Abstract
Free full text

A transgene-encoded cell surface polypeptide for selection, in vivo tracking, and ablation of engineered cells
Abstract
An unmet need in cell engineering is the availability of a single transgene encoded, functionally inert, human polypeptide that can serve multiple purposes, including ex vivo cell selection, in vivo cell tracking, and as a target for in vivo cell ablation. Here we describe a truncated human EGFR polypeptide (huEGFRt) that is devoid of extracellular N-terminal ligand binding domains and intracellular receptor tyrosine kinase activity but retains the native amino acid sequence, type I transmembrane cell surface localization, and a conformationally intact binding epitope for pharmaceutical-grade anti-EGFR monoclonal antibody, cetuximab (Erbitux). After lentiviral transduction of human T cells with vectors that coordinately express tumor-specific chimeric antigen receptors and huEGFRt, we show that huEGFRt serves as a highly efficient selection epitope for chimeric antigen receptor+ T cells using biotinylated cetuximab in conjunction with current good manufacturing practices (cGMP)-grade anti-biotin immunomagnetic microbeads. Moreover, huEGFRt provides a cell surface marker for in vivo tracking of adoptively transferred T cells using both flow cytometry and immunohistochemistry, and a target for cetuximab-mediated antibody-dependent cellular cytotoxicity and in vivo elimination. The versatility of huEGFRt and the availability of pharmaceutical-grade reagents for its clinical application denote huEGFRt as a significant new tool for cellular engineering.
Introduction
Cell-based therapies, including genetically manipulated cell products, are an emerging area in applied biotechnology. Foremost in the cell therapy field, and now a component of standard medical practice, is hematopoietic stem cell transplantation. Likewise, adoptive transfer of T cells for infectious and neoplastic disease is the subject of intense clinical research. In both of these cell therapy applications, a variety of genetic engineering approaches are being studied to endow hematopoietic or lymphoid cells with novel attributes, to increase either their therapeutic potency and/or safety.1 Genetic engineering of cellular therapeutics frequently is accompanied by the need to purify cells that express therapeutic transgene(s) and to cull out nonexpressing cells that either lack transgene-endowed therapeutic activity or safety features. Once cell products are administered to patients, having the ability to track the frequency and distribution of these cells, and, if need be, facilitate their elimination, are all desirable features of their genetic engineering. Constraints in vector capacity, unintended transgene function, and immunogenicity dictate that a single functionally inert polypeptide of a native human sequence that fulfills each of the selection/tracking/suicide purposes would be ideal, particularly if this could be achieved with a commercially available, Food and Drug Administration (FDA)-approved pharmaceutical.
Although a variety of xenogenic enzymes have been used (eg, bacterial neomycin and hygromycin phosphotransferases) for in vitro selection of genetically modified cells, these systems suffer from the often prolonged culture necessary to achieve selection, and from the high likelihood that the expressed xenogenic protein will be immunogenic.2,3 Alternately, human cell surface transmembrane or glycosylphosphatidylinositol-linked proteins are a logical choice for antibody-based physical separation platforms, such as fluorescence-activated cell sorting and immunomagnetic selection. To date, a variety of human cell surface polypeptides have been described for cell marking, including the low affinity nerve growth factor receptor (ΔLNGFR),4 CD34,5 CD19,6 CD20 and CD4,7 and the glycosylphosphatidylinositol-anchored CD90.8 However, none of these candidates simultaneously satisfies the criteria of being unique to the genetically modified hematopoietic/lymphoid cell, lacking functionality, and having a commercially available pharmaceutical-grade monoclonal antibody specific for an epitope present in the extracellular portion of the polypeptide.
Human epidermal growth factor receptor (EGFR; ErbB-1, HER1 in humans) is a receptor tyrosine kinase of the ErbB family of growth factor receptors that is not expressed by cells of the hematopoietic and lymphopoietic systems. Ligand (EGF, TGF-α) binding occurs within N-terminal extracellular domains I and II of EGFR resulting from transition of receptor tyrosine kinase inactive monomers to active homodimers.9 Extracellular domain III contains the binding site of cetuximab (Erbitux), an IgG1 chimeric antibody licensed by the FDA for the treatment of metastatic colorectal cancer and head and neck cancer.10 We hypothesized that human EGFR could be rendered incapable of binding ligands by removal of domains I and II, and devoid of signaling activity by deletion of its cytoplasmic tail, while retaining an intact cetuximab binding site in extracellular domain III. Here we demonstrate the utility of such a truncated EGFR (huEGFRt) expressed by transduced T cells for immunomagnetic purification using biotinylated cetuximab, cell tracking by flow cytometry and immunohistochemistry, and in vivo cell ablation after systemic cetuximab administration.
Methods
Antibodies and flow cytometry
Fluorochrome-conjugated isotype controls, anti-CD3, anti-CD4, anti-CD8, anti-CD28, anti-CD45, anti–granzyme B, and anti–T-cell receptor-αβ (TCRαβ), and streptavidin were obtained from BD Biosciences. Biotinylated anti-Fc was purchased from Jackson ImmunoResearch Laboratories. Phycoerythrin (PE)-conjugated anti-biotin was purchased from Miltenyi Biotec. Biotinylated EGF was purchased from Invitrogen. PE-conjugated anti-EGFR was purchased from Abcam. All were used according to the manufacturer's instructions. Data acquisition was performed on a FACSCalibur (BD Biosciences), and the percentage of cells in a region of analysis was calculated using FCS Express, Version 3 (De Novo Software).
For generation of the biotinylated cetuximab, all materials were purchased from the COH pharmacy. Briefly, 200 mg cetuximab (Erbitux) was buffer exchanged to PBS (D-PBS, pH 7.5 ± 0.1) using a MidGee Hoop Cartridge (UFP-30-E-H42LA). The material (2 mg/mL) was modified with Sulfo-NHS-LC-biotin (20:1) in a 1-hour room temperature reaction and then diafiltered to remove the excess biotin. The biotinylated cetuximab was then buffer exchanged to PBS, glycerol added to a final concentration of 20%, and the material was frozen. Product purity was confirmed on NuPAGE Novex Bis-Tris gels with or without SDS reduction.
