Abstract
Free full text

Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming
SUMMARY
The bioenergetics of somatic dedifferentiation into induced pluripotent stem cells remains largely unknown. Here, stemness factor mediated nuclear reprogramming reverted mitochondrial networks into cristae-poor structures. Metabolomic footprinting and fingerprinting distinguished derived pluripotent progeny from parental fibroblasts according to elevated glucose utilization and production of glycolytic end products. Temporal sampling demonstrated glycolytic gene potentiation prior to induction of pluripotent markers. Functional metamorphosis of somatic oxidative phosphorylation into acquired pluripotent glycolytic metabolism conformed to an embryonic-like archetype. Stimulation of glycolysis promoted, while blockade of glycolytic enzyme activity blunted, reprogramming efficiency. Metaboproteomics resolved upregulated glycolytic enzymes and downregulated electron transport chain complex I subunits underlying cell fate determination. Thus, the energetic infrastructure of somatic cells transitions into a required glycolytic metabotype to fuel induction of pluripotency.
INTRODUCTION
Stemness transcription factors reprogram somatic cell fate to achieve embryonic-like pluripotency, the hallmark of induced pluripotent stem cells (iPSC) (Takahashi and Yamanaka, 2006; Meissner et al., 2007; Nelson et al., 2010). Developmental metabolic plasticity is associated with pluripotent stem cell fate specification (Dzeja et al., 2011), yet the metabolic dynamics that match the bioenergetic demand of nuclear reprogramming have not been established.
Somatic cells largely depend on mitochondrial oxidative phosphorylation as the primary source of energy production (DeBerardinis et al., 2008). In contrast, embryonic stem cells (ESC) rely on glycolytic ATP generation regardless of oxygen availability (Chung et al., 2007; Kondoh et al., 2007). The reliance on glycolysis reflects a low copy number of mitochondrial DNA (mtDNA) as well as low numbers of nascent mitochondria (St John et al., 2005; Cho et al., 2006). Differentiation increases mtDNA abundance and promotes mitochondrial biogenesis to form networks of elongated and cristae-rich mitochondria in support of competent oxidative metabolism (Facucho-Oliveira et al., 2007; Chung et al., 2008). Divergent energetic requirements of somatic versus pluripotent phenotypes implicate a distinguishing metabolic profile characterizing dedifferentiation.
We here demonstrate by high-resolution metabolomics that in nuclear reprogramming energy metabolism of iPSC progeny is transformed away from the parental source. Upregulated glycolytic enzymes and downregulated electron transport chain subunits enabled a metabolic switch converting somatic oxidative metabolism into a glycolytic flux-dependent, mitochondria-independent state underlying pluripotent induction.
RESULTS
Transformed Mitochondrial Infrastructure and Metabolomic Profile Define Nuclear Reprogramming
Reprogramming by four stemness transcription factors (4F) restructured mouse embryonic fibroblasts (MEF), characterized by organized mitochondrial networks, to a primordial cytotype featuring an increased nuclear-to-cytosol ratio with few perinuclear mitochondria (Figures 1A-D). Mature tubular and cristae-rich somatic mitochondria transitioned into immature spherical and cristae-poor structures in 4F iPSC, indicative of bioenergetic remodeling (Figures 1B and D). High-resolution 1H NMR quantification of 18 extracellular metabolites (Figures (Figures1E1E and S1) decoded the metabolic consequences of dedifferentiation by separating the 4F iPSC metabolomic footprint from the parental MEF landscape (Figure 1F). Accordingly, glucose and lactate were identified as distinguishing metabolites (Figure 1G). Rates of glucose utilization (2.3 ± 0.1 and 2.2 ± 0.1 nmol/μg protein/h) and lactate production (4.4 ± 0.1 and 4.8 ± 0.1 nmol/μg protein/h) were significantly elevated in two 4F iPSC lines (4F iPS1 and 4F iPS2) compared to MEF (1.8 ± 0.1 and 3.1 ± 0.1 nmol/μg protein/h, respectively, n=6, p<0.05; Figure 2H and 2I). Thus, nuclear reprogramming induces mitochondrial regression and extracellular metabolome resetting.
