Abstract
Purpose
Prostate specific antigen and free prostate specific antigen have limited specificity to detect clinically significant, curable prostate cancer, leading to unnecessary biopsy, and detection and treatment of some indolent tumors. Specificity to detect clinically significant prostate cancer may be improved by [-2]pro-prostate specific antigen. We evaluated [-2]pro-prostate specific antigen, free prostate specific antigen and prostate specific antigen using the formula, ([-2]pro-prostate specific antigen/free prostate specific antigen × prostate specific antigen(1/2)) to enhance specificity to detect overall and high grade prostate cancer.Materials and methods
We enrolled 892 men with no history of prostate cancer, normal rectal examination, prostate specific antigen 2 to 10 ng/ml and 6-core or greater prostate biopsy in a prospective multi-institutional trial. We examined the relationship of serum prostate specific antigen, free-to-total prostate specific antigen and the prostate health index with biopsy results. Primary end points were specificity and AUC using the prostate health index to detect overall and Gleason 7 or greater prostate cancer on biopsy compared with those of free-to-total prostate specific antigen.Results
In the 2 to 10 ng/ml prostate specific antigen range at 80% to 95% sensitivity the specificity and AUC (0.703) of the prostate health index exceeded those of prostate specific antigen and free-to-total prostate specific antigen. An increasing prostate health index was associated with a 4.7-fold increased risk of prostate cancer and a 1.61-fold increased risk of Gleason score greater than or equal to 4 + 3 = 7 disease on biopsy. The AUC of the index exceeded that of free-to-total prostate specific antigen (0.724 vs 0.670) to discriminate prostate cancer with Gleason 4 or greater + 3 from lower grade disease or negative biopsy. Prostate health index results were not associated with age and prostate volume.Conclusions
The prostate health index may be useful in prostate cancer screening to decrease unnecessary biopsy in men 50 years old or older with prostate specific antigen 2 to 10 ng/ml and negative digital rectal examination with minimal loss in sensitivity.Free full text

A Multi-Center Study of [−2]Pro-Prostate-Specific Antigen (PSA) in Combination with PSA and Free PSA for Prostate Cancer Detection in the 2.0 to 10.0 ng/mL PSA Range
Abstract
Purpose
PSA and free PSA (fPSA) have limited specificity for detecting clinically significant, curable prostate cancer (PCa), leading to unnecessary biopsies and detection and treatment of some indolent tumors. [−2]proPSA (p2PSA) may improve specificity for detecting clinically significant PCa. Our objective was to evaluate p2PSA, fPSA, and PSA in a mathematical formula (prostate health index [phi] = [−2]proPSA / fPSA) × PSA1/2) to enhance specificity for detecting overall and high-grade PCa.
Materials and Methods
We enrolled 892 men in a prospective multi-institutional trial with no history of PCa, normal rectal examination, a PSA of 2–10 ng/mL, and ≥6- core prostate biopsy. We examined the relationship of serum PSA, %fPSA and phi with biopsy results. The primary endpoints were the specificity and AUC using phi to detect overall and Gleason ≥7 prostate cancer on biopsy compared with %fPSA.
Results
For the 2–10 ng/mL PSA range, at 80–95% sensitivity, the specificity and AUC (0.703) of phi exceeded those of PSA and %fPSA. Increasing phi was associated with a 4.7-fold increased risk of PCa and 1.61-fold increased risk of Gleason ≥7 disease on biopsy. The AUC for phi (0.724) exceeded that of %fPSA (0.670) in discriminating between PCa with Gleason ≥ 4+3 vs. lower grade disease or negative biopsies. Phi results were not associated with age and prostate volume.
Conclusions
Phi may be useful in PCa screening to reduce unnecessary biopsies in men age ≥50 years with PSA 2–10 ng/mL and negative DRE, with minimal loss in sensitivity.
INTRODUCTION
PSA testing was approved by the FDA using a 4.0 ng/mL cutoff for recommending prostate biopsy. Lower cutoffs further enhance early prostate cancer (PCa) detection,1 since PSA correlates with the risk of overall and high-grade PCa at PSA concentrations <4 ng/mL.2 However, PSA testing may be confounded by benign conditions.
The low specificity at PSA <10.0 ng/mL has created a diagnostic gray zone in which PCa is found on biopsy in ~25% of patients. This is important, since most PCa is curable at PSA <10.0 ng/mL; whereas, PSA >10 ng/mL often portends advanced disease.3
PSA in serum is either complexed with proteins or in an unbound form called free PSA (fPSA).4 At PSA levels of 4.0–10.0 ng/mL, the ratio of fPSA to PSA (%fPSA) significantly improves discrimination between PCa and benign conditions.5
Different regions of the prostate contain varying proportions of fPSA isoforms, including proPSA that is associated with PCa. [−2]proPSA (p2PSA) is the primary form in PCa tissue.6–8 At PSA of 2.0–10.0 ng/mL, p2PSA further improves specificity for PCa detection relative to %fPSA.9–13
The utility of p2PSA at PSA <4.0 ng/mL and its relationship to PCa aggressiveness are relevant to the PCa screening debate, including concerns about overdiagnosis and overtreatment.13–19 Preliminary evidence suggests that a higher percentage of p2PSA may be associated with more aggressive PCa.10, 12, 13, 19
Selecting thresholds for clinical use of p2PSA has received limited study. We evaluated the relationship of p2PSA** combined with fPSA and PSA in a mathematical formula called Prostate Health Index (phi) with prostate cancer detection and tumor features.