Cell lines
Unless otherwise indicated, all cell lines were maintained in RPMI 1640 (Irvine Scientific) supplemented with 2mM l-glutamine (Irvine Scientific), 25mM HEPES (Irvine Scientific), and 10% heat-inactivated FCS (Hyclone), hereafter referred to as culture media (CM).
To generate T cells, human peripheral blood mononuclear cells (PBMCs) were isolated over Ficoll-Paque (Pharmacia Biotech) from heparinized peripheral blood obtained from consenting healthy donors participating on a COH NMC Internal Review Board-approved protocol in accordance with the Declaration of Helsinki. For generation of OKT3 blasts, washed PBMCs were stimulated with 25 U/mL interleukin-2 (IL-2) and a 1:1 (cell/bead) ratio of Dynabeads Human T expander CD3/CD28 (Invitrogen). For generation of central memory T (TCM) cell-derived effector cells, washed PBMCs were first AutoMACS depleted using anti-CD45RA beads (Miltenyi Biotec) with or without CD14 beads (Miltenyi Biotec) per the manufacturer's protocol, and then positively selected on the AutoMACS using biotinylated DREG56 (anti-CD62L) and antibiotin beads (Miltenyi Biotec) to produce purified CD62L+CD45RO+ TCM cells. The CD19CAR-T2A-EGFRt_epHIV7 lentiviral construct contains: (1) the chimeric antigen receptor (CAR) sequence consisting of the VH and VL gene segments of the CD19-specific FMC63 monoclonal antibody (mAb), an IgG4 hinge-CH2-CH3, the transmembrane, and cytoplasmic signaling domains of the costimulatory molecule CD28, and the cytoplasmic domain of the CD3ζ chain11; (2) the self-cleaving T2A sequence12; and (3) the truncated EGFR sequence as indicated (Figure 1). The huEGFRt was synthesized by polymerase chain reaction splice overlap extension to fuse in frame the human granulocyte-macrophage colony-stimulating factor (GM-CSF) receptor's leader peptide to domains III, IV, and the transmembrane spanning components of huEGFR (base pairs 1000-2004). This fusion product was then cloned into the epHIV7 vector (in which the cytomegalovirus promoter of pHIV713 had been replaced with an EF-1 promoter) along with the CD19CAR and T2A sequences, and the final construct was confirmed by sequence analysis. Lentiviral transduction was carried out on TCM stimulated with CD3/CD28 Dynabeads (1:3) and 25 U/mL IL-2 on retronectin (50 μg/mL)-coated plates at a multiplicity of infection of 3 with 5 μg/mL polybrene. After 4 hours, warm medium was added to double volume, and cultures were maintained in CM for 14 days until the T cells were rapidly expanded with anti-CD3 mAb (OKT3) as previously described.14 The huEGFRt-expressing cells were purified on the AutoMACS with biotinylated cetuximab and anti-biotin microbeads (Miltenyi Biotec) as per the manufacturer's instructions.
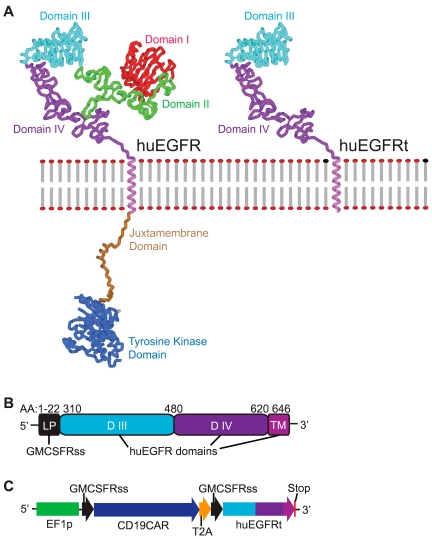
Generation of huEGFRt expressing construct. (A) Molecular model of human EGFR versus EGFRt proteins. Models were created using Accelrys Discovery Studio, Version 2.0 software and are based on the human EGFR crystal structure files 1YY9 (http://www.pdb.org/pdb/explore/explore.do?structureId = 1YY9), which provides the structure of the 4 extracellular domains (red, green, teal, and purple), and 3GOP (http://www.pdb.org/pdb/explore/explore.do?structureId = 3GOP), which provides the crystal structure of the intracellular juxtamembrane domain (brown) and the tyrosine kinase domain (blue). The transmembrane helix (magenta) is modeled on known helix structure. The structure of the last approximately 190 amino acids of the C-terminal intracellular domain is not known, so the placeholder for this is indicated (gray). (B) Schematic of the huEGFRt amino acid sequence. Amino acid ranges that were used from the GM-CSF receptor-α chain signal sequence (GMCSFRss, which directs surface expression) and the huEGFR sequence (domains III, IV and the transmembrane domain) are indicated. (C) Schematic of the CD19CAR-T2A-EGFRt construct contained in the lentiviral vector. Codon optimized sequence portions of the CD19-specific, CD28 costimulatory CAR (CD19CAR), followed by the self-cleavable T2A, and huEGFRt genes are indicated, along with the Elongation Factor 1 promoter sequences (EF-1p), the GM-CSF receptor-α chain signal sequences (GMCSFRss), and the 3 nucleotide stop codon.
H9 T lymphoblast, SupB15 leukemia, A431 epidermoid carcinoma, and CTLL-2 mouse cytotoxic T lymphoblast cell lines (ATCC) were grown in the corresponding ATCC recommended media. Epstein-Barr virus-transformed lymphoblastoid cell lines (LCLs) were made from PBMCs as previously described.15 LCL-OKT3 cells were generated by electroporating LCLs with an OKT3–2A-hygromycin_pEK plasmid at 5 μg/107 cells, followed by selection in CM with 0.4 mg/mL hygromycin. NS0-IL15 cells were generated as previously described.16 CTLL-2 cells, which had been engineered and selected to express both eGFP-T2A-IL2_ pcDNA3.1(+) and ffLuc-zeocin_pcDNA3.1(+), were subsequently transduced and selected for huEGFRt expression as described in the previous paragraph, resulting in IL-2–secreting ffLuc+huEGFRt+ CTLL-2 cells. All DNA construct and construction associated polymerase chain reaction primer sequences are available on request.
