Abstract
Free full text

Physical association of HDAC1 and HDAC2 with p63 mediates transcriptional repression and tumor maintenance in squamous cell carcinoma
Associated Data
Abstract
Squamous cell carcinoma (SCC) is a treatment-refractory subtype of human cancer arising from stratified epithelium of the skin, lung, esophagus, oropharynx and other tissues. A unifying feature of SCC is high-level expression of the p53-related protein p63 (TP63) in 80% of cases. The major protein isoform of p63 expressed in SCC is ΔNp63α, an N-terminally truncated form which functions as a key SCC cell survival factor by mechanisms that are unclear. In this study we demonstrate that ΔNp63α associates with HDAC1 and HDAC2 to form an active transcriptional repressor complex that can be targeted to therapeutic advantage. Repression of pro-apoptotic Bcl-2 family member genes including PUMA by p63/HDAC is required for survival of SCC cells. Cisplatin chemotherapy, a mainstay of SCC treatment, promotes dissociation of p63 and HDAC from the PUMA promoter, leading to increased histone acetylation, PUMA activation and apoptosis. These effects are recapitulated upon targeting the p63/HDAC complex selectively with class I/II HDAC inhibitors using both in vitro and in vivo models. Sensitivity to HDAC inhibition is directly correlated with p63 expression and is abrogated in tumor cells that overexpress endogenous Bcl-2. Together, our results elucidate a mechanism of p63-mediated transcriptional repression and they identify the ΔNp63α/HDAC complex as an essential tumor maintenance factor in SCC. Additionally, our findings offer a rationale to apply HDAC inhibitors for SCC treatment.
Introduction
Understanding the biochemical basis for tumor maintenance is critical to the rational application of targeted therapeutic agents. In squamous cell carcinoma (SCC), the p53 family member p63 is a key survival factor whose inhibition by RNA interference induces apoptosis, and whose degradation by cisplatin-based chemotherapy is thought to be important for the therapeutic response to this agent (1-4). The p63 gene is expressed through two promoters as two distinct isoform classes which either contain (TAp63) or lack (ΔNp63) an N-terminal transactivation domain. Additional isoform heterogeneity is generated through alternative C-terminal splicing (5). Consistently, the major p63 isoform overexpressed in SCC is ΔNp63α, a protein which has been shown to function as a positive and negative transcriptional regulator of different target gene subsets (5, 6).
Given its potential therapeutic relevance, precisely how ΔNp63α mediates tumor cell survival is under intensive investigation. We previously demonstrated that ΔNp63α functions in part by binding and suppressing the pro-apoptotic activity of the related p53 family member p73 (1, 7). Whether binding to p73 is sufficient for tumor cell survival in this setting is unresolved. Additionally, we and others have observed localization of p63 to the promoters of pro-apoptotic Bcl-2 family members including PUMA, raising the possibility that ΔNp63α functions as an active transcriptional repressor (1, 8). Here, we use biochemical approaches to identify an endogenous repressor complex involving ΔNp63α, HDAC1 and HDAC2, and we demonstrate the potential relevance of p63/HDAC-mediated transcriptional repression in the response to cisplatin chemotherapy and HDAC inhibitor therapy in SCC.
Materials and Methods
Cell Lines and Xenograft Assays
Cell lines JHU-029, JHU-011 (1), and HO1N1 (9); KYSE-30, KYSE-150 (10); and FaDU (11) were the generous gifts of David Sidransky (Johns Hopkins University), S. Michael Rothenberg (MGH), and James Rocco (MGH), respectively. Each line was maintained by the MGH Center for Molecular Therapeutics cell line bank and underwent high-density SNP typing, revealing that each was unique compared to >800 other banked lines. Xenograft tumors were generated by subcutaneous injection of 2 × 106 JHU-029 tumor cells and 106 NIH 3T3 cells suspended in 1:1 matrigel (BD Biosciences): RPMI.
Lentiviral and retroviral production, Luciferase assays, and mRNA QRT-PCR
Production of virus, luciferase assays, and mRNA analysis were performed as described (1). Primers used for QRT-PCR are shown in Table S1.