METHODS
Study Design
We conducted a multi-center, double-blind, case-control clinical trial to validate phi in the 2.0–10.0 ng/mL PSA range. This formula was developed from an independent dataset,20 and is calculated as (p2PSA pg/mL / fPSA ng/mL) × (PSA ng/mL) ½. Intuitively, higher [−2] proPSA and PSA with a lower fPSA has greater likelihood of PCa. The study protocol was approved by the IRB of each participating institution, and all participants provided informed consent.
Study population
We evaluated 1372 men from October 2003 through June 2009 from 8 medical centers. The study cohort included men age ≥50 years of all ethnic backgrounds who met the following criteria: (1) no history of PCa, (2) non-suspicious digital rectal examination (DRE) findings, (3) pre-study PSA of 1.5–11.0 ng/mL (all PSA concentrations were re-tested in the Access Hybritech assay, and only those 2–10 ng/mL were included), (4) ≥6 core biopsy within 6 months of blood draw, and (5) a histologic diagnosis from prostate biopsy.
Exclusion criteria were: (1) treatment with medications that alter PSA levels or interventions such as transurethral resection of the prostate prior to blood draw, (2) acute prostatitis or urinary infection at blood draw, (3) a final Access Hybritech PSA value outside the 2.0–10.0 ng/mL range, (4) no blood draw or biopsy at the appropriate time interval, or (5) prior androgen-replacement therapy.
Seven men were excluded due to unevaluable tests from hemolyzed or lipemic samples or p2PSA duplicate results with >15% coefficient of variation at p2PSA concentrations ≤ 20 pg/mL, for which samples could not be retested. Finally, one site enrolled only men aged 55–75 years (our study enrolled men aged ≥ 50 years), and our study-specific sample storage limit (≤ 5 years) further limited the evaluable population to men aged 62–74. Because the age distribution from this site may not be representative of the target population, we performed separate analyses excluding and including these men.
The final study population of 892 men included: (1) 121 (13.6%) prospectively enrolled, (2) 743 (83.3%) prospectively enrolled under separate protocols, and (3) 28 (3.1%) retrospective samples. The study population included 706 (79.2%) initial biopsies, 159 (17.8%) repeat biopsies, and 27 (3%) with unknown history of prior biopsy. Each institution enrolled an approximately equal number of men with or without PCa, for a total of 430 (48.2%) men with PCa and 462 (51.8%) without. Participants and investigators were blinded to p2PSA results, and testing sites were blinded to individual clinical information.
Test Methods
Access Hybritech p2PSA, PSA, and fPSA assays were measured on the Beckman Coulter Access 2 Immunoassay Analyzer***. Serum samples were collected and processed within 8 hours, then stored frozen at ≤−70°C prior to testing (≤5 years from the date of blood draw), conditions that allowed accurate measurement of phi.21 Samples were tested at one of 3 laboratories. PSA and fPSA assays were run using one-sample replicate. The p2PSA assay was run in duplicate (first replicate used for data analysis, consistent with the proposed product labeling) according to the testing protocol. Evaluation of the first replicate compared to the mean of duplicates using Passing-Bablock regression analyses showed no difference (Spearman R=0.9985). The p2PSA assay is a two-site immunoenzymatic sandwich assay using specific monoclonal antibodies and 6 calibrators from 0- 5000 pg/mL.
Statistical Methods
The minimum sample size was estimated as 295 patients without cancer to detect a 10% difference in specificity between phi and % fPSA at α = 0.05 and β = 0.10. In addition, a minimum sample size of 350 cancer patients was determined to accurately estimate sensitivity at 95% with a 95% confidence interval of ± <3%. The target sample size was then increased to 400 participants in each group.
The primary null hypothesis was that phi has no greater specificity than %fPSA at 95% sensitivity. This hypothesis was tested using bootstrap-based receiver operating characteristic (ROC) analysis.22 Briefly, 1000 datasets of benign and PCa patients were generated to repetitively sample the study population.23–25 Differences in the specificity between phi and %fPSA at 95% sensitivity were calculated for the 1000 pairs of replicate datasets. The standard error of the difference in specificities was then estimated with adjustment for correlation between the results of the two tests. Finally, the bootstrap-estimated standard error was used to evaluate whether the difference in specificities is >0 assuming normal distribution of the differences. A one-sided statistical test was performed for this analysis. This method was also used to compare the specificities of phi and %fPSA at 90%, 85%, and 80% sensitivities.