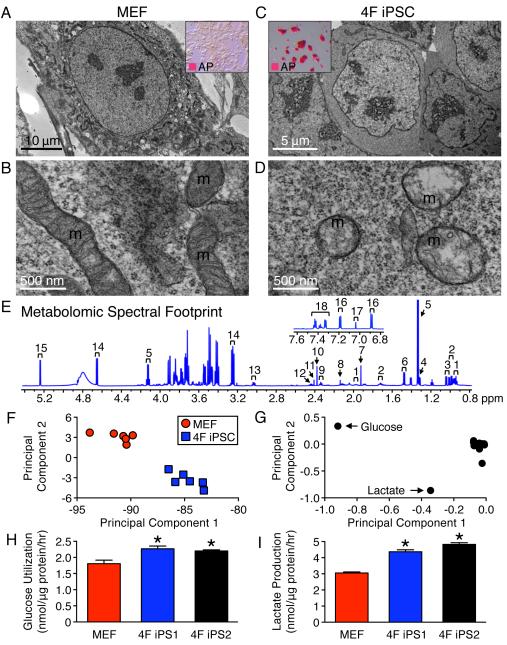
Nuclear reprogramming induced regression from elongated and cristae-rich mitochondria (m) of MEF (A, B) to spherical and cristae-poor remnant structures in four stemness factor derived iPSC (4F iPSC) (D). Insets demonstrate conversion of fibroblast monolayers (A) into compact clusters (C), which stained for the pluripotency marker AP. 1H NMR spectra of extracellular metabolites from 4F iPSC: 1 – isoleucine, 2 – leucine, 3 – valine, 4 – threonine, 5 – lactate, 6 – alanine, 7 – acetate, 8 – methionine, 9 – glutamate, 10 – pyruvate, 11 – succinate, 12 – glutamine, 13 – lysine, 14 – β-glucose, 15 – α-glucose, 16 – tyrosine, 17 – histidine and 18 – phenylalanine (E). Principal component analysis segregated 4F iPSC metabolomic phenotypes away from the MEF profile with principal component 1 accounting for 88% and 2 for 8% of the total variance (F). The loading plot assigned glucose and lactate as key metabolites contributing to segregation (G). Increased utilization of glucose and production of glycolytic end products in excess of MEF (H, I) were reproduced in independent iPSC lines (4F iPS1 and 4F iPS2). Values are mean ± SEM, n=6. * P<0.05 versus MEF. See also Figure S1.
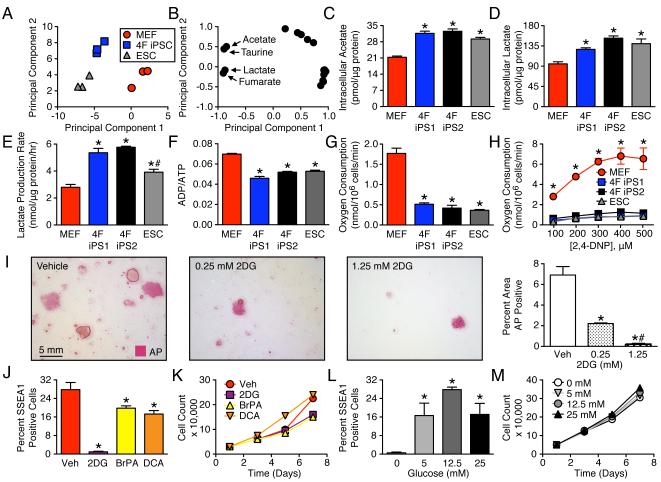
1H NMR fingerprinting of intracellular metabolites segregated 4F iPSC away from MEF and towards ESC (A). First principal component accounts for 67% and the second for 25% of the total variance. Acetate, taurine, lactate, and fumarate were identified as differentiating metabolites (B). Intracellular concentrations of glycolytic end products were distinct in 4F iPSC compared to MEF, and were similar to ESC patterns (C, D). Nuclear reprogramming elevated lactate efflux rates (E) and reduced energy turnover in 4F iPSC, similar to that of ESC (F). Compared to MEF, iPSC and ESC had reduced basal oxygen consumption (G) and lower maximal uncoupled oxidative capacity (H). During reprogramming the glycolytic inhibitor, 2-deoxyglucose (2DG), blunted induction of the pluripotency marker alkaline phosphatase (I). Glycolytic inhibitors, 1.25 mM 2DG and 100 μM 3-bromopyruvic acid (BrPA) and alternatively a stimulator of oxidative pyruvate disposal, 5 mM dichloroacetate (DCA), reduced reprogramming efficiency, assessed by SSEA1 FACS analysis (J), without altering the growth pattern of MEF (K). Stimulation of glycolysis by elevating extracellular glucose promoted the number of cells achieving the reprogrammed state (L) without altering growth of the parental population (M). Values are mean ± SEM, n=3 except for lactate efflux where n=6. In all panels except H, *P<0.05 versus MEFs and #P<0.05 versus 4F iPSC. In H, *P<0.05 versus 4F iPS and ESC. See also Figure S2.