Preparation of nuclear extracts and glycerol density gradient fractionation
Nuclear extracts were prepared by suspending cells in hypotonic buffer (10mM Tris-HCl pH 7.5, 1.5mM MgCl2, 10mM KCl) for 20 minutes, followed by douncing. Pelleted nuclei were suspended in 1 volume 20mM KCl nuclear buffer (20mM Tris-HCl pH 7.5, 1.5mM MgCl2, 0.2 mM EDTA, 25% glycerol). One volume 1.2M KCl nuclear buffer was added dropwise then incubated for 30 minutes at 4°C with rotation. Cleared supernatant was dialyzed against BC-100 buffer (100mM KCl, 20mM Tris-HCl pH 7.5, 0.2 mM EDTA, 20% glycerol). Glycerol density gradient fractionation was performed as previously described (12).
Tandem affinity purification
Cells were stably infected with pMSCV-ΔNp63α-FLAG-HA (C-terminal) or pMSCV-GFP-FLAG-HA plasmids, and cleared lysates from nuclear extracts were incubated for 4 hours with α-FLAG conjugated beads. Beads were washed with 100mM, 250mM, 500mM, 250mM, and 100mM KCl wash buffer (50mM Tris-HCl pH 7.5, 5mM MgCl2, 0.2 mM EDTA, 0.1% NP-40, 10% glycerol). Immune complexes were eluted with 0.5mg/mL FLAG peptide in 150mM KCl wash buffer. Eluate was incubated 12 hours at 4°C with α-HA conjugated beads. Beads were washed with 100mM, 200mM, 250mM, 200mM, and 100mM KCl wash buffer and boiled in Laemmli buffer. Proteins were visualized using the SilverQuest Silver Staining Kit (Invitrogen).
Immunoprecipitation and Chromatin Immunoprecipitation
Cleared nuclear lysates were incubated with antibody and protein A beads for 3 hours at 4°C, and immunocomplexes were washed with 100mM, 250mM, 400mM, 250mM, 100mM KCl wash buffer. For transient transfections, 293T cells were transfected with pcDNA-ΔNp63α–FLAG (C-terminal) mutants and pcDNA3-HDAC1. 40 hours post-transfection, cells were washed with cold PBS and incubated in hypotonic buffer for 20 minutes at 4°C. Following sonication, 3M KCl was added dropwise to a final concentration of 150mM and proteins were immunoprecipitated as above. ChIP was performed as previously described (13) with modifications detailed in Supplementary Methods.
Statistics
P values were determined using the student’s unpaired t test unless indicated otherwise. For correlation between ΔNp63α levels and TSA sensitivity, Pearson’s Product-moment Correlation Coefficient (R2) was calculated and a two tailed P-value was generated from a probability table.
Results and Discussion
Interaction between endogenous p63 and HDAC1/2
In order to uncover the biochemical basis for p63-dependent transcriptional regulation, we isolated p63-associated nuclear proteins from JHU-029, a human squamous cell carcinoma (SCC)-derived cell line in which endogenous p63 functions as an essential suppressor of apoptosis (1, 7). Using tandem affinity purification (TAP), we purified complexes from nuclear extracts of cells expressing either ΔNp63α-FLAG/HA or control nuclear GFP-FLAG/HA. Expected p63-associated proteins, including endogenous p63 and p73, were identified on silver-stained gels and subsequently confirmed by mass spectrometry (Figure 1A) (1, 14). The next most abundant silver-stained band, observed consistently following ΔNp63α but not GFP purification, contained HDAC1 and HDAC2 proteins (Figure 1A). To confirm the specificity of their interactions with p63, we performed western analysis for HDAC1 and HDAC2 following TAP for tagged ΔNp63α or nuclear GFP control. Consistent with our mass spectrometry findings, endogenous HDAC1 and HDAC2 specifically interacted with ΔNp63α but not with nuclear GFP (Figure 1A).