The secondary null hypothesis was that the area under the ROC curve (AUC) for phi equals that of %fPSA. This hypothesis was tested by evaluating whether the difference between the estimated AUCs for the two tests equals 0 using empirical methods.26, 27 The standard error of the difference was calculated accounting for the correlation in AUCs as appropriate for comparison of paired data. The difference between the two estimated AUCs has been shown to have a Chi-square distribution with one degree of freedom. The AUCs for phi and %fPSA were also estimated for each prostate volume tertile to determine whether the observed trend in AUCs differed by prostate volume.
The validity of pooling data across sites was evaluated by fitting a logistic regression model with cancer status as the dependent variable, with phi (dichotomized at the estimated cutoff for 95% sensitivity) and site as independent predictors including interaction terms for site and phi. A statistically significant parameter estimates for this interaction terms was considered evidence of heterogeneity in phi performance by site.
Comparisons between participant subgroups were performed using the Wilcoxon Rank-Sum test for continuous variables and the χ2 test for categorical variables. Two-sided statistical tests were used on all analyses except as noted above, and statistical significance was defined as p<0.05. All analyses were performed using SAS version 9.2 (SAS Institute, Cary, North Carolina).
Individual Patient Risk Assessment
A 25% PCa detection rate has been previously reported in men with PSA of 2.0–10.0 ng/mL.3 For this study, cancer patients were over-sampled by design, resulting in 48.2% of study participants with PCa. Since the proportion of PCa was determined by design, direct calculation of PCa probability would result in inflated estimates for detecting PCa. Therefore, to obtain more accurate risk estimates for PCa, we adjusted the proportion of PCa to 25% by repetitively sampling the study population 1000 times with each replicate dataset consisting of 462 (75%) benign and 154 (25%) cancer participants.23–25 The mean probability of cancer in the bootstrapped datasets for each phi range was used as the point estimate, and bootstrap-estimated standard errors were used to calculate 95% confidence intervals. Likewise, relative risk estimates were calculated for each replicate dataset by dividing the probability of PCa in each phi range to that of phi 0–24.9. The mean relative risk and bootstrap-estimated standard errors were used to calculate the risk estimate and 95% confidence intervals. In addition, age-stratified probability estimates for PCa were calculated to determine whether observed trends persist in all age groups.
Association of phi with Gleason Score
Among participants with PCa, the probability of a Gleason score ≥7 was calculated directly from the proportion of participants in each phi range with Gleason score ≥7. Risk ratios were estimated by dividing the probability of Gleason score ≥7 in each phi range to that of phi 0–24.9. Confidence intervals were calculated using the normal approximation of the binomial distribution. The Cochrane-Armitage test for trend was used to determine whether increasing phi ranges corresponds to increasing probability of PCa with Gleason score ≥7. ROC analysis was used to evaluate the clinical utility of phi in detecting PCa with Gleason scores 4+3 or higher.
RESULTS
Participants
Table 1 shows the demographics and results for each assay. Both phi and p2PSA were significantly higher in PCa than controls; whereas, fPSA and %fPSA were lower in PCa than controls. Total PSA and age were comparable between groups.
