mitomycin C treatment.
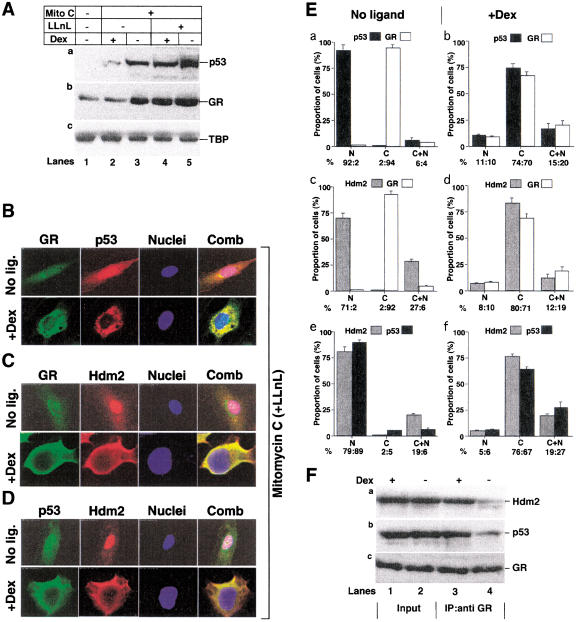
Dexamethasone induces degradation, cytoplasmic accumulation, and trimeric complex formation of p53, GR, and Hdm2 following mitomycin C treatment. (A) HUVEC cells were grown for 24 h with vehicle (ethanol), 10−7 M Dex, and 50 μM LLnL, as indicated, and with (lane 1) or without (lanes 2–5) 10 μM mitomycin C. Lysates (50 μg) were Western blotted with antibodies against (a) p53 (DO-1); (b) GR (E-20 and P-20); (c) TBP (3G3). (Lanes 1,3,5) (−,vehicle); (lanes 2,4) +Dex. (B–D) HUVEC were grown with 10 μM mitomycin C, 50 μM LLnL, vehicle (no ligand), or 10−7 M Dex for 24 h. The cells were fixed, incubated with antibodies [(B) GR (E-20 and P-20) and p53 (DO-1); (C) GR (E-20 and P-20) and Hdm2 (2A10); (D) p53 (588) and Hdm2 (2A10)], secondary antibodies, and analyzed by confocal microscopy. (E) Quantitation of (B–D). The cells were grown without (a,c,e) or with (b,d,f) 10−7 M Dex. At least 500 cells were counted for each graph. The percentage of cells with protein in the nucleus (N), cytoplasm (C), and both (N +
+ C) are shown. (a,b) p53
C) are shown. (a,b) p53 +
+ GR; (c,d) Hdm2
GR; (c,d) Hdm2 +
+ GR; (e,f) Hdm2
GR; (e,f) Hdm2 +
+ p53. (F) HUVEC were incubated with 50 μM LLnL, vehicle (−, ethanol) or 10−7 M Dex, and 10 μM mitomycin C for 24 h. Lysates (500 μg) were immunoprecipitated with antibodies against GR (E-20 and P-20). The IPs were Western blotted with antibodies against Hdm2 and p53 (a,b). The efficiency of IP was verified with self-antibodies (c). (Lanes 1,3) +Dex; (lanes 2,4) (−, vehicle). Input, 10% lysate used for IP.
p53. (F) HUVEC were incubated with 50 μM LLnL, vehicle (−, ethanol) or 10−7 M Dex, and 10 μM mitomycin C for 24 h. Lysates (500 μg) were immunoprecipitated with antibodies against GR (E-20 and P-20). The IPs were Western blotted with antibodies against Hdm2 and p53 (a,b). The efficiency of IP was verified with self-antibodies (c). (Lanes 1,3) +Dex; (lanes 2,4) (−, vehicle). Input, 10% lysate used for IP.
We determined whether the cellular localization of p53 and GR are the same in mitomycin C as in hypoxia-treated cells. HUVEC cells were treated with mitomycin C in the presence of LLnL, and the cells examined by indirect immunofluorecence. Typical cells are shown in Figure Figure7B–D7B–D and the quantitation in Figure Figure7E.7E. In the absence of Dex, mitomycin C treatment resulted in nuclear accumulation of p53, whereas GR remained cytoplasmic. However, in the presence of Dex, p53 as well as GR were cytoplasmic and colocalized extensively (Fig. (Fig.7B).7B). Similar Dex-dependent colocalization was observed when cells were examined for GR and Hdm2 (Fig. (Fig.7C)7C) or for p53 and Hdm2 (Fig. (Fig.7D).7D). These results show that p53, GR, and Hdm2 are colocalized in the presence of Dex during mitomycin C treatment, and suggest that they could form a triple complex. Indeed, GR antibody immunoprecipitates from mitomycin C-treated HUVEC cells grown in the presence of LLnL were found to contain both Hdm2 and p53, to a much larger extent in the presence than in the absence of Dex (Fig. (Fig.7F,7F, cf. lanes 3 and 4).