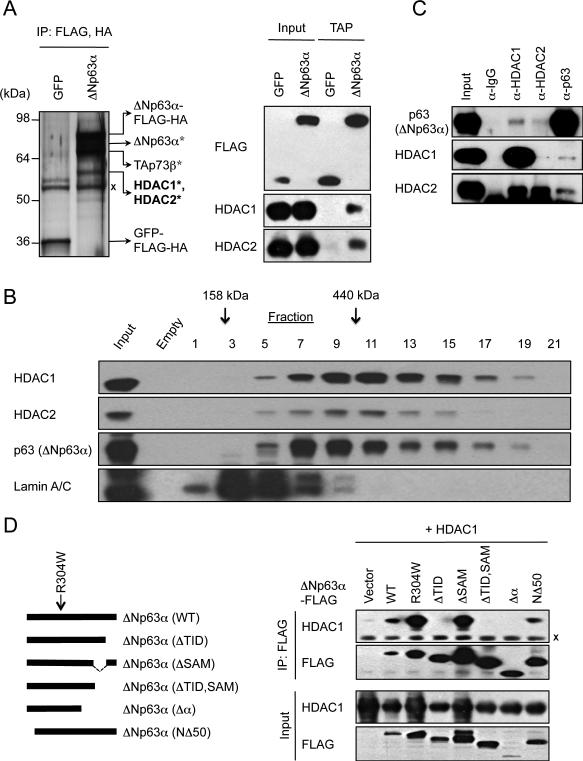
A, Left, silver-stained gel following TAP of C-terminal FLAG/HA-tagged ΔNp63α or control GFP proteins in JHU-029 cells. *Indicates proteins identified by mass spectrometry. X Ig heavy chain. Right, TAP/western blot confirmation of specific HDAC1 and HDAC2 binding to ΔNp63α. B, Co-fractionation of ΔNp63α HDAC1 and HDAC2 on a 10-40% glycerol density gradient in JHU-029 cells. Fraction numbers and molecular weight standards are indicated. Lamin A/C serves as a negative control. C, Association of endogenous proteins in JHU-029 nuclear extracts, assessed by IP/western analysis. D, The p63 TID domain is required for HDAC interaction. Left, schematic of ΔNp63α deletion mutants. Right, HDAC1 and ΔNp63α–FLAG were co-expressed in 293T cells, followed by α-FLAG IP. Details of p63 deletion constructs are shown in Figure S1B.
Using glycerol density gradient fractionation, we observed co-fractionation of endogenous ΔNp63α, HDAC1, and HDAC2 in complexes greater than 440kDa, potentially suggesting the presence of a complex involving these three proteins (Figure 1B). To confirm the endogenous association we performed reciprocal coimmunoprecipitations for p63, HDAC1 and HDAC2 in JHU-029 cells (Figure 1C) and a second HNSCC line, FaDU (Figure S1A), and observed a specific interaction between these three proteins. Finally, in order to examine these interactions in more detail we mapped the domain of p63 required for HDAC association. We transfected a series of FLAG-tagged p63 deletion mutants (Figure S1B) together with HDAC1 into 293T cells, and performed immunoprecipitations using either α-FLAG (Figure 1D) or α-HDAC1 (Figure S1C) antibodies. Remarkably, only the transactivation inhibitory domain (TID) of ΔNp63α was required for HDAC binding, while the sterile alpha motif (SAM) domain, a putative protein interaction domain, was entirely dispensable (5). Given the well-established association between HDAC1 and HDAC2 (15), our findings collectively suggest that ΔNp63α, HDAC1 and HDAC2 exist in a trimeric complex in SCC cells.
Requirement for p63 promoter association in p63-mediated repression
We hypothesized that p63 mediates direct transcriptional repression in SCC cells through recruitment of HDACs to the promoters of pro-apoptotic genes including PUMA. This hypothesis requires that p63 and HDACs are localized to this promoter, and that promoter binding by p63 is essential for its ability to repress transcription. We therefore performed chromatin immunoprecipitation (ChIP) for p63 and HDAC1 in SCC cells, and observed specific binding of both endogenous proteins to the PUMA locus (Figure 2A). Binding of p63 and HDAC1 was also observed within the regulatory regions of other p63-repressed genes (Figure S2A). To address the functional contribution of promoter binding by ΔNp63α we first used a PUMA promoter reporter assay (1). We co-expressed either wild-type ΔNp63α (WT) or a naturally-occurring DNA binding-deficient point mutant, ΔNp63α (R304W) (5), together with TAp73β or p53 and examined luciferase activity. Wild-type ΔNp63α was a potent suppressor of both p73 and p53-dependent PUMA reporter activation, while the non-DNA binding mutant ΔNp63α (R304W) was defective in suppressing activation (Figure 2B). Of note, the p63 mutant was expressed at similar levels as the wild-type (Figure 2B) and exhibits comparable binding to p73 (Figure S2B) but not to p53 (Figure S2C).

A, ChIP showing endogenous p63 and HDAC1 preferentially localized to the p53-family binding site in the PUMA promoter versus the control (ACTB) promoter. **p<0.01; ***p<.001. B, Repression of p53/p73-dependent transactivation by ΔNp63α requires promoter binding, assessed using a PUMA promoter reporter as described in Materials and Methods. Partial repression of p73 activity by ΔNp63α R304W reflects its binding to p73 but not p53. C, Binding of tagged wild-type (WT) but not mutant (R304W) ΔNp63α to the endogenous PUMA promoter assessed by ChIP in JHU-029 cells. # Denotes shRNA-resistant construct. D, Repression of endogenous PUMA by wild-type but not mutant ΔNp63α following lentiviral shRNA knockdown of endogenous p63 in JHU-029 cells, assessed by real-time quantitative RT-PCR (QRT-PCR) at 72 hours. Values are normalized to ACTB and expressed relative to cells transduced with control (GFP-directed) shRNA. *p<0.05. All error bars +/- SEM for triplicate experiments.