Protein analysis
Cell lysis was carried out in 1% Triton-X lysis buffer containing phosphatase inhibitor cocktail II (Sigma-Aldrich; 1:20 by volume). Western blots (50 μg protein per lane) were probed with the Phospho-EGF receptor antibody sampler kit (Cell Signaling Technology), and IRDye 800 conjugated anti–β-actin antibody (LI-COR) as per the manufacturer's instructions. Blots were imaged on the Odyssey Infrared Imaging System (LI-COR).
Cytotoxicity and cytokine assays
Four-hour chromium release assays were performed as previously described.17 Antibody-dependent cell-mediated cytotoxicity (ADCC) was determined using up to 2.5 × 105 freshly isolated PBMCs that had been prestimulated overnight with 10 ng/mL huGM-CSF (R&D Systems) as effector cells in cocultures with 5 × 103 51Cr-labeled huEGFRt+ T cells as targets, with or without 1 μg/mL of either Erbitux or rituximab (Rituxan Genentec/Biogen IDEC).
Cytokine production was measured by coculturing T cells (5 × 105) overnight in 96-well tissue culture plates with 5 × 105 of LCL-OKT3, LCL, or SupB15. Supernatants were analyzed by cytometric bead array using a Bio-Plex Human Cytokine Panel (Bio-Rad) according to the manufacturer's instructions.
In vivo T-cell engraftment
All mouse experiments were approved by the COHNMC Institute Animal Care and Use Committee. Six- to 10-week old NOD/Scid IL-2RγCnull mice were injected intravenously on day 0 with 2 × 107 of either EGFRt-negative control or EGFRt-selected T cells and subcutaneously with 5 × 106 live NS0-IL15 cells to provide a systemic supply of human IL-15 in vivo. Bone marrow was harvested from killed animals, and cell suspensions were analyzed by flow cytometry. Alternatively, femurs were fixed in 10% formalin for 24 hours, decalcified for 2 hours (Richard-Allan Scientific), and embedded in paraffin for immunohistochemical staining with anti-CD45 (DAKO) or anti-EGFR (clone 31G7; Invitrogen) according to the manufacturer's instructions, followed by counterstain with hematoxylin. Bright-field images were acquired using an Olympus AX70 automated microscope (40× with UPlanApo, NA of 1; and 100× with PlanAchormat, NA of 1.25), a Retiga EXi camera, and ImagePro Plus v6.3 (MediaCybernetic), high-end image acquisition software.
For in vivo ADCC, 3 × 106 ffLuc+huEGFRt+ CTLL-2 cells were administered intravenously to 8- to 10-week old NOD/scid mice, and engraftment was monitored by Xenogen imaging of luciferase activity.18 After 11 days, Erbitux versus Rituxcan was administered at 1 mg/mouse intraperitoneally daily for 6 days. The 1-mg/mouse dose correlates with 0.02 to 0.03 mg/cm2 in a 20- to 25-g mouse, which falls within the dose of < 0.04 mg/cm2 for human use as described in the Erbitux package insert. The use of body surface area in dose translation from animal to human studies is supported by Reagan-Shaw et al.19
Results
Design of a trifunctional polypeptide based on native sequence of human EGFR
We considered a combined N-terminal/C-terminal truncated version of human EGFR (huEGFRt) that lacks the domains essential for ligand binding and tyrosine kinase activity but retains the cetuximab-binding epitope as a candidate molecule for cell selection, tracking, and ablation. Accordingly, we synthesized a DNA consisting of only the extracellular domains III/IV and the transmembrane domain of the human EGFR. A comparison of molecular models of the full-length human EGFR and huEGFRt is shown in Figure 1A. To facilitate the surface expression of the huEGFRt, we used the signal sequence of the human GM-CSF receptor-α chain, the 22 amino acids of which are immediately upstream of EGFR domain III (Figure 1B). To evaluate the utility of huEGFRt as a selection marker for purifying transduced T cells functionalized for tumor targeting, we constructed a multidomain DNA construct composed of the coding sequence for a CD19-specific chimeric antigen receptor (CD19CAR)11 placed in frame with huEGFRt but separated by the coding sequence of the self-cleaving/ribosome skip T2A sequence (Figure 1C). This cDNA was then incorporated into the lentiviral vector packaging plasmid epHIV-7 under a modified human EF-1α promoter for subsequent production of VSV-g pseudotyped self-inactivating lentivirus.
Cetuximab (Erbitux) retains huEGFRt reactivity after biotinylation
Based on published work detailing the EGFR binding epitope location of cetuximab, we evaluated whether cetuximab reactivity with EGFR domain III was retained in the conformation adopted in the absence of N-terminal domains I and II (Figure 2A). To detect and/or select for huEGFRt-expressing cells that bound cetuximab, we generated biotinylated cetuximab from pharmaceutical-grade antibody via a standard amine-specific biotinylation reaction using sulfo-NHS-LC-biotin. The resulting purified biotinylated product retained high affinity binding to EGFR (Figure 2B-C). Titration experiments revealed that as little as 14.5 ng biotinylated cetuximab was sufficient for maximal staining of 106 huEGFRt+ cells. Next, human H9 T cells were transduced with our CD19CAR-T2A-huEGFRt lentiviral vector. Consistent with a predicted coordinate expression of the CAR and huEGFRt based on their T2A linkage, the frequency of cells that expressed surface huEGFRt determined by staining with biotinylated cetuximab/PE-conjugated anti-biotin mAb correlated with the frequency of cells that expressed the CAR detected with a PE-conjugated anti–human Fc-specific mAb (Figure 2D).