Discussion
Glucocorticoids and their receptors play important roles in both apoptotic cell death and cellular proliferation. An agonist of the glucocorticoid receptor, Dex, is known to induce apoptosis in T cells, B lymphocytes, and multiple myoloma cells (Martin and Green 1995). However, it also represses apoptosis and increases proliferation in cancer cell lines (Chang et al. 1997), neutrophils (Daffern et al. 1999), retinal pigment cells (He et al. 1994), fibroblasts (Li et al. 1998), rat hepatoma (Yamamoto et al. 1998), and serum-depleted T lymphocytes that express Bcl2 (Huang and Cidlowski 1999). The divergent functions of glucocorticoids and GR in different contexts can be attributed to interactions with different pro- and antiproliferation factors. For example, GR interacts with and affects the functions of the proapoptotic protein DAP3 (Hulkko et al. 2000). Here we show that GR and the tumor supressor p53 regulate, by novel mechanisms, each other's functions in normal cells in response to a physiological stress.
We have demonstrated that Dex induces mutual down-regulation of p53 and GR-dependent downstream target genes, both under hypoxia in HUVEC and under normoxia in HepG2 cells. HUVEC are normal human cells with typical GR-dependent functions. Under normoxic conditions, in which p53 levels are very low, HUVEC display a ligand-dependent translocation of the receptor to the nucleus and transactivation of GR downstream genes such as PEPCK and G-6-Pase. However, under hypoxia, which induces a large increase in p53 levels, GR functions are seriously impaired. p53 induction may not be required in HepG2 because its levels are constitutively high. Down-regulation of GR, which occurs within 3 h in HepG2, takes about 12–18 h in HUVEC, probably due to the time required to increase p53 levels. The constitutively high level of p53 in HepG2 could account for the reported insensitivity of HepG2 to Dex induction of GR activity (Xing and Quinn 1993).
We have shown, in vitro and in vivo, that GR and p53 interact weakly in the absence of ligand and more strongly in the presence of Dex. Interestingly, GR interacts with the NLS of p53, and apparently only in the presence of ligand. This interaction could result in ligand-dependent masking of the NLS, sequestration in the cytoplasm, and in consequence, inhibition of transactivation. However, we cannot exclude that the complex accelerates nuclear export of both proteins.
We have found that GR and p53 inhibit each other's activities by cytoplasmic sequestration in response to hypoxia and DNA damage in a ligand-dependent manner. Hypoxia and DNA damage can be expected to have similar effects. Hypoxia is a potent inducer of Nitric Oxide Synthase in rats and endothelial cells of bovine origin (Arnet et al. 1996; Gess et al. 1997). Nitric oxide oxy-radicals damage DNA (Xu et al. 2000), and up-regulate p53 (Forrester et al. 1996). We have previously shown that neuroblastoma cells represent a pathological situation in which p53 is inhibited by cytoplasmic sequestration (Sengupta et al. 2000b). There is growing evidence that p53 sequestration in the cytoplasm may be a common mechanism of p53 inactivation. For example, both Mot-2 (Wadhwa et al. 1999) and hepatitis B virus X protein (Elmore et al. 1997) have been shown to physically interact with p53 and repress its transcriptional activity by sequestration in the cytoplasm. Estradiol inactivates p53 by intracellular redistribution (Molinari et al. 2000). The results from experiments with in vitro translated proteins and overexpression in cells suggest that p53 down-regulates ER-responsive genes by interfering with the binding of ER to ERE (Liu et al. 1999). Moreover, ER protects p53 from Hdm2-mediated degradation through the formation of a triple complex (Liu et al. 2000). Cytoplasmic sequestration and functional repression of p53 in mammary epithelium can be reversed with a hormone cocktail (Kuperwasser et al. 2000). However, in none of these studies was the subcellular localization of the complex and the relevance of the interactions to physiological conditions addressed.
We have established that Dex stimulates the formation of a triple complex that contains Hdm2 in addition to p53 and GR. This triple complex apparently forms by GR and Hdm2 binding to different domains of p53. Hdm2 binds to the N-terminal transactivation domain, whereas GR binds to the DNA binding domain and the NLS. Interestingly, Hdm2 recruitment is associated with increased ubiquitylation and proteasome-mediated degradation of p53 and GR. In contrast, ER apparently inhibits Hdm2-mediated degradation of p53 (Liu et al. 2000). Dex induces cytoplasmic accumulation of Hdm2, triple complex formation, and degradation, suggesting that degradation is mediated by cytoplasmic proteasomes. Hdm2-mediated degradation of p53 is generally thought to occur in the cytoplasm. However, a recent study suggests that it may also occur in the nucleus (Yu et al. 2000). In neuroblastoma cells both p53–Hdm2 (Zaika et al. 1999) and p53–GR (Sengupta et al. 2000b) complexes are found in the cytoplasm, suggesting that the cytoplasmic degradation function of Hdm2 is defective in these cells. Thus, lack of degradation as well as cytoplasmic sequestration distinguish neuroblastoma from normal cells.
We have discovered that GR is a new substrate for Hdm2, thereby extending the range of targets for this E3 ubiquitin ligase. Mdm2 mediates degradation of the cell fate regulator Numb (Juven-Gershon et al. 1998) and E2F (Loughran and La Thangue 2000) in a p53-independent manner. In contrast, it targets GR (this study) and HIF-1α (Ravi et al. 2000) in a p53-dependent manner. It is conceivable, therefore, that p53-mediated recruitment of Mdm2 is an important mechanism for the degradation of proteins that interact with p53 under different physiological conditions.