Since transient reporter assays lack chromatin context, we next tested whether suppression of endogenous PUMA transcription required DNA-bound p63. We expressed retroviral FLAG-tagged wild-type or mutant ΔNp63α (R304W) in SCC cells, then performed ChIP using an anti-FLAG antibody. As expected, wild-type ΔNp63α showed significant binding to the PUMA promoter, while the mutant showed little or no binding over background (Figure 2C). As a functional test we then ablated expression of endogenous p63 in these cells, having engineered the ectopic ΔNp63α constructs to contain silent point mutants which made them resistant to the lentiviral shRNA (Figure S2D). Ectopic wild-type ΔNp63α nearly completely suppressed PUMA induction following endogenous p63 knockdown, while mutant ΔNp63α-expressing cells showed dramatic PUMA induction (Figures 2D and S2E) and cell death (Figure S2F) in this setting. Taken together, these data demonstrate the requirement for promoter-bound p63 in suppression of endogenous PUMA transcription and cell death in SCC cells.
HDAC and p63-dependent regulation of PUMA and chemotherapy response in SCC cells
Having documented the presence of endogenous HDAC1 and p63 at the PUMA promoter (Figure 2A) we wished to test the biochemical requirement for HDAC activity in PUMA regulation. Treatment with the potent class I/II HDAC inhibitor trichostatin A (TSA) caused a dose-dependent induction of PUMA mRNA in three different SCC cell lines (Figure 3A). A similar dose-dependent induction of PUMA was observed following treatment with vorinostat (SAHA), a second generation inhibitor which is currently FDA-approved for treatment of cutaneous T cell lymphoma (CTCL) (Figure S3A) (16). PUMA mRNA induction by TSA corresponded temporally with increased histone H4 acetylation at the p63 binding site within the PUMA promoter (Figures 3B and S3B), consistent with a direct effect of the inhibitor at this promoter. In order to demonstrate directly a connection between the presence of p63 and HDAC activity at the PUMA promoter we assayed histone acetylation following p63 knockdown in SCC cells. Indeed, histone H4 acetylation was significantly induced following ablation of p63, concurrent with endogenous PUMA up-regulation (Figures 3C and S3C). Thus, HDAC activity controls PUMA expression in SCC cells in a p63-dependent manner.
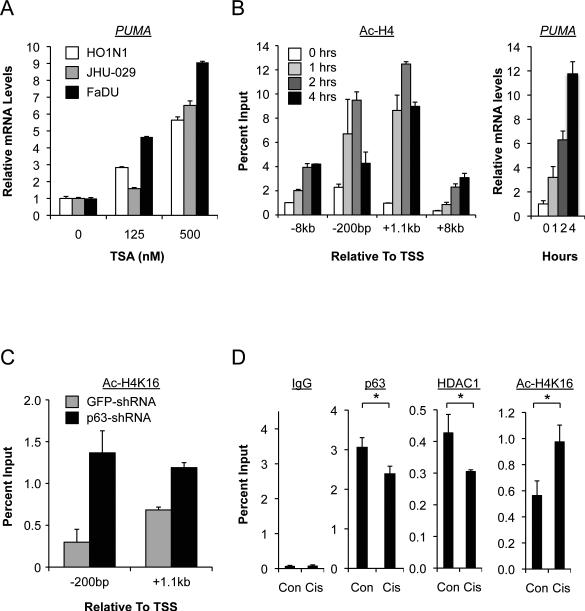
A, HDAC inhibition induces PUMA in SCC cells, assessed by QRT-PCR at 4 hours. B, Left, ChIP showing TSA (500nM) increases histone H4 acetylation which coincides temporally with induction of endogenous PUMA, right. Note that -200bp relative to the Transcriptional Start Site represents the p63 binding site in the PUMA promoter. C, ChIP showing histone H4 deacetylation of the PUMA locus is reversed by p63 knockdown using lentiviral shRNA at 48 hours. D, ChIP showing cisplatin (4μM, 24 hours) causes coordinate reversal of p63/HDAC1 occupancy and histone H4 deacetylation at the PUMA promoter (-200bp), which coincides with PUMA induction (Figure S3D). *p<0.05.