TABLE 1
Clinical Characteristics of the Study Population
| Characteristic | Benign N=462 | Cancer N=430 | p-value | Total N=892 | |
|---|---|---|---|---|---|
| Age | Median | 63.0 | 63.0 | 63.0 | |
| Mean ± SD | 62.6 ± 7.0 | 63.0 ± 7.1 | 62.8 (7.0) | ||
| Range | 50 – 84 | 50 – 84 | 50 – 84 | ||
| 0.477 | |||||
| Race, n(%) | Caucasian | 361 (78.1) | 365 (84.9) | 726 (81.4) | |
| African-American | 24 (5.2) | 22 (5.1) | 46 (5.2) | ||
| Other | 22 (4.8) | 9 (2.1) | 31 (3.5) | ||
| Unknown | 55 (11.9) | 34 (7.9) | 89 (10.0) | ||
| 0.025 | |||||
| Ethnicity, n(%) | Hispanic | 14 (3.0) | 6 (1.4) | 20 (2.2) | |
| Not Hispanic | 187 (40.5) | 153 (35.6) | 340 (38.1) | ||
| Unknown | 261 (56.5) | 271 (63.0) | 532 (59.6) | ||
| 0.059 | |||||
| Prostate Volume | Median | 51.0 | 40.0 | 45.0 | |
| Mean ± SD | 55.1 ± 23.2 | 44.3 ± 19.4 | 50.1 ± 22.2 | ||
| Range | 16 – 209 | 14 – 120 | 14 – 209 | ||
| <0.001 | |||||
| Prior Biopsy, n(%) | No prior biopsy | 345 (74.7) | 361 (84.0) | 706 (79.2) | |
| Prior biopsy | 105 (22.7) | 54 (12.6) | 159 (17.8) | ||
| Unknown | 12 (2.6) | 15 (3.5) | 27 (3.0) | ||
| <0.001 | |||||
| Gleason Score, n(%) | 5 | Not Applicable | 1 (0.2) | 1 (0.2) | |
| 6 | 289 (67.2) | 289 (67.2) | |||
| 7 | 119 (27.7) | 119 (27.7) | |||
| 8 | 9 (2.0) | 9 (2.0) | |||
| 9 | 11 (2.6) | 11 (2.6) | |||
| Unknown | 1 (0.2) | 1 (0.2) | |||
| Not Applicable | |||||
| PSA (ng/mL) | Median | 5.1 | 5.3 | 5.1 | |
| Mean ± SD | 5.3 ± 1.9 | 5.4 ± 1.9 | 5.4 ± 1.9 | ||
| Range | 2.0 – 10.0 | 2.0 – 9.8 | 2.0 – 10.0 | ||
| 0.199 | |||||
| fPSA (ng/mL) | Median | 1.0 | 0.7 | 0.9 | |
| Mean ± SD | 1.0 ± 0.5 | 0.9 ± 0.5 | 1.0 ± 0.5 | ||
| Range | 0.1 – 4.3 | 0.2 – 3.9 | 0.1 – 4.3 | ||
| <0.001 | |||||
| [−2]proPSA (pg/mL) | Median | 12.9 | 14.1 | 13.3 | |
| Mean ± SD | 14.4 ± 7.1 | 16.8 ± 11.1 | 15.5 ± 9.3 | ||
| Range | 2.9 – 43.5 | 2.9 – 93.5 | 2.9 – 93.5 | ||
| 0.003 | |||||
| %fPSA | Median | 18.8 | 15.1 | 17.0 | |
| Mean ± SD | 20.0 ± 8.0 | 16.4 ± 7.6 | 18.3 ± 8.0 | ||
| Range | 3.1 – 53.2 | 3.7 – 51.1 | 3.1 – 53.2 | ||
| <0.001 | |||||
| phi | Median | 30.3 | 42.2 | 34.7 | |
| Mean ± SD | 33.9 ± 15.0 | 49.2 ± 31.3 | 41.3 ± 25.5 | ||
| Range | 13.7 – 98.2 | 10.2 – 325.8 | 10.2 – 325.8 | ||
| <0.001 | |||||
Of the participants, 89.8% had ≥12-core biopsy, and 98% had ≥10 cores. Overall, 30.6%, 49.9%, and 19.6% of participants were aged 50–59, 60–69 and 70–84 years, respectively. Mean age and PSA were similar across the 7 clinical sites. In addition, none of the interaction terms in the statistical model for evaluating heterogeneity by site was significant, supporting data pooling across sites. There were no significant differences in age (P=0.123), PSA (P=0.106), p2PSA (P=0.088), %fPSA (P=0.125), or phi (P=0.848) between Caucasians and African-Americans.
Receiver Operating Characteristic (ROC) Results
Figure 1 shows the sensitivity and specificity for all observed PSA, fPSA, p2PSA, %fPSA, and phi cutoffs in the 2.0–10.0 ng/mL PSA range. At a given sensitivity, phi demonstrated greater specificity than the other analytes (Table 2). At 95% sensitivity, the specificity of phi was 16.0% compared to 8.4% for %fPSA (P=0.015), 7.6% for p2PSA, 6.5% for PSA, and 3.5% for fPSA, rejecting the primary null hypothesis. Moreover, at lower sensitivities (90%, 85%, and 80%) for PCa detection, the specificity of phi was significantly greater than %fPSA (i.e., unnecessary biopsies possibly avoided: 26% vs. 18%, P= 0.036; 39% vs. 28%, P= 0.006; 45% vs. 37%, P= 0.031, respectively).