It is interesting to consider why p53 and GR cross talk under physiological conditions. Both p53 and GR mediate stress responses, but with intrinsic differences in their actions. GR, when complexed to glucocorticoids, is involved in fight-or-flight responses, and maintains homeostasis during internal or environmental changes. Hence, GR is generally involved in a survival response. On the other hand, p53, which has been described as the “guardian of the genome,” responds to extensive genomic and other stresses by inducing cell death. p53 can be considered to be a death response. Hence, p53 and GR can be considered to be protagonists in the question of life or death, and should, therefore, have antagonistic effects on each other's functions. Hypoxia and the subsequent DNA damage represent such a situation where p53 and GR have opposite effects. The death response via p53 is held in check by ligand-activated GR, which induces p53 degradation. GR itself is also restrained by increased degradation induced by p53. Mutual negative cross talk between these factors thereby functions as a balance between the cellular responses to conflicting signals.
In summary, we have demonstrated for the first time that p53 and GR functionally cross talk under physiological conditions. We have also identified p53 and GR downstream genes that are down-regulated during this process. The repression of each other's functions is enhanced by ligand. The core domain and NLS of p53 bind to GR in a ligand-enhanced manner, resulting in the cytoplasmic relocalization of both proteins. The p53–GR complex in the cytoplasm also contains the E3 ubiquitin ligase Hdm2, which mediates degradation of both p53 and GR. Although GR degradation by the ubiquitin proteasome machinery has been postulated, our studies provide the first direct evidence for it in a physiological situation.
Material and methods
Recombinants
GST constructs
GST–p53(305–321) was generated by ligating oligonucleotides into the BamHI and EcoRi sites of pGEX-2T (Pharmacia Biotech). The other p53 mutants were generated by PCR and cloning into the same sites of pGEX-2T.
Reporters, expression vectors, and cDNA probes
p53CON (Funk et al. 1992); CTS1 (Conseiller et al. 1998); pcDNA3GR (Sengupta et al. 2000b); CMV–LacZ, IGBMC core facility; pBC–IP3.2 (Wasylyk et al. 1999); HA-Ub (Treier et al. 1994); pBS–β-actin (IGBMC core facility); pBS–glucose-6-phosphate (Lange et al. 1994); pBS–PEPCK (Bartels et al. 1989); pRCp53(WT) and pRCp53(22,23) (Lin et al. 1994); pxMDM2, pxΔR, pxC462A (Argentini et al. 2000); pEGGFP-C1 (Clonetech).
Cells
HSC-2 (Momose et al. 1989); HUVEC, ATCC (CRL-1730); HepG2, ECACC (Ref. 85011430); HCT116 p53(WT) and HCT116 p53(−/−) (Bunz et al. 1998).
Protein expression, purification, and GST interactions
interactions
GST proteins were induced with 1 mM IPTG in E. coli DH5α grown at 25°C, and extracted by sonication in STE buffer (10 mM Tris-HCl at pH 8.0; 150 mM NaCl; 1 mM EDTA supplemented with 5 mM DTT, 1mM PMSF, 2.5 μg/mL proteinase inhibitors and 1.5% Sarkosyl). The sonicate was adjusted to 2% Triton X-100, agitated for 30 min at 4°C, centrifuged, and the crude extract stored in aliquots at −70°C. Glutathione-S-Sepharose beads were equilibrated in GST buffer (50 mM Tris-HCl at pH 7.5, 100 mM KCl, 10 mM MgCl2, 5% glycerol, 0.5% NP-40 supplemented with 0.3 mM DTT, 1 mM PMSF and 2.5 μg/mL proteinase inhibitors), incubated with the crude extracts at 4°C for 90 min, and washed. Bound proteins were analyzed by SDS-PAGE and Coomassie staining. GR was produced by in vitro translation with pcDNA3GR and the TNT Quick coupled transcription/translation system (Promega) for 90 min at 30°C. Equivalent amounts of bound protein were incubated with 5 μL of in vitro translated GR for 1 h at 4°C in the presence of vehicle (ethanol), Dex, Cortisol, or RU486 (Sigma). The bound proteins were washed and analyzed by SDS-PAGE and fluorography.
Cell culture and transfection
transfection
Cells were maintained in DMEM plus 10% serum and antibiotics at 37°C with 5% CO2. HUVEC medium was supplemented with ECGF (50 μg/mL), Heparin (50 μg/mL), and Glutamine (1 mM). The HCT116 cell lines were grown in McKoy's 5A medium plus 10% serum, antibiotics, and 2 mM glutamine. HSC-2 were transfected by calcium phosphate precipitation (Chen and Okayama 1987) with 4 μg of plasmid in 36-mm plates (six-well cluster, Costar 3516). After 16 h the cells were washed with medium and incubated in full medium with ligands or vehicle (ethanol). To express HA-Ub and IP3, low-passage HUVEC (<15) was transfected with Superfect (QIAGEN) according to the manufacturer's instructions. After 3 h with Superfect the cells were incubated in normal medium for 24 h. HCT116 cells were transfected with Lipfectamine 2000 (Life Technologies) in the presence of serum for 6 h, washed, and incubated in normal medium for 12 h before hypoxic treatment. Post-transfection is defined as after the removal of the facilitator. Hypoxia experiments were carried out in anaerobic chambers (Anaerocult, Merck, Cat. no. 1.13829) at 37°C according to the manufacturer's instructions; 10 μM mitomycin C (Sigma) was used.