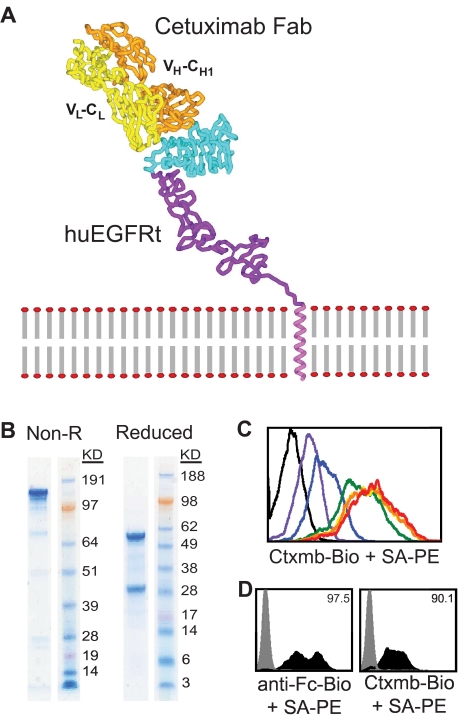
Generation and utility of a biotinylated cetuximab. (A) Model is based on the crystal structure file 1YY9 (http://www.pdb.org/pdb/explore/explore.do?structureId = 1YY9), which provides the structure of the 4 extracellular domains (red, green, teal, and purple), and the bound cetuximab Fab (yellow and orange). The transmembrane helix (magenta) is modeled on known helix structure. (B) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels of nonreduced (left) and reduced (right) final biotinylated cetuximab product. (C) Titration of biotinylated cetuximab. A total of 106 EGFR+ cells were stained with 0 μg (black), 1.45 μg (red), 0.145 μg (orange), 14.5 ng (yellow), 1.45 ng (green), 0.145 ng (blue), or 14.5 pg (purple) of biotinylated cetuximab followed by 0.5 μg PE-conjugated streptavidin and analyzed by flow cytometry. A total of 14.5 ng of biotinylated cetuximab was deemed sufficient for future staining of 106 cells. (D) Biotinylated cetuximab can detect huEGFRt on transduced H9 cells. H9 cells were transduced with the CD19CAR-T2A-EGFRt containing lentivirus at a multiplicity of infection of 10 and analyzed 2 days later by flow cytometry using biotinylated cetuximab or biotinylated anti-Fc Ab (to detect the CD19CAR) followed by SA-PE (black histograms). Gray histograms represent nontransduced parental H9. Percentage positive staining is indicated in each histogram.
Immunomagnetic enrichment of huEGFRt+ human T cells after lentiviral transduction
The biotinylated cetuximab could in principle be used for either immunomagnetic selection or FACS of huEGFRt+ cells. We first evaluated the use of biotinylated cetuximab in conjunction with commercially available antibiotin microbeads for the immunomagnetic selection of human T cells transduced with a self-inactivating lentivirus that directs the coexpression of CD19CAR and huEGFRt (Figure 3). A panel of primary human T-cell lines that contained from 2.6%-40% huEGFRt+ cells was generated by lentiviral transduction after stimulation of PBMCs or purified central memory (CD45RO+CD62L+ TCM) or effector memory (CD45RO+CD62L− TEM) T-cell subsets with anti-CD3/anti-CD28 beads (Figure 3B; supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Immunomagnetic selection of these T-cell lines using a 2-step labeling process (biotinylated cetuximab followed by anti-biotin microbeads) consistently resulted in a population of cells that were > 90% huEGFRt+ (Figure 3B; supplemental Figure 1). A proliferative advantage of huEGFRt− cells over huEGFRt+ cells was observed in cultures of unselected transduced T cells subjected to OKT3-mediated expansion (Figure 3C). After immunomagnetic selection, the level of huEGFRt expression and the frequency of expressing cells remained stable over 3 consecutive 14-day cycles of OKT3-based expansion14 (Figure 3D). Of interest, the fold expansion of EGFRt+ cells after immunomagnetic selection was significantly enhanced over that of huEGFRt+ cells in the unselected cultures (compare Figure 3D parentheses and Figure 3C). These data demonstrate that huEGFRt can serve as a cell surface marker unique to transduced human T cells and enable subsequent cetuximab-based immunomagnetic purification of stable huEGFRt-expressing cell populations.
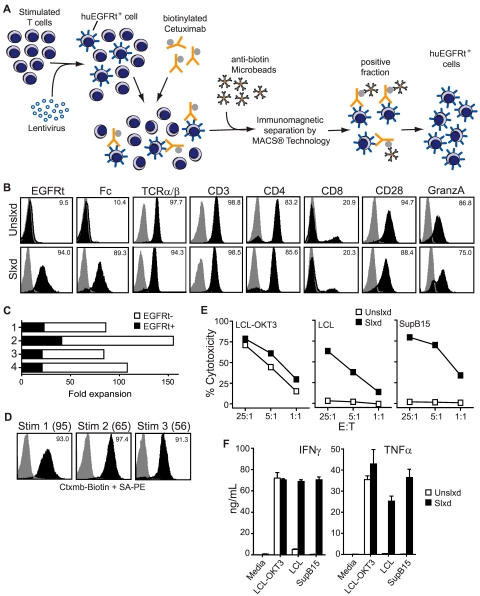
Selected EGFRt+ CD19CAR+ T cells can be expanded with maintenance of effector phenotype. (A) Schematic of the immunomagnetic huEGFRt selection procedure. (B) After 1 rapid expansion cycle, nonselected (Unslxd) and huEGFRt-selected (Slxd) T cells were phenotyped for surface EGFR (ie, EGFRt, with biotinylated cetuximab), Fc (ie, CAR), and effector T-cell markers TCRαβ, CD3, CD4, CD8, CD28, or intracellular granzyme A (black histogram) versus isotype control Ab (gray histogram) by flow cytometry. Percentage of positive staining is indicated in each histogram. (C) Fold expansion of nontransduced (EGFRt-) and transduced (EGFRt+) T cells in unselected cultures of 4 different transduced cell lines was determined after OKT3-mediated stimulation. (D) huEGFRt expression of selected T cells was determined by flow cytometry after each of 3 rounds of OKT3-mediated stimulation (Stim 1, 2, and 3). Percentage of cetuximab-mediated staining (black) compared with that of SA-PE alone (gray) is indicated in each histogram. (E) Nonselected (Unslxd) and huEGFRt-selected (Slxd) T cells were incubated for 4 hours with 51Cr-labled CD19+ LCL or SupB15 cells, or OKT3-epressing LCL cells as targets at the indicated effector to target ratios. Percentage cytotoxicity (mean ± SE) of triplicate wells is depicted. (F) Nonselected (Unslxd) and EGFRt-selected (Slxd) T cells were incubated with CD19+ LCL or SupB15 cells, or OKT3-epressing LCL cells and supernatants were analyzed by Bioplex. IFN-γ and TNF-α levels (mean ± SE) are shown.