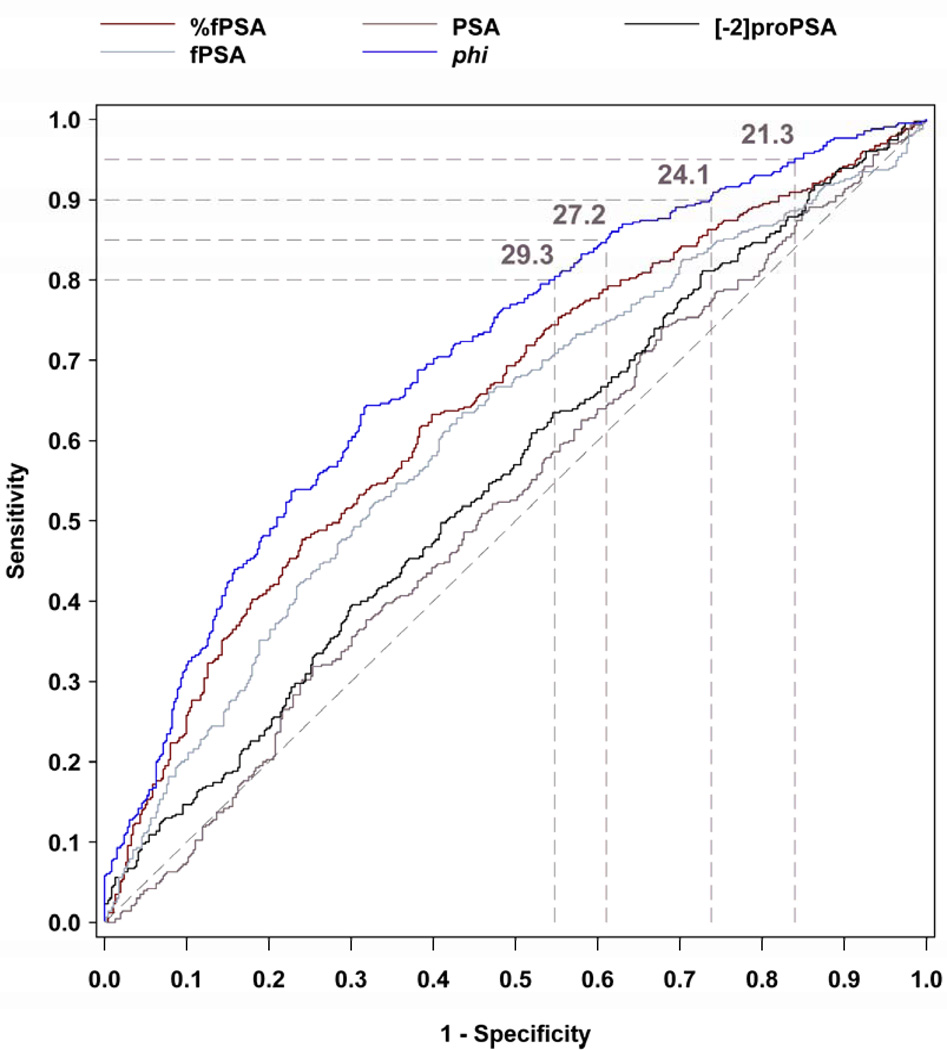
PSA, fPSA, [−2]proPSA, %fPSA, and Phi ROC Curves in the 2–10 ng/mL PSA Range Sensitivity × 1-Specificity for Sequential Cutpoints
TABLE 2
Sensitivity and Specificity for PCa Using Various phi Cutoffs in Men with Non-Suspicious DRE
| % Sensitivity | phi Cutoff | % Specificity (n) |
|---|---|---|
| 99 | 17.2 | 5.2 (24) |
| 98 | 18.4 | 8.4 (39) |
| 95 | 21.3 | 16.0 (74) |
| 90 | 24.1 | 26.2 (121) |
| 89.1 | 25.0 | 29.4 (136) |
| 85 | 27.2 | 39.0 (180) |
| 80 | 29.3 | 45.2 (209) |
| 75 | 31.1 | 52.6 (243) |
| 70 | 33.4 | 60.0 (277) |
| 65 | 35.0 | 65.2 (301) |
| 60 | 37.5 | 70.3 (325) |
| 55 | 39.1 | 74.2 (343) |
| 50 | 42.2 | 79.0 (365) |
| 45 | 44.3 | 82.7 (382) |
| 40 | 46.7 | 85.7 (396) |
| 35 | 49.3 | 87.4 (404) |
| 30 | 52.6 | 90.7 (419) |
| 25 | 55.9 | 91.8 (424) |
| 20 | 61.9 | 93.7 (433) |
| 15 | 67.6 | 95.2 (440) |
| 10 | 78.1 | 97.6 (451) |
| 5 | 104.2 | 100 (462) |
The AUC for PCa detection was significantly greater for phi (AUC=0.703) than for %fPSA (0.648, P=0.004), fPSA (0.615), p2PSA (0.557), or PSA (0.525), rejecting the secondary null hypothesis.
Individual Patient Risk Assessment
Higher phi values were associated with an increased risk of PCa detection based upon the adjusted 25% proportion of PCa cases (Table 3). Of the study population, 25%, 33%, 30%, and 13% had phi values of 0–24.9, 25.0–34.9, 35.0–54.9, and ≥ 55.0, respectively. Compared to phi < 25.0, the relative risk of PCa detection on biopsy was 1.6-, 3.0-, and 4.7-fold higher at phi values of 25.0–34.9, 35.0–54.9, and ≥ 55.0, respectively. Overall, a phi≥ 55.0 was associated with a 52.1% probability of PCa.