Transcriptional activity
Cell lysates were analyzed for luciferase and β-galactosidase activities as described previously (Sengupta et al. 2000a).
Western blots
Equal quantities of protein, estimated by the Bio-Rad assay on RIPA buffer lysates, were fractionated by SDS-PAGE, transferred to nitrocellulose membranes (Schleicher and Schuell), and Western blotted. Primary and secondary antibodies: Bax, N20 (Santa Cruz Biotechnology); p21WAF1/CIP1, 1WA-IC581 (Sengupta et al. 2000a); TBP, 3G3 (Brou et al. 1993); p53, DO-1 (Vojtesek et al. 1992); p53, PAb 421 (Wade-Evans and Jenkins 1985); p53, #588, raised against a GST–human p53(320–393) (C. Wasylyk and the IGBMC core facility); GR, E-20 and P-20 (Santa Cruz Biotechnology); PARP (Kaufmann et al. 1993); HA, 12CA5 (Boerhinger); PEPCK (Zimmer and Magnuson 1990); Hdm2, 2A10 (Chen et al. 1993); HIF-1α, C-19 (Santa Cruz Biotechnology); peroxidase-coupled anti-rabbit or anti-mouse antibodies (Jackson Laboratories); p53, 1801 (Ab-2) (Oncogene Research Products); GFP (Clontech). The blots were revealed by chemiluminescence (Pierce Super Signal).
Northern blots
Forty micrograms of total RNA, extracted with Trizol (GIBCO BRL), was electrophoresed on 6% formaldehyde–1% agarose gels, transferred to Hybond N+ membranes, hybridized with random-primed probes at 55°C (for PEPCK and G-6-Pase) or 65°C (for p53, GR, and actin), washed, and autoradiographed.
Immunofluorescence
Cells were fixed with acetone:methanol (1:1), blocked with 10% goat serum, incubated with primary antibodies for 2 h, secondary antibodies linked to either Cy3 or FITC (Moll et al. 1996) and Hoechst (to stain nuclei), then visualized by confocal microscopy (Leica TCS 4D).
Immunoprecipitations
Cells were lysed on ice in NP-40 buffer (150 mM NaCl, 1% NP-40, 50 mM Tris-HCl at pH 8.0) and the cleared supernatants used for immunoprecipitation. The antigen–antibody complex was allowed to form on ice for 1 h, then immobilized on protein G beads, washed three times, boiled in Laemmli buffer, and fractionated by SDS-PAGE.
Acknowledgments
We thank Julia Young for critical reading of the manuscript; D. Bohmann, E. Conseiller, M.R. el-Maghrabi, T. Kietzmann, and J. Shay for the gifts of recombinants; N. Tsuchida for HSC-2 cells; D. Granner, G. de Murcia, and L. Tora for the gift of antibodies; the IGBMC core facilities for help and support; and BioAvenir (Aventis, Rhone-Poulenc), the Centre National de la Recherche Scientifique, the Institut National de la Santé et de la Recherche Médicale, the Hôpital Universitaire de Strasbourg, the Association pour la Recherche sur le Cancer, the Fondation pour la Recherche Médicale, the Ligue Nationale Française contre le Cancer (Equipe labellisée), the Ligue Régionale (Haut-Rhin) contre le Cancer, and the Ligue Régionale (Bas-Rhin) contre le Cancer for financial assistance.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL rf.gbsarts-u.cmbgi@hob; FAX 33-3-88-65-32-01.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.202201.
References
- An WG, Kanekal M, Simon MC, Maltepe E, Blagosklonny MV, Neckers LM. Stabilization of wild-type p53 by hypoxia-inducible factor 1α Nature. 1998;392:405–408. [Abstract] [Google Scholar]
- Argaud D, Zhang Q, Pan W, Maitra S, Pilkis SJ, Lange AJ. Regulation of rat liver glucose-6-phosphatase gene expression in different nutritional and hormonal states: Gene structure and 5′-flanking sequence. Diabetes. 1996;45:1563–1571. [Abstract] [Google Scholar]
- Argentini M, Barboule N, Wasylyk B. The contribution of the RING finger domain of MDM2 to cell cycle progression. Oncogene. 2000;19:3849–3857. [Abstract] [Google Scholar]
- Arnet UA, McMillan A, Dinerman JL, Ballermann B, Lowenstein CJ. Regulation of endothelial nitric-oxide synthase during hypoxia. J Biol Chem. 1996;271:15069–15073. [Abstract] [Google Scholar]
- Ashcroft M, Taya Y, Vousden KH. Stress signals utilize multiple pathways to stabilize p53. Mol Cell Biol. 2000;20:3224–3233. [Europe PMC free article] [Abstract] [Google Scholar]
- Bartels H, Linnemann H, Jungermann K. Predominant localization of phosphoenolpyruvate carboxykinase mRNA in the periportal zone of rat liver parenchyma demonstrated by in situ hybridization. FEBS Lett. 1989;248:188–194. [Abstract] [Google Scholar]