We next asked whether huEGFRt expression perturbed the phenotypic and/or functional attributes of CAR redirected T cells. Expression of T-cell markers TCRαβ, CD3, CD4, CD8, CD28, and intracellular granzyme A were equivalent in CD19-specific CAR+ huEGFRt+ T-cell lines before and after cetuximab selection (Figure 3B). Moreover, because of the T2A-mediated linkage of CAR with huEGFRt expression, the frequency and level of expression of each transgene were increased proportionally after immunomagnetic selection (Figure 3B). Thus, both CD19-specific cytolytic potency (Figure 3E) and the quantities of secreted IFN-γ/TFN-α (Figure 3F) by selected CAR+ huEGFRt+ effector T cells after antigen engagement were superior compared with unselected CAR+ huEGFRt+ effector T cells. The CD19-specific cytotoxicity of selected CAR+ huEGFRt+ effector T cells was also found to be similar to that of CAR+ huEGFRt-negative effector T cells (supplemental Figure 2).
huEGFRt expressed on T cells is functionally inert
As shown in our molecular model (Figure 1A), huEGFRt is predicted to be incapable of binding ligands because of the elimination of extracellular domains I and II, and to be devoid of receptor tyrosine kinase signaling because of the removal of its cytoplasmic tail.20 Flow cytometric analysis confirmed that biotinylated huEGF does not bind to T cells that express huEGFRt but binds to control A431 epidermoid carcinoma cells that express full-length EGFR (Figure 4A). Western immunoblot analyses for EGFR phosphorylation were carried out on cetuximab-selected huEGFRt+ T cells after culture with EGF. As expected, and in contrast to the induction of phospho-EGFR seen when EGFR+ A431 tumor cells were incubated with EGF, phospho-EGFR was not detected in lysates of huEGFRt+ T cells under identical conditions (Figure 4B).
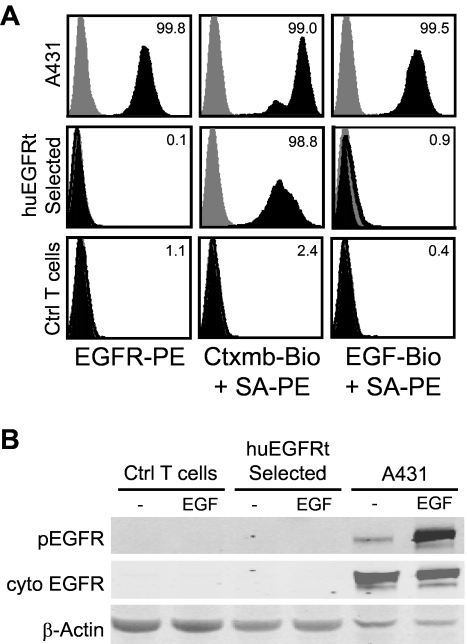
The huEGFRt expressed on selected T cells is inert. (A) EGF does not bind to the surface of huEGFRt-expressing T cells. A431, EGFRt-selected T cells, and negative control T cells were analyzed by flow cytometry after incubation with PE-conjugated anti-EGFR, or either biotinylated cetuximab or biotinylated EGF followed by PE-conjugated streptavidin (black histogram) versus PE-conjugated isotype control antibody or streptavidin alone (gray histogram). Percentage positive staining is indicated in each histogram. (B) huEGFRt expressed on T cells is not phosphorylated on coincubation with EGF. Negative control T cells, huEGFRt-selected T cells, or A431 cells were incubated for 5 minutes with or without 100 ng/mL EGF and then lysed in the presence of phosphatase inhibitor. Lysates run on Western blots were then probed using antibodies specific for β-actin, the cytoplasmic domain of EGFR, or the phosphorylated tyrosine at position 1068 of EGFR.
Tracking of adoptively transferred huEGFRt+ T cells using flow cytometry and immunohistochemistry
To test the utility of huEGFRt for tracking the engraftment of adoptively transferred T cells, we harvested blood and bone marrow specimens from NOD/Scid IL-2RγC null mice engrafted with CD19CAR+EGFRt+ human T cells. First, we subjected unfixed peripheral blood and bone marrow mononuclear cell samples to flow cytometric analysis after staining with biotinylated cetuximab and PE-conjugated streptavidin (Figure 5A; and data not shown). Although the level of human CD45+ T-cell engraftment (20%-25%) was similar in animals administered either EGFRt-negative or -positive T cells, double staining for human CD45 and EGFR allowed for the resolution of huEGFRt+ (ie, transgene-expressing) human T cells from their huEGFRt-negative counterparts. Second, we sought to determine whether standard paraffin-embedded fixed tissue specimens were amenable to detection of huEGFRt+ T-cell infiltrates using FDA-approved EGFR-specific diagnostic kits. We performed immunohistochemical analysis of paraffin-embedded femurs from engrafted mice and could detect huEGFRt+ cells in the bone marrow (Figure 5B). These data support the utility of huEGFRt to serve as a tracking marker for quantifying the frequency and tissue distribution of adoptively transferred T cells. Of note, despite the significant levels of human T-cell engraftment observed, neither group of mice exhibited overt signs of graft versus host disease (ie, weight loss, diarrhea, and ruffled fur were not observed).