TABLE 3
Risk Assessment Probability of PCa using phi
| phi Range | Probability of Cancer (95% Confidence Interval) | Relative Risk (95% Confidence Interval) | Percent of patients in phi range |
|---|---|---|---|
| 0–24.9 | 11.0% (6.5% – 15.8%) | 1.0 | 24.9% |
| 25.0–34.9 | 18.1% (13.7% – 22.6%) | 1.6 (1.0 – 3.1) | 32.8% |
| 35.0–54.9 | 32.7% (27.3% – 38.0%) | 3.0 (1.9 – 5.3) | 29.5% |
| 55.0+ | 52.1% (42.0% – 62.1%) | 4.7 (3.0 – 8.3) | 12.8% |
Age and Probability of PCa
Higher phi values were also associated with higher bootstrapped risk estimates of PCa within each age group. The probability (and relative risk [RR]) of PCa ranged from 10.9% (phi 0–24.9) to 53.4% (phi ≥ 55) (RR 4.9) for the 50–59 age group, 12.5% (phi 0–24.9) to 54.5% (phi ≥ 55) (RR 4.4) for the 60–69 age group, and 5.8% (phi 0–24.9) to 44.8% (phi ≥ 55) (RR 7.7) for the > 70 age group.
Association of phi with Gleason Score
Phi also had a significant relationship with biopsy Gleason score (r=0.138, P=0.004). Among participants with PCa, biopsy Gleason score was <7 in 290 (67.6%) and ≥7 in 139 (32.4%) Compared to phi < 25.0, the relative risk of Gleason ≥ 7 PCa increased to 1.08 for phi values from 25.0–34.9, 1.15 for phi values from 35.0–54.9, and 1.61 for phi ≥ 55.0. The corresponding proportion of cancers with a Gleason score ≥ 7 increased from 26.2% to 28.2%, 30.1%, and 42.1% at phi values of 0–24.9, 25.0–34.9, 35.0–54.9, and ≥ 55.0, respectively (Cochran-Armitage test for trend, P=0.013) (Table 4). The AUC for phi (0.724) exceeded that of %fPSA (0.670) in discriminating between Gleason ≥ 4+3 vs. lower Gleason grade PCa or negative biopsies.
TABLE 4
Relationship of phi with Biopsy Gleason Score
| Gleason Score on Biopsy | |||
|---|---|---|---|
| phi Range | Less than 7 n (%) | ≥7 n (%) | Risk Ratio (95% CI) |
| 0–24.9 | 34 (73.9) | 12 (26.1) | 1.0 |
| 25.0–34.9 | 74 (71.8) | 29 (28.2) | 1.08 (0.61, 1.92) |
| 35.0–54.9 | 116 (69.9) | 50 (30.1) | 1.15 (0.67, 1.98) |
| 55.0+ | 66 (57.9) | 48 (42.1) | 1.61 (0.95, 2.75) |
Note: One participant excluded with missing Gleason score.
Cochran-Armitage test for trend, p=0.01
Relationship of TRUS volume and phi
The AUCs for phi exceeded those of %fPSA in all three prostate volume tertiles (≤38, 39–53, and ≥54cc): 1st tertile: AUC 0.693 for phi vs. 0.614 for %fPSA; 2nd tertile: 0.707 vs. 0.593; 3rd tertile: 0.642 vs. 0.559.
Evaluation of Excluded Participants
AUCs for phi with and without the excluded site were 0.696 and 0.703, respectively. Similarly, AUCs for %fPSA were 0.634 and 0.648, respectively.
COMMENT
Prostate biopsy is routinely recommended for suspicious DRE results regardless of PSA.3 Biopsy is also recommended using PSA thresholds ranging from 2.5 to 4.0 ng/mL.1, 2, 15 However, this has led to unnecessary biopsies and possible over-detection of some cancers.15–17 To elucidate whether phi PSA-isoform measurement can improve PCa early detection, we examined a large, prospective cohort to predict biopsy findings in patients with moderate PSA elevations (2.0–10.0 ng/mL) and benign DRE findings. Such men are at higher risk of PCa (25% cancer detection rate compared with 4% in the general male population aged ≥50 years).3 Our bootstrapped population was designed to mirror this 25% incidence of PCa on biopsy.
Prostate biopsy may be associated with discomfort, anxiety, and financial costs. Minor complications occur frequently, and major complications are possible, underscoring the need for more specific markers to reduce unnecessary biopsies. We sought to determine the utility of p2PSA and phi for this clinical goal.
Precursor forms of PSA have been shown to improve the accuracy of PSA for detecting PCa.5, 6, 9–12, 28, 29 Specifically, preliminary reports suggest that p2PSA may be useful at PSA concentrations from 2.0–10.0 ng/mL.6, 9–12, 28, 29 Some, but not all, studies have suggested an association between proPSA and PCa aggressiveness.10, 12, 20 Thus, p2PSA and phi are being investigated in active surveillance programs to help overtreatment of insignificant PCa.19, 30
Catalona et al. previously reported in the PSA range of 2.0–10.0 ng/mL, the proPSA-to-fPSA ratio (%proPSA) yielded a higher specificity than %fPSA.9 Results from a separate multi-site study also supported the role of p2PSA, in combination with PSA and fPSA, in reducing unnecessary biopsies.12, 13
In the current study, the specificity for phi was higher than %fPSA at all pre-specified sensitivities, and PCa risk increased directly with increasing phi values. This suggests a role for phi as a patient monitoring tool, since increasing phi values reflect PCa risk.19 For example, at 95% sensitivity, the specificity of phi was 16.0% compared to 8.4% for %fPSA. Moreover, at lower sensitivities (90%, 85%, and 80%) for PCa detection that might be preferred to reduce the detection of possibly “insignificant” tumors, phi had a significantly greater specificity than %fPSA. These results were consistent across age groups, PSA concentrations, and ethnic groups, suggesting that they are representative of the intended-use population.