- Bauer A, Tronche F, Wessely O, Kellendonk C, Reichardt HM, Steinlein P, Schutz G, Beug H. The glucocorticoid receptor is required for stress erythropoiesis. Genes & Dev. 1999;13:2996–3002. [Europe PMC free article] [Abstract] [Google Scholar]
- Boyd SD, Tsai KY, Jacks T. An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat Cell Biol. 2000;2:563–568. [Abstract] [Google Scholar]
- Brou C, Chaudhary S, Davidson I, Lutz Y, Wu J, Egly JM, Tora L, Chambon P. Distinct TFIID complexes mediate the effect of different transcriptional activators. EMBO J. 1993;12:489–499. [Europe PMC free article] [Abstract] [Google Scholar]
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. [Abstract] [Google Scholar]
- Chandel NS, Vander Heiden MG, Thompson CB, Schumacker PT. Redox regulation of p53 during hypoxia. Oncogene. 2000;19:3840–3848. [Abstract] [Google Scholar]
- Chang TC, Hung MW, Jiang SY, Chu JT, Chu LL, Tsai LC. Dexamethasone suppresses apoptosis in a human gastric cancer cell line through modulation of bcl-x gene expression. FEBS Lett. 1997;415:11–15. [Abstract] [Google Scholar]
- Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. [Europe PMC free article] [Abstract] [Google Scholar]
- Chen J, Marechal V, Levine AJ. Mapping of the p53 and mdm-2 interaction domains. Mol Cell Biol. 1993;13:4107–4114. [Europe PMC free article] [Abstract] [Google Scholar]
- Cheung J, Smith DF. Molecular chaperone interactions with steroid receptors: An update. Mol Endocrinol. 2000;14:939–946. [Abstract] [Google Scholar]
- Conseiller E, Debussche L, Landais D, Venot C, Maratrat M, Sierra V, Tocque B, Bracco L. CTS1: A p53-derived chimeric tumor suppressor gene with enhanced in vitro apoptotic properties. J Clin Invest. 1998;101:120–127. [Europe PMC free article] [Abstract] [Google Scholar]
- Daffern PJ, Jagels MA, Hugli TE. Multiple epithelial cell-derived factors enhance neutrophil survival. Regulation by glucocorticoids and tumor necrosis factor-alpha. Am J Respir Cell Mol Biol. 1999;21:259–267. [Abstract] [Google Scholar]
- Elmore LW, Hancock AR, Chang SF, Wang XW, Chang S, Callahan CP, Geller DA, Will H, Harris CC. Hepatitis B virus X protein and p53 tumor suppressor interactions in the modulation of apoptosis. Proc Natl Acad Sci. 1997;94:14707–14712. [Europe PMC free article] [Abstract] [Google Scholar]
- Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem. 2000;275:8945–8951. [Abstract] [Google Scholar]
- Forrester K, Ambs S, Lupold SE, Kapust RB, Spillare EA, Weinberg WC, Felley-Bosco E, Wang XW, Geller DA, Tzeng E, et al. Nitric oxide-induced p53 accumulation and regulation of inducible nitric oxide synthase expression by wild-type p53. Proc Natl Acad Sci. 1996;93:2442–2447. [Europe PMC free article] [Abstract] [Google Scholar]
- Fritsche M, Haessler C, Brandner G. Induction of nuclear accumulation of the tumor-suppressor protein p53 by DNA-damaging agents. Oncogene. 1993;8:307–318. [Abstract] [Google Scholar]
- Funk WD, Pak DT, Karas RH, Wright WE, Shay JW. A transcriptionally active DNA-binding site for human p53 protein complexes. Mol Cell Biol. 1992;12:2866–2871. [Europe PMC free article] [Abstract] [Google Scholar]
- Gess B, Schricker K, Pfeifer M, Kurtz A. Acute hypoxia upregulates NOS gene expression in rats. Am J Physiol. 1997;273:R905–R910. [Abstract] [Google Scholar]
- Geyer RK, Yu ZK, Maki CG. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat Cell Biol. 2000;2:569–573. [Abstract] [Google Scholar]
- He S, Wang HM, Ye J, Ogden TE, Ryan SJ, Hinton DR. Dexamethasone induced proliferation of cultured retinal pigment epithelial cells. Curr Eye Res. 1994;13:257–261. [Abstract] [Google Scholar]
- Honda R, Yasuda H. Activity of MDM2, a ubiquitin ligase, toward p53 or itself is dependent on the RING finger domain of the ligase. Oncogene. 2000;19:1473–1476. [Abstract] [Google Scholar]
- Huang ST, Cidlowski JA. Glucocorticoids inhibit serum depletion-induced apoptosis in T lymphocytes expressing Bcl-2. FASEB J. 1999;13:467–476. [Abstract] [Google Scholar]
- Hulkko SM, Wakui H, Zilliacus J. The pro-apoptotic protein death-associated protein 3 (DAP3) interacts with the glucocorticoid receptor and affects the receptor function. Biochem J. 2000;349:885–893. [Europe PMC free article] [Abstract] [Google Scholar]
- Imai E, Stromstedt PE, Quinn PG, Carlstedt-Duke J, Gustafsson JA, Granner DK. Characterization of a complex glucocorticoid response unit in the phosphoenolpyruvate carboxykinase gene. Mol Cell Biol. 1990;10:4712–4719. [Europe PMC free article] [Abstract] [Google Scholar]