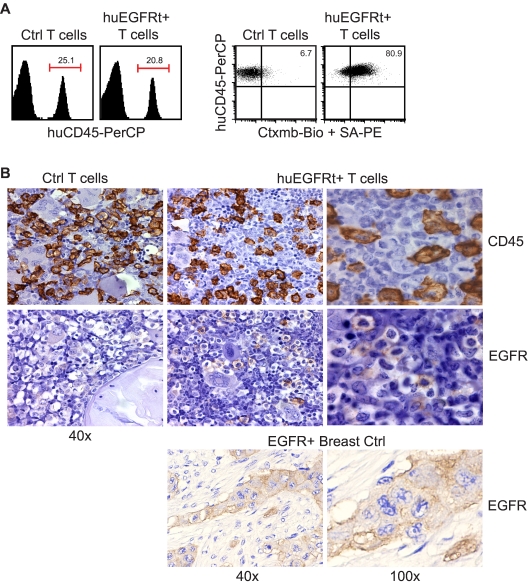
Use of EGFRt as a marker for in vivo detection of engineered cells. (A) Day 20 bone marrow cells harvested from mice that were engrafted with either unmodified huEGFRt-negative control T cells (Ctrl T cells) or huEGFRt-selected T cells (EGFRt+ T cells) were stained using peridinin chlorophyll protein-conjugated anti–human CD45 and biotinylated cetuximab followed by PE-conjugated streptavidin. Total percentage of CD45+ cells in bone marrow are indicated in the left-hand panels. Percentage of EGFRt+ cells within the CD45-gated population are indicated in the right-hand panels, using quadrants that were created based on isotype control staining. (B) Day 20 femurs were also analyzed by immunohistochemistry for CD45 versus EGFR expression. EGFR expression on paraffin-embedded breast tumor tissue was used as a positive control for EGFR staining. Representative ×40 and ×100 images are shown as indicated.
Cetuximab binding to huEGFRt sensitizes human T cells to ADCC
A valuable feature of a cell surface selection/tracking marker would be its capacity to serve as a target for in vivo cell ablation, should a clinical situation warrant this maneuver. We therefore evaluated the extent to which Erbitux bound to huEGFRt on T cells activates ADCC in vitro, and whether the engraftment of adoptively transferred huEGFRt+ T cells in NOD/scid mice could be attenuated by Erbitux administration (Figure 6). The addition of Erbitux to cultures containing 51Cr-labeled huEGFRt+ T cells and human GM-CSF activated fresh PBMCs as a source of effectors specifically sensitized T cells to cytolysis as measured by 4-hour chromium release assay. The addition of the CD20-specific mAb Rituxan had no effect on triggering ADCC of huEGFRt+ T cells in this assay. We next derived huEGFRt+ CTLL-2 murine T cells that were additionally modified to secrete autocrine IL-2 and express the firefly luciferase biophotonic reporter and adoptively transferred these cells via intravenous injection to NOD/scid mice that subsequently received Erbitux or Rituxan. NOD/scid mice are an established animal model for ADCC21–23 because of their residual innate immunity, including NK-cell activity.24 The in vivo engraftment of transferred CTLL-2, as measured by in vivo biophotonic imaging, was significantly inhibited (97%, P < .05) in mice that received Erbitux (1 mg intraperitoneally daily; Figure 6B-C). The Erbitux-mediated elimination of the ffLuc+huEGFRt+ CTLL-2 cells took between 4 and 6 days. These data support the use of Erbitux administration as a suicide strategy for patients receiving huEGFRt+ T cells should toxicity warrant this intervention.
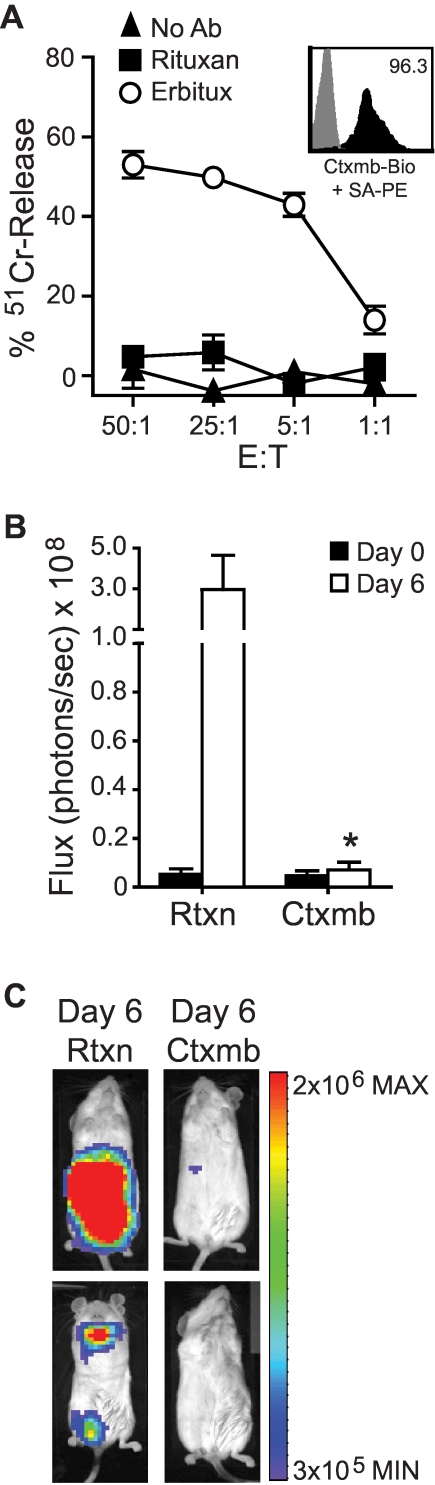
Use of EGFRt as a target for Erbitux-mediated ADCC of engineered cells. (A) 51Cr-labeled huEGFRt-selected T cells (inset depicts huEGFRt expression) were mixed with 1 μg/mL of Erbitux or Rituxan as a negative control before addition of GM-CSF–stimulated human PBMCs as effectors. The percentage 51Cr-release was then determined after a 4-hour incubation. Data are representative of 2 separate experiments. (B) NOD/scid mice were inoculated intravenously with ffLuc+huEGFRt+ CTLL-2 cells. On successful engraftment (day 0), the mice were divided into 2 groups (n = 5) and then treated with either Erbitux (Ctxmb) or Rituxan (Rtxn) intraperitoneally daily for 6 days. Data are ± SE; total flux levels of luciferase activity were measured by Xenogen imaging. *P = .0159, the flux of Erbitux versus Rituxan-treated mice at day 6 using the Mann-Whitney test. (C) Representative bioluminescence images of NOD/scid mice from (B) at day 6.