For individual risk assessment, the probability of PCa varied considerably based upon phi values. For example, a man with a phi ≥ 55 (13% of the study population) had a > 52% probability of PCa and 4.7-fold increased relative risk of positive biopsy. In contrast, at approximately 90% sensitivity, a patient with a phi < 25 had an 11% probability of PCa.
For the PCa group, higher phi values were also significantly associated with a higher percentage of biopsy Gleason grade ≥ 7, ranging from 26% to 42% for phi concentrations < 25 and ≥ 55, respectively. For the entire study population, the AUC for phi (0.724) exceeded that of %fPSA (0.670) in discriminating Gleason ≥ 4+3 PCa vs. lower Gleason grade PCa or negative biopsies. Using a phi cutoff of 21.3 (95% sensitivity), 25% of missed cancers were Gleason score ≥7; therefore, careful surveillance is necessary. The AUCs for phi also exceeded those of %fPSA in all three prostate volume tertiles, suggesting that phi provides better discrimination of PCa from benign disease than %fPSA across the spectrum of prostate volumes. Because phi did not differ by age and race these results suggest that phi may be applicable to a broad spectrum of men as an adjunct to predict clinically-significant PCa.
The large number of subjects in the present validation study provides confidence in the phi cutoffs determined. Phi is highly effective when used in patients with moderately elevated PSA concentrations who may be most likely to benefit from early diagnosis and curative PCa treatment. A physician might recommend biopsy for a patient with a phi ≥ 55.0 (risk = 52.1%) and surveillance for some men with a phi <25.0 (risk = 11.0%). For patients reluctant to undergo prostatic biopsy, a high phi might increase compliance with the appropriate follow-up.
We conclude that the phi measurement ([−2]proPSA / fPSA) × PSA1/2) may be useful to reduce unnecessary biopsies with improved specificity at various sensitivities for PCa detection in men age ≥50 years with PSA concentrations from 2.0–10.0 ng/mL, and negative DRE findings.****
Acknowledgments
Funding/Support:
This work was funded by Beckman Coulter Incorporated, Carlsbad, California; and supported in part by the National Institutes of Health/National Cancer Institute (NIH/NCI) Johns Hopkins Prostate SPORE Grant #P50CA58236, the Early Detection Research Network NIH/NCI Grant #U01-CA86323, and NIH/NCI U01 CA86323 to Dr Partin; NIH/NCI U24 CA115102 to Dr Chan; NIH/NCI U01CA113913 to Dr Sanda; the Urological Research Foundation, Northwestern-University of Chicago Prostate SPORE grant (NIH/NCI P50 CA90386-05S2), the Robert H. Lurie Comprehensive Cancer Center grant (NIH/NCI P30 CA60553), and Beckman Coulter Incorporated to Dr Catalona; the Mayo Clinic Prostate SPORE grant NIH/NCI CA091956 to Dr Klee.
Role of the Sponsor:
Funding for the study was provided by Beckman Coulter, Inc., which contributed to the design, collection and analysis of the study data. Beckman Coulter authors and the clinical investigators jointly developed the manuscript content.
Footnotes
Presented at the Annual Meeting of the American Urological Association, San Francisco, California, June 2, 2010
*Not intended as off-label promotion of any Beckman Coulter, Inc. product.
**Pending FDA approval.
***All trademarks are the property of their respective owners.
****Our results apply to the Access Hybritech p2PSA, PSA and fPSA assays on the Beckman Coulter Access Immunoassay Systems, as studies have shown that results differ when assays from different manufacturers or standardization are used.31
Author Contributions:
Mizrahi, Broyles, Shin and Cruz had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.Study concept and design: Catalona, Mizrahi, Broyles, Shin.
Acquisition of the data: Catalona, Partin, Sanda, Wei, Klee, Bangma, Slawin, Marks, Broyles, Chan, Sokoll, Roberts, van Schaik, Mizrahi.
Analysis and interpretation of the data: Catalona, Partin, Sanda, Klee, Slawin, Marks, Chan, Sokoll, Roberts, van Schaik, Wei, Bangma, Broyles, Shin, Cruz, Loeb, Mizrahi. An independent statistical analysis was performed by Edward F. Vonesh, PhD of the Department of Preventive Medicine, Northwestern University.
Drafting of the manuscript: Catalona, Mizrahi, Broyles, Loeb.
Critical revision of the manuscript for important intellectual content: Catalona, Mizrahi, Broyles, Partin, Sanda, Wei, Loeb, Bangma, van Schaik, Vonesh.
Statistical analysis: Shin. Obtained funding: Catalona, Partin, Sanda, Wei, Klee, Bangma, Slawin, Marks, Chan, Sokoll, Roberts, van Schaik.