- Jackson MW, Berberich SJ. MdmX protects p53 from Mdm2-mediated degradation. Mol Cell Biol. 2000;20:1001–1007. [Europe PMC free article] [Abstract] [Google Scholar]
- Jenq W, Rabb H, Wahe M, Ramirez G. Hypoxic effects on the expression of mineralocorticoid and glucocorticoid receptors in human renal cortex epithelial cells. Biochem Biophys Res Commun. 1996;218:444–448. [Abstract] [Google Scholar]
- Jimenez GS, Khan SH, Stommel JM, Wahl GM. p53 regulation by post-translational modification and nuclear retention in response to diverse stresses. Oncogene. 1999;18:7656–7665. [Abstract] [Google Scholar]
- Juven-Gershon T, Shifman O, Unger T, Elkeles A, Haupt Y, Oren M. The Mdm2 oncoprotein interacts with the cell fate regulator Numb. Mol Cell Biol. 1998;18:3974–3982. [Europe PMC free article] [Abstract] [Google Scholar]
- Kaufmann SH, Desnoyers S, Ottaviano Y, Davidson NE, Poirier GG. Specific proteolytic cleavage of poly(ADP-ribose) polymerase: An early marker of chemotherapy-induced apoptosis. Cancer Res. 1993;53:3976–3985. [Abstract] [Google Scholar]
- Kellendonk C, Tronche F, Reichardt HM, Schutz G. Mutagenesis of the glucocorticoid receptor in mice. J Steroid Biochem Mol Biol. 1999;69:253–259. [Abstract] [Google Scholar]
- Koumenis C, Alarcon R, Hammond E, Sutphin P, Hoffman W, Murphy M, Derr J, Taya Y, Lowe SW, Kastan M, et al. Regulation of p53 by hypoxia: Dissociation of transcriptional repression and apoptosis from p53-dependent transactivation. Mol Cell Biol. 2001;21:1297–1310. [Europe PMC free article] [Abstract] [Google Scholar]
- Kuperwasser C, Pinkas J, Hurlbut GD, Naber SP, Jerry DJ. Cytoplasmic sequestration and functional repression of p53 in the mammary epithelium is reversed by hormonal treatment. Cancer Res. 2000;60:2723–2729. [Abstract] [Google Scholar]
- Lakin ND, Jackson SP. Regulation of p53 in response to DNA damage. Oncogene. 1999;18:7644–7655. [Abstract] [Google Scholar]
- Lange AJ, Argaud D, el-Maghrabi MR, Pan W, Maitra SR, Pilkis SJ. Isolation of a cDNA for the catalytic subunit of rat liver glucose-6-phosphatase: Regulation of gene expression in FAO hepatoma cells by insulin, dexamethasone and cAMP. Biochem Biophys Res Commun. 1994;201:302–309. [Abstract] [Google Scholar]
- Li S, Mawal-Dewan M, Cristofalo VJ, Sell C. Enhanced proliferation of human fibroblasts, in the presence of dexamethasone, is accompanied by changes in p21Waf1/Cip1/Sdi1 and the insulin-like growth factor type 1 receptor. J Cell Physiol. 1998;177:396–401. [Abstract] [Google Scholar]
- Lin J, Chen J, Elenbaas B, Levine AJ. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes & Dev. 1994;8:1235–1246. [Abstract] [Google Scholar]
- Liu G, Schwartz JA, Brooks SC. p53 down-regulates ER-responsive genes by interfering with the binding of ER to ERE. Biochem Biophys Res Commun. 1999;264:359–364. [Abstract] [Google Scholar]
- Liu G, Schwartz JA, Brooks SC. Estrogen receptor protects p53 from deactivation by human double minute-2. Cancer Res. 2000;60:1810–1814. [Abstract] [Google Scholar]
- Loughran O, La Thangue NB. Apoptotic and growth-promoting activity of E2F modulated by MDM2. Mol Cell Biol. 2000;20:2186–2197. [Europe PMC free article] [Abstract] [Google Scholar]
- Lu W, Pochampally R, Chen L, Traidej M, Wang Y, Chen J. Nuclear exclusion of p53 in a subset of tumors requires MDM2 function. Oncogene. 2000;19:232–240. [Abstract] [Google Scholar]
- Martin SJ, Green DR. Apoptosis and cancer: The failure of controls on cell death and cell survival. Crit Rev Oncol Hematol. 1995;18:137–153. [Abstract] [Google Scholar]
- Mathupala SP, Heese C, Pedersen PL. Glucose catabolism in cancer cells. The type II hexokinase promoter contains functionally active response elements for the tumor suppressor p53. J Biol Chem. 1997;272:22776–22780. [Abstract] [Google Scholar]
- Molinari AM, Bontempo P, Schiavone EM, Tortora V, Verdicchio MA, Napolitano M, Nola E, Moncharmont B, Medici N, Nigro V, et al. Estradiol induces functional inactivation of p53 by intracellular redistribution. Cancer Res. 2000;60:2594–2597. [Abstract] [Google Scholar]
- Moll UM, Ostermeyer AG, Haladay R, Winkfield B, Frazier M, Zambetti G. Cytoplasmic sequestration of wild-type p53 protein impairs the G1 checkpoint after DNA damage. Mol Cell Biol. 1996;16:1126–1137. [Europe PMC free article] [Abstract] [Google Scholar]