Cisplatin-based chemotherapy, a mainstay for treatment of advanced head and neck SCC (HNSCC), promotes degradation of ΔNp63α and induction of PUMA which have been linked to the therapeutic response in this disease (3, 4). We found that disruption of the p63/HDAC complex contributes to the response to cisplatin, as PUMA expression induced by cisplatin (Figures S3D and S3E) was accompanied by a loss of endogenous p63 and HDAC1 at the PUMA promoter, and by an increase in histone acetylation (Figures 3D and S3D). Thus p63/HDAC-mediated PUMA transcriptional repression is mitigated in the physiological response to cisplatin chemotherapy.
Targeting p63/HDAC-dependent transcriptional repression in SCC
We have demonstrated previously that some SCC cell lines are able to bypass the requirement for ΔNp63α as a survival factor through overexpression of endogenous Bcl-2 itself (1, 17). Consistent with this observation, we found that SCC lines which exhibit low expression of ΔNp63α showed high-level expression of Bcl-2, and vice-versa (Figure 4A and reference (1)). We therefore hypothesized that lines with high ΔNp63α expression are “addicted” to ΔNp63α/HDAC function and therefore would be sensitive to HDAC inhibition, while lines with low ΔNp63α would exhibit HDAC inhibitor resistance. Indeed, we observed a direct correlation between ΔNp63α protein levels and sensitivity to TSA in SCC cells (Figures 4B and S4A). Additionally, we found that ectopic Bcl-2 expression was sufficient to confer remarkable in vitro TSA resistance in the TSA-sensitive line JHU-029 (Figure S4B). Thus, although multiple pathways may contribute to effects of HDAC inhibition in SCC cells (18), these data support a prominent role for the p63-dependent pathway we describe here.

A, Inverse correlation between ΔNp63α and Bcl-2 levels in the indicated SCC lines, assessed by western blot. GAPDH serves as a loading control. B, Direct correlation between endogenous ΔNp63α protein levels, assessed by densitometry, and sensitivity to TSA in SCC cells, assessed by 8-point standard curve at 3 days (Figure S4A). All values are relative to FaDU cells. C, Vorinostat blocks tumor progression in SCC in vivo, but Bcl-2 induces complete resistance. JHU-029/GFP (left) or JHU-029/Bcl-2 (right) xenografts in Nude mice were treated either with DMSO vehicle (n=22, 22) or 50mg/kg vorinostat (n=22, 16) daily by IP injection starting at day 9. *** p<0.001 by multiple measures ANOVA. Error bars indicate +/- SEM. D, Apoptosis is induced by HDAC inhibition in vivo. Left, lysates from the indicated xenograft tumors were examined for cleaved PARP1. Right, cleaved Caspase 3 was detected by IHC in sections from the indicated tumors. *p<0.05. Error bars indicate +/- SEM for representative fields (500 cells/field) from 32 tumors (GFP) or 14 tumors (Bcl-2).
Finally, we sought to model HDAC inhibition for treatment of SCC in vivo and to determine the contribution of Bcl-2 expression in this setting. Notably, we recently showed that Bcl-2 expression in primary HNSCC is an intrinsic resistance factor and a powerful predictor of relapse following cisplatin-based therapy (17). We established a xenograft assay using JHU-029 cells, which form tumors in 100% of Nude mice when injected subcutaneously. Mice bearing palpable tumors, derived from JHU-029 cells expressing either a retroviral control (GFP) or Bcl-2 vector, were treated by IP injection with vorinostat or vehicle control. Vorinostat treatment substantially and consistently blocked tumor progression in mice with GFP-expressing tumors (Figure 4C). Remarkably, however, expression of Bcl-2 induced complete resistance to vorinostat treatment (Figure 4C). To determine the physiological basis for the response to HDAC inhibition in vivo we assayed markers of proliferation and apoptosis in these tumors. We observed no difference in proliferation following vorinostat treatment in any tumors, as assessed by Ki67 staining (Figures S4C,D). In contrast, control vorinostat-treated tumors showed substantial cleaved PARP-1 (Figure 4D) and activated Caspase 3 (Figures 4D), which were completely absent in Bcl-2-expressing tumors. All together, our findings demonstrate the presence of a functional p63/HDAC complex which serves as a direct repressor of the apoptotic transcriptional program in SCC. HDAC inhibitors target this complex to induce tumor cell killing through up-regulation of pro-apoptotic Bcl-2 family members, while sensitivity to these drugs can be abrogated in tumor cells that overexpress Bcl-2.