Discussion
Here we describe a truncated cell surface huEGFRt polypeptide having 3 key uses for genetic engineering of cell-based therapies: ex vivo cell purification, in vivo cell tracking, and cell ablation. Human EGFR was selected because of lack of expression by hematopoietically derived cells, in particular T lymphocytes, and the potential to truncate it to derive a molecule that is devoid of the ligand binding and receptor tyrosine kinase signaling associated with the intact EGFR while retaining the epitope recognized by the commercially available FDA-approved mAb, cetuximab (Erbitux).
huEGFRt is composed of only 336 of the 1185 amino acids in the full-length EGFR and lacks both the extracellular ligand binding domains I and II and the entire cytoplasmic tail necessary for signaling. We fused the 22-amino acid amino terminal leader peptide of the human GM-CSF receptor-α chain based on its ability to sort type I transmembrane proteins to the plasma membrane in T cells.25 Thus, the huEGFRt is encoded by a relatively small cDNA that is amenable to incorporation into most vector types, leaving ample capacity to include additional transgene coding sequences. We demonstrate that a self-inactivating lentivirus vector containing a cDNA coding for a tumor-reactive CAR situated 5′ to huEGFRt with an intervening 2A cleavable/ribosomal skip linker directs the coordinate expression of both cell surface proteins in transduced cells. More complex modular designs are also possible, as exemplified by expressing a CAR-T2A-huEGFRt-T2A-mycophenolic acid-resistant human inosine monophosphate dehydrogenase (IMPDH2dm; supplemental Figure 1). Using 2A cleavable linkers is advantageous when balanced transgene expression levels are desired, as would be the case when expressing transgenic TCRαβ pairs, for example.26
The extracellular and cytoplasmic truncations of EGFR serve to reduce the size of the huEGFRt open reading frame and also render huEGFRt unable to bind EGF and mediate EGFR signaling. These features would prevent transferred T cells from acting as a sink for EGFR ligands at local sites, the consequences of which are difficult to predict. The residual stalk of EGFR's extracellular domains III and IV are not anticipated to have adhesive interactions with other cell membrane proteins or act as a dominant negative inhibitor of other cytokine receptors. Consistent with this assertion, the expression of huEGFRt by human CD8+ T cells does not result in any overt phenotypic, proliferative, or functional impairment. Further studies in which huEGFRt is expressed by hematopoietic stem cells, NK cells, and other therapeutic cell populations will be needed to determine the generalization of our observations in T cells. Deletion of the EGFR cytoplasmic tail eliminates the potential that the receptor can signal in transduced cells. Because the truncations of EGFR that result in huEGFRt involve the N- and C-termini, novel junctional amino acid sequences are not created that could provide immunogenic epitopes for antitransgene antibody or T-cell responses in recipients of cell products. It remains possible that the conformation of the extracellular huEGFRt could elicit an antibody response or that the fusion site of the GM-CSF leader and EGFR could be immunogenic, and this will need to be addressed by in vivo studies in humans. It is noteworthy, however, that the junctional amino acid sequence between the GM-CSF leader and EGFR does not correspond to sequences that are predicted to bind to common class I human leukocyte antigen alleles (ie, using the MHC-I antigenic peptide processing prediction algorithm available at http://www.mpiib-berlin. mpg.de/MAPPP27 to run the junction spanning sequence MLLLVTSLLLCELPHPAFLLIPRKVCNGIGIGE).
A key feature of the huEGFRt transgene is that it traffics to the cell surface as a type I transmembrane protein retaining extracellular domains III and IV. The N-terminal domain III of huEGFRt is conformationally intact in as much as it retains the epitope for cetuximab binding. Pharmaceutical-grade cetuximab, a chimeric mouse Fab-human IgG1 monoclonal antibody, is amenable to biotinylation through chemical conjugation and reformulation. In addition, we found that Erbitux, as formulated for commercial use, has no carriers or additives that interfere with biotinylation and that, after this modification, the antibody retains EGFR-specific high affinity binding. The availability of cGMP grade anti-biotin microbeads (Miltenyi Biotec) enables selection of huEGFRt+ cells on the CliniMACS device. We show that, even when starting populations of huEGFRt+ T cells are < 5%, immunomagnetic selection can enrich huEGFRt+ cells to > 90% purity. The selection system might be further refined by the availability of a peptide capable of competing off Erbitux bound to huEGFRt.28 This would be advantageous when selection is timed close to patient infusion where residual cell bound Erbitux could elicit human anti-chimeric antibody responses that would limit the survival of subsequent cell doses.