- Momose F, Araida T, Negishi A, Ichijo H, Shioda S, Sasaki S. Variant sublines with different metastatic potentials selected in nude mice from human oral squamous cell carcinomas. J Oral Pathol Med. 1989;18:391–395. [Abstract] [Google Scholar]
- Mori N, Yamate J, Stassen AP, Oka S, Okumoto M, Tsubura A, Akamatsu T, Sakuma S, Demant P. Modulations of glucocorticoid-induced apoptosis linked to the p53 deletion and to the apoptosis susceptibility gene Rapop1 (radiation-induced apoptosis 1) Oncogene. 1999;18:4282–4285. [Abstract] [Google Scholar]
- Pison CM, Chauvin C, Fontaine E, Catelloni F, Keriel C, Paramelle B, Leverve XM. Mechanism of gluconeogenesis inhibition in rat hepatocytes isolated after in vivo hypoxia. Am J Physiol. 1995;268:E965–E973. [Abstract] [Google Scholar]
- Ravi R, Mookerjee B, Bhujwalla ZM, Sutter CH, Artemov D, Zeng Q, Dillehay LE, Madan A, Semenza GL, Bedi A. Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1α Genes & Dev. 2000;14:34–44. [Europe PMC free article] [Abstract] [Google Scholar]
- Reichardt HM, Umland T, Bauer A, Kretz O, Schutz G. Mice with an increased glucocorticoid receptor gene dosage show enhanced resistance to stress and endotoxic shock. Mol Cell Biol. 2000;20:9009–9017. [Europe PMC free article] [Abstract] [Google Scholar]
- Riva C, Chauvin C, Pison C, Leverve X. Cellular physiology and molecular events in hypoxia-induced apoptosis. Anticancer Res. 1998;18:4729–4736. [Abstract] [Google Scholar]
- Sasson R, Tajima K, Amsterdam A. Glucocorticoids protect against apoptosis induced by serum deprivation, cyclic adenosine 3′,5′-monophosphate and p53 activation in immortalized human granulosa cells: Involvement of Bcl-2. Endocrinology. 2001;142:802–811. [Abstract] [Google Scholar]
- Segnitz B, Gehring U. The function of steroid hormone receptors is inhibited by the hsp90-specific compound geldanamycin. J Biol Chem. 1997;272:18694–18701. [Abstract] [Google Scholar]
- Sengupta S, Ralhan R, Wasylyk B. Tumour regression in a ligand inducible manner mediated by a chimeric tumour suppressor derived from p53. Oncogene. 2000a;19:337–350. [Abstract] [Google Scholar]
- Sengupta S, Vonesch JL, Waltzinger C, Zheng H, Wasylyk B. Negative cross-talk between p53 and the glucocorticoid receptor and its role in neuroblastoma cells. EMBO J. 2000b;19:6051–6064. [Europe PMC free article] [Abstract] [Google Scholar]
- Sherr CJ, Weber JD. The ARF/p53 pathway. Curr Opin Genet Dev. 2000;10:94–99. [Abstract] [Google Scholar]
- Sionov RV, Haupt Y. The cellular response to p53: The decision between life and death. Oncogene. 1999;18:6145–6157. [Abstract] [Google Scholar]
- Stad R, Ramos YF, Little N, Grivell S, Attema J, van Der Eb AJ, Jochemsen AG. Hdmx stabilizes Mdm2 and p53. J Biol Chem. 2000;275:28039–28044. [Abstract] [Google Scholar]
- Stempien-Otero A, Karsan A, Cornejo CJ, Xiang H, Eunson T, Morrison RS, Kay M, Winn R, Harlan J. Mechanisms of hypoxia-induced endothelial cell death. Role of p53 in apoptosis. J Biol Chem. 1999;274:8039–8045. [Abstract] [Google Scholar]
- Stommel JM, Marchenko ND, Jimenez GS, Moll UM, Hope TJ, Wahl GM. A leucine-rich nuclear export signal in the p53 tetramerization domain: Regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18:1660–1672. [Europe PMC free article] [Abstract] [Google Scholar]
- Tao W, Levine AJ. Nucleocytoplasmic shuttling of oncoprotein Hdm2 is required for Hdm2-mediated degradation of p53. Proc Natl Acad Sci. 1999;96:3077–3080. [Europe PMC free article] [Abstract] [Google Scholar]
- Tian H, McKnight SL, Russell DW. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes & Dev. 1997;11:72–82. [Abstract] [Google Scholar]
- Treier M, Staszewski LM, Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994;78:787–798. [Abstract] [Google Scholar]
- Vojtesek B, Bartek J, Midgley CA, Lane DP. An immunochemical analysis of the human nuclear phosphoprotein p53. New monoclonal antibodies and epitope mapping using recombinant p53. J Immunol Methods. 1992;151:237–244. [Abstract] [Google Scholar]
- Wade-Evans A, Jenkins JR. Precise epitope mapping of the murine transformation-associated protein, p53. EMBO J. 1985;4:699–706. [Europe PMC free article] [Abstract] [Google Scholar]