Administrative, technical, or material support: Broyles, Mizrahi.
Study supervision: Mizrahi and Broyles.
Financial Disclosures:
Neither Dr Klee nor The Mayo Clinic have received royalties of greater than the federal threshold for significant financial interest from Beckman Coulter for the licensing of a technology unrelated to this research. Dr Wei receives research grant support from Sanofi Aventis and Beckman Coulter Incorporated and is on the advisory board of Envisioneering, Inc; Dr Catalona receives research support from Beckman Coulter Incorporated, deCODE Genetics, Inc, and OHMX.
Additional Contributions:
We thank Alain Artus PhD, Jessica Banks, Willeke Bolle, Jerardina Bueti, Janna Chamberlin, Phillip Cooper, Claude Darte PhD, Renu Dua, Willard Dunn, Debra Elliott, Bianca Gago, MD, Marcia Goodmanson, Robin Gurganus RN, Donghui Kan MS, Joep Kurstjens, Maureen Lemens RN, Lisa Ledebuhr, Lori Lofaro, Kathleen Loveland, Jiuliu Lu, Malu Macairan MD, Leslie Mangold MS, Patricia Nunnelly, Daniel O’Brien, Kellie Paich, Mindy Rawlins, Simpa Salami MD MPH, Javed Siddiqui MS, Edward F. Vonesh PhD, Mark Wildhagen PhD, and Sara Wyness for their assistance.REFERENCES
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.juro.2010.12.032
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3140702?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1016/j.juro.2010.12.032
Article citations
Integrative proteogenomic profiling of high-risk prostate cancer samples from Chinese patients indicates metabolic vulnerabilities and diagnostic biomarkers.
Nat Cancer, 5(9):1427-1447, 06 Sep 2024
Cited by: 0 articles | PMID: 39242942
Risk factors for Gleason score upgrade from prostate biopsy to radical prostatectomy.
Explor Target Antitumor Ther, 5(4):981-996, 30 Jul 2024
Cited by: 0 articles | PMID: 39280242 | PMCID: PMC11390291
Review Free full text in Europe PMC
phi and phiD predict adverse pathological features after radical prostatectomy for prostate cancer in Chinese population.
Cancer Med, 13(15):e70085, 01 Aug 2024
Cited by: 1 article | PMID: 39119746 | PMCID: PMC11310664
Prostate cancer biomarkers: from early diagnosis to precision treatment.
Clin Transl Oncol, 26(10):2444-2456, 14 May 2024
Cited by: 1 article | PMID: 38744755
Review
Blood-Based DNA Methylation Analysis by Multiplexed OBBPA-ddPCR to Verify Indications for Prostate Biopsies in Suspected Prostate Cancer Patients.
Cancers (Basel), 16(7):1324, 28 Mar 2024
Cited by: 0 articles | PMID: 38611002 | PMCID: PMC11010987
Go to all (254) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Diagnostic Impacts of Clinical Laboratory Based p2PSA Indexes on any Grade, Gleason Grade Group 2 or Greater, or 3 or Greater Prostate Cancer and Prostate Specific Antigen below 10 ng/ml.
J Urol, 203(1):83-91, 20 Aug 2019
Cited by: 2 articles | PMID: 31430244
Operating characteristics of prostate-specific antigen in men with an initial PSA level of 3.0 ng/ml or lower.
JAMA, 294(1):66-70, 01 Jul 2005
Cited by: 386 articles | PMID: 15998892
Percent free prostate specific antigen in the total prostate specific antigen 2 to 4 ng./ml. range does not substantially increase the number of biopsies needed to detect clinically significant prostate cancer compared to the 4 to 10 ng./ml. range.
J Urol, 168(2):504-508, 01 Aug 2002
Cited by: 23 articles | PMID: 12131298
Significance of serum free prostate specific antigen in the screening of prostate cancer.
J Urol, 156(6):1964-1968, 01 Dec 1996
Cited by: 23 articles | PMID: 8911366
Review
Funding
Funders who supported this work.
NCI NIH HHS (22)
Grant ID: P30 CA60553
Grant ID: P50 CA058236
Grant ID: P50 CA091956-01
Grant ID: U01-CA86323
Grant ID: CA091956
Grant ID: P30 CA060553
Grant ID: P50 CA058236-09A1
Grant ID: U01 CA086323
Grant ID: P30 CA006973
Grant ID: P50 CA090386
Grant ID: U01 CA086323-06
Grant ID: U01 CA113913
Grant ID: U24 CA115102-01
Grant ID: P30 CA060553-07
Grant ID: P50 CA091956
Grant ID: U24 CA115102
Grant ID: P50 CA090386-01
Grant ID: U01 CA086323-01
Grant ID: P50 CA90386-05S2
Grant ID: P50CA58236
Grant ID: U01 CA113913-01
Grant ID: U01CA113913
NCRR NIH HHS (1)
Grant ID: UL1 RR025741