Hematopoietically derived cells do not express EGFR; as a consequence, cells marked by the expression of huEGFRt can be tracked in vivo. Flow cytometry permits multiparameter analysis when huEGFRt detection is combined with other markers (eg, CD34, CD4, and CD8), and can readily quantify genetically modified cells present in a mixed population in blood, bone marrow, cerebrospinal fluid, or other bodily fluid. Tracking the tissue distribution of transferred gene-marked cells in biopsy specimens is considerably more challenging. We show that FDA-approved immunohistochemical diagnostic kits for detecting EGFR expression in paraffin-embedded biopsy samples can readily detect huEGFRt+ T cells at a staining intensity similar to that of EGFR+ breast cancer samples. Unlike PCR-based vector detection, which only quantifies the number of gene modified cells, detection of surface huEGFRt coupled using a 2A linker to the expression of a biologically active transgene, such as a CAR, provides for an assessment of gene-modified cells that express the CAR.
The antiproliferative and proapoptotic effects of cetuximab on EGFR+ tumors are in part the result of blocking EGFR ligand-dependent homodimerization and heterodimerization with HER2.10 In addition, cetuximab has the ability to fix complement and mediate ADCC, which are also correlated with antitumor activity in vivo.29–32 We demonstrated that Erbitux bound to huEGFRt expressed on T cells directs the cytotoxicity of ADCC effector cells and that in vivo engraftment of huEGFRt+ T cells can be inhibited by systemic administration of Erbitux. Unlike the suicide strategy based on CD20 expression and the use of rituximab,33–36 our suicide system would not ablate B cells and would be compatible for T-cell transfer in patients who are receiving rituximab as part of the lymphoma/leukemia therapy. It is also important to note that CD20 is a tetraspan transmembrane protein, and there have not been CD20 truncations described that retain rituximab binding. The safety and kinetics of T-cell elimination using Erbitux will require careful delineation, ideally in a nonhuman primate model. As cellular therapy is refined and cell products with the capacity for extended duration and high-level engraftment are developed, cell ablation maneuvers that use clinical reagents will become an increasingly useful feature of the therapeutic platform.
Acknowledgments
The authors thank Brenda Aguilar and Mahesh Jonnalagadda for their technical assistance.
This work was supported by the National Institutes of Health (P01 CA30206, P50 CA107399, P50 CA138293, R01 CA136551) and the General Clinical Research Center (M01 RR0004), as well as the Tim Nesvig Lymphoma Research Foundation, and the Marcus Foundation.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: X.W. and D.C. designed and performed research, collected, analyzed, and interpreted data, and cowrote the manuscript; W.-C.C. and C.W.W. designed and performed research and collected and analyzed data; M.S. collected, analyzed, and interpreted data and cowrote the manuscript; J.R.O. analyzed and interpreted data and cowrote the manuscript; S.J.F. and S.R.R. interpreted data and cowrote the manuscript; and M.C.J. designed research, analyzed and interpreted data, and cowrote the manuscript.
Conflict-of-interest disclosure: M.C.J. is an inventor of licensed patents and equity holder in ZetaRx Inc, a licensee of these patents. The remaining authors declare no competing financial interests.
Correspondence: Michael C. Jensen, Center for Immunity and Immunotherapies, Seattle Children's Research Institute, M/S C90S-6, 1900 Ninth Ave, Seattle, WA 98101; e-mail: [email protected].
References
Articles from Blood are provided here courtesy of The American Society of Hematology
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1182/blood-2011-02-337360
Article citations
Antitumor effects of intracranial injection of B7-H3-targeted Car-T and Car-Nk cells in a patient-derived glioblastoma xenograft model.
Cancer Immunol Immunother, 73(12):256, 05 Oct 2024
Cited by: 0 articles | PMID: 39367952 | PMCID: PMC11456075
Unlocking the potential of iPSC-derived immune cells: engineering iNK and iT cells for cutting-edge immunotherapy.
Front Immunol, 15:1457629, 30 Aug 2024
Cited by: 0 articles | PMID: 39281684 | PMCID: PMC11392856
Review Free full text in Europe PMC
A New Chimeric Antibody against the HIV-1 Fusion Inhibitory Peptide MT-C34 with a High Affinity and Fc-Mediated Cellular Cytotoxicity.
Biology (Basel), 13(9):675, 29 Aug 2024
Cited by: 0 articles | PMID: 39336102 | PMCID: PMC11428423
Manufacturing CD20/CD19-targeted iCasp9 regulatable CAR-TSCM cells using a Quantum pBac-based CAR-T engineering system.
PLoS One, 19(8):e0309245, 27 Aug 2024
Cited by: 1 article | PMID: 39190688 | PMCID: PMC11349195
A platform to deliver single and bi-specific Cas9/guide RNA to perturb genes in vitro and in vivo.
Mol Ther, 32(10):3629-3649, 31 Jul 2024
Cited by: 0 articles | PMID: 39091030
Go to all (399) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Protein structures in PDBe (2)
-
(4 citations)
PDBe - 1YY9View structure
-
(2 citations)
PDBe - 3GOPView structure
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
A Cetuximab-Mediated Suicide System in Chimeric Antigen Receptor-Modified Hematopoietic Stem Cells for Cancer Therapy.
Hum Gene Ther, 30(4):413-428, 01 Apr 2019
Cited by: 22 articles | PMID: 30860401 | PMCID: PMC6479239
Antibody with Infinite Affinity for In Vivo Tracking of Genetically Engineered Lymphocytes.
J Nucl Med, 59(12):1894-1900, 14 Jun 2018
Cited by: 27 articles | PMID: 29903928 | PMCID: PMC6278895
A transgene-encoded truncated human epidermal growth factor receptor for depletion of anti- B-cell maturation antigen CAR-T cells.
Cell Immunol, 363:104342, 14 Mar 2021
Cited by: 12 articles | PMID: 33765541
Funding
Funders who supported this work.
NCI NIH HHS (5)
Grant ID: P50 CA138293
Grant ID: P01 CA030206
Grant ID: P01 CA30206
Grant ID: P50 CA107399
Grant ID: R01 CA136551
NCRR NIH HHS (1)
Grant ID: M01 RR0004
 2,3
2,3