These findings reveal a tumor-specific context for HDAC function in SCC which will inform the rational and effective application of these agents. For example, a recent clinical trial of late-stage, chemotherapy-refractory HNSCC patients treated with vorinostat did not show clinical responses (19). This finding is consistent with our data demonstrating that a common transcriptional and apoptotic response pathway involving p63 and HDAC1/2 appears to participate in the response to both cisplatin and HDAC inhibitors. Conceivably, treating patients earlier in the course of disease may improve the efficacy of HDAC inhibition in SCC. Our study also provides insight into a specific resistance mechanism, suggesting that HDAC inhibitors may not be useful as single agents in Bcl-2 positive SCCs. An attractive approach for these tumors might instead include Bcl-2 inhibitors, which are currently in clinical trials, either alone or in combination with HDAC inhibitors (20). If successful, such a stratified and targeted approach based on an understanding of tumor-selective biology would represent a significant advance against this disease.
Acknowledgements
We thank Jonathan Whetstine, David Sweetser, and Anders Naar for helpful advice and reagents; Kristine Torres-Lockhart, Mary Lynch, Zach Nash, and Catherine Wilson for technical assistance, and the Taplin Mass Spectrometry facility (Harvard Medical School) for protein identification.
Grant Support: NIH R01 DE-015945 (L.W.E., L.H.) ; ACS/Mass Biotech Council Cancer Research Challenge-AstraZeneca Pharmaceuticals LP Fellowship PF-09-100-01 MGO (M.R.R); Tracey Davis Memorial Fund (N.F.); Fondation pour la Recherche Medicale (B.O.).
Footnotes
Conflicts of interest: The authors declare no competing conflicts of interest.
References
Full text links
Read article at publisher's site: https://doi.org/10.1158/0008-5472.can-11-0046
Read article for free, from open access legal sources, via Unpaywall:
https://aacrjournals.org/cancerres/article-pdf/71/13/4373/2655658/4373.pdf
Citations & impact
Impact metrics
Citations of article over time
Article citations
Type I interferon signaling induces melanoma cell-intrinsic PD-1 and its inhibition antagonizes immune checkpoint blockade.
Nat Commun, 15(1):7165, 26 Aug 2024
Cited by: 0 articles | PMID: 39187481 | PMCID: PMC11347607
Histone modifications in head and neck squamous cell carcinoma.
Front Oncol, 14:1427725, 25 Jun 2024
Cited by: 0 articles | PMID: 38983924 | PMCID: PMC11231198
Review Free full text in Europe PMC
Identification of HTRA4 as a Transcriptional Target of p63 in Trophoblast.
Am J Pathol, 194(7):1162-1170, 01 Jul 2024
Cited by: 0 articles | PMID: 38880601
Targeting histone deacetylases in head and neck squamous cell carcinoma: molecular mechanisms and therapeutic targets.
J Transl Med, 22(1):418, 03 May 2024
Cited by: 0 articles | PMID: 38702756
Review
HDAC2 as a target for developing anti-cancer drugs.
Comput Struct Biotechnol J, 21:2048-2057, 13 Mar 2023
Cited by: 13 articles | PMID: 36968022 | PMCID: PMC10030825
Review Free full text in Europe PMC
Go to all (77) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
ΔNp63α utilizes multiple mechanisms to repress transcription in squamous cell carcinoma cells.
Cell Cycle, 12(3):409-416, 16 Jan 2013
Cited by: 12 articles | PMID: 23324337 | PMCID: PMC3587441
p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis.
Cancer Cell, 9(1):45-56, 01 Jan 2006
Cited by: 303 articles | PMID: 16413471
Identifying pathways regulating the oncogenic p53 family member ΔNp63 provides therapeutic avenues for squamous cell carcinoma.
Cell Mol Biol Lett, 27(1):18, 23 Feb 2022
Cited by: 4 articles | PMID: 35196980 | PMCID: PMC8903560
TP63 links chromatin remodeling and enhancer reprogramming to epidermal differentiation and squamous cell carcinoma development.
Cell Mol Life Sci, 77(21):4325-4346, 23 May 2020
Cited by: 36 articles | PMID: 32447427 | PMCID: PMC7588389
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NIDCR NIH HHS (3)
Grant ID: R01 DE-015945
Grant ID: R01 DE015945-07
Grant ID: R01 DE015945