- Wadhwa R, Takano S, Mitsui Y, Kaul SC. NIH 3T3 cells malignantly transformed by mot-2 show inactivation and cytoplasmic sequestration of the p53 protein. Cell Res. 1999;9:261–269. [Abstract] [Google Scholar]
- Wasylyk C, Salvi R, Argentini M, Dureuil C, Delumeau I, Abecassis J, Debussche L, Wasylyk B. p53 mediated death of cells overexpressing MDM2 by an inhibitor of MDM2 interaction with p53. Oncogene. 1999;18:1921–1934. [Abstract] [Google Scholar]
- Whitesell L, Cook P. Stable and specific binding of heat shock protein 90 by geldanamycin disrupts glucocorticoid receptor function in intact cells. Mol Endocrinol. 1996;10:705–712. [Abstract] [Google Scholar]
- Xing L, Quinn PG. Involvement of 3′,5′-cyclic adenosine monophosphate regulatory element binding protein (CREB) in both basal and hormone-mediated expression of the phosphoenolpyruvate carboxykinase (PEPCK) gene. Mol Endocrinol. 1993;7:1484–1494. [Abstract] [Google Scholar]
- Xu W, Liu L, Smith GC, Charles I, G. Nitric oxide upregulates expression of DNA-PKcs to protect cells from DNA-damaging anti-tumour agents. Nat Cell Biol. 2000;2:339–345. [Abstract] [Google Scholar]
- Yamamoto M, Fukuda K, Miura N, Suzuki R, Kido T, Komatsu Y. Inhibition by dexamethasone of transforming growth factor β1-induced apoptosis in rat hepatoma cells: A possible association with Bcl-xL induction. Hepatology. 1998;27:959–966. [Abstract] [Google Scholar]
- Yu ZK, Geyer RK, Maki CG. MDM2-dependent ubiquitination of nuclear and cytoplasmic P53. Oncogene. 2000;19:5892–5897. [Abstract] [Google Scholar]
- Zaika A, Marchenko N, Moll UM. Cytoplasmically “sequestered” wild type p53 protein is resistant to Mdm2-mediated degradation. J Biol Chem. 1999;274:27474–27480. [Abstract] [Google Scholar]
- Zimmer DB, Magnuson MA. Immunohistochemical localization of phosphoenolpyruvate carboxykinase in adult and developing mouse tissues. J Histochem Cytochem. 1990;38:171–178. [Abstract] [Google Scholar]
Articles from Genes & Development are provided here courtesy of Cold Spring Harbor Laboratory Press
Full text links
Read article at publisher's site: https://doi.org/10.1101/gad.202201
Read article for free, from open access legal sources, via Unpaywall:
http://genesdev.cshlp.org/content/15/18/2367.full.pdf
Citations & impact
Impact metrics
Article citations
Mouse liver sinusoidal endothelial cell responses to the glucocorticoid receptor agonist dexamethasone.
Front Pharmacol, 15:1377136, 08 Oct 2024
Cited by: 0 articles | PMID: 39439887 | PMCID: PMC11494038
Class 3 phosphoinositide 3-kinase promotes hepatic glucocorticoid receptor stability and transcriptional activity.
Acta Physiol (Oxf), 235(1):e13793, 07 Feb 2022
Cited by: 1 article | PMID: 35094500 | PMCID: PMC9539506
Effect of dexamethasone treatment at variable therapeutic windows in reversing nitrogen mustard-induced corneal injuries in rabbit ocular in vivo model.
Toxicol Appl Pharmacol, 437:115904, 30 Jan 2022
Cited by: 10 articles | PMID: 35108561 | PMCID: PMC8849585
Acquired Glucocorticoid Resistance Due to Homologous Glucocorticoid Receptor Downregulation: A Modern Look at an Age-Old Problem.
Cells, 10(10):2529, 24 Sep 2021
Cited by: 13 articles | PMID: 34685511 | PMCID: PMC8533966
Review Free full text in Europe PMC
Overcoming Glucocorticoid Resistance in Acute Lymphoblastic Leukemia: Repurposed Drugs Can Improve the Protocol.
Front Oncol, 11:617937, 11 Mar 2021
Cited by: 24 articles | PMID: 33777761 | PMCID: PMC7991804
Review Free full text in Europe PMC
Go to all (83) article citations
Other citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Nuclear and cytoplasmic degradation of endogenous p53 and HDM2 occurs during down-regulation of the p53 response after multiple types of DNA damage.
FASEB J, 17(12):1622-1630, 01 Sep 2003
Cited by: 30 articles | PMID: 12958168
Negative cross-talk between p53 and the glucocorticoid receptor and its role in neuroblastoma cells.
EMBO J, 19(22):6051-6064, 01 Nov 2000
Cited by: 67 articles | PMID: 11080152 | PMCID: PMC305812
A small synthetic peptide, which inhibits the p53-hdm2 interaction, stimulates the p53 pathway in tumour cell lines.
J Mol Biol, 299(1):245-253, 01 May 2000
Cited by: 69 articles | PMID: 10860736
Ser/Thr protein phosphatase type 5 (PP5) is a negative regulator of glucocorticoid receptor-mediated growth arrest.
Biochemistry, 38(28):8849-8857, 01 Jul 1999
Cited by: 73 articles | PMID: 10413457