Transition from Somatic Oxidative Metabolism to Pluripotent Glycolysis Supports iPSC Derivation
Intracellular metabolite fingerprinting validated the glycolytic capacity of 4F iPSC, segregating the acquired metabolomic pattern away from parental MEF and towards the pluripotent ESC standard (Figures (Figures2A2A and S1B). The resolved 18 intracellular metabolite panel distinguished iPSC based upon acetate, lactate, fumarate and taurine concentrations (Figure (Figure2B2B and S1C). 4F iPSC accumulation of acetate was similar to ESC (31.6 ± 0.9, 32.5 ± 1.0 and 29.2 ± 0.7 pmol/μg protein, n=3), and distinct from MEF (21.3 ± 0.5 pmol/μg protein, n=3 populations, p<0.05, Figure 2C). Comparably, lactate was equivalent in 4F iPSC and ESC (127 ± 3, 152 ± 5 and 140 ± 10 pmol/μg protein, n=3), yet significantly different from MEF (95 ± 5 pmol/μg protein, n=3, p<0.05; Figure 2D). Lactate efflux (5.3 ± 0.3 and 5.8 ± 0.1 nmol/μg protein/h) at a rate double that of MEF (2.8 ± 0.2 nmol/μg protein/h, n=6, p<0.05; Figure 2E) indicated functional glycolysis in 4F iPSC. Consistent with the lower efficiency of glycolytic ATP production compared to oxidative phosphorylation, the ADP/ATP ratio, an index of cellular energy turnover, was reduced in 4F iPSC and ESC compared to MEF (0.046 ± 0.002, 0.052 ± 0.001 and 0.053 ± 0.001 versus 0.070 ± 0.001, respectively, n=3, p<0.05, Figure 2F). Overall, oxygen consumption was low in 4F iPSC and ESC, compared to MEF both at baseline (0.51 ± 0.04, 0.42 ± 0.07 and 0.36 ± 0.02 versus 1.8 ± 0.13 nmol/106 cells/min, n=3, p<0.05, Figure 2G) and under electron transport chain uncoupling (0.98 ± 0.10, 1.22 ± 0.09 and 0.95 ± 0.13 versus 6.51 ± 1.05 nmol/106 cells/min, n=3, p<0.05, Figure 2H). 4F iPSC preserved the ability to generate mitochondrial membrane potential and demonstrated mitochondrial hyperpolarization (Figure S3A-C), indicative of reduced ATP utilization.
Treatment of MEF undergoing reprogramming with 2-deoxyglucose (2DG), a general inhibitor of glycolysis, blunted induction of the pluripotent marker alkaline phosphatase (Figure 2I), implicating a glycolytic requirement for iPSC generation. Beyond 2DG, the hexokinase 2 inhibitor 3-bromopyruvic acid (BrPA) (Ko et al., 2001) and pyruvate dehydrogenase kinase inhibitor dichloroacetate (DCA) (Stacpoole, 1989) reduced reprogramming efficiency (0.9 ± 0.3 in 2DG, 19.8 ± 1.0 in BrPA, 17.3 ± 1.5 in DCA versus 27.8 ± 3.1% SSEA1 positive cells in vehicle, n=3, p<0.05, Figure 2J) without impairing cell growth (Figure 2K). Extracellular metabolite profiles of MEF treated with 2DG or BrPA were consistent with inhibition of glycolysis and stimulation of oxidative metabolism, while DCA treatment stimulated mitochondrial function and accelerated consumption of oxidative substrates (Figure S2A-C). Glucose supplementation stimulated glycolytic flux, reduced mitochondrial function (Figure S2 D-F) and increased SSEA1 positive cell density (0.5 ± 0.3, 16.6 ± 5.3, 27.8 ± 1.1 and 17.1 ± 4.7% SSEA1 positive cells in 0, 5, 12.5 and 25 mM glucose, n=3, p<0.05). Conversely, few MEF underwent nuclear reprogramming in the absence of glucose (Figure 2L-M). Transition from somatic oxidative metabolism to pluripotent glycolysis thus contributes to nuclear reprogramming efficiency.
Glycolytic Flux Fuels Pluripotent Induction
Mitochondrial membrane potential served as a surrogate of metabolic transition during nuclear reprogramming. Prospective live cell imaging distinguished ESC-like compact clusters with high tetramethylrhodamine methyl ester (TMRM) fluorescence from transfected non-reprogrammed MEF with low mitochondrial membrane fluorescence (Figure 3A). FACS sampling of high TMRM cells at day 7 and 14 selected an emergent subpopulation within small cell clusters, consistent with mitochondrial membrane potential polarization during dedifferentiation (Figure S3A-D). At day 7, high TMRM cells had significantly elevated glycolytic gene expression (Glut1, Hxk2, Pfkm and Ldha), which met that of ESC by 2-weeks of reprogramming (Figure 3B). In contrast, at 1-week of reprogramming pluripotent gene expression (Fgf4, Nanog, Oct4 and Sox2) remained low in high TMRM cells, similar to starting MEF, with pluripotent gene induction apparent only after 2-weeks (Figure 3C). Metabolic reprogramming thus precedes pluripotent gene expression during somatic cell dedifferentiation from an oxidative to a glycolytic iPSC phenotype (Figure 3D).
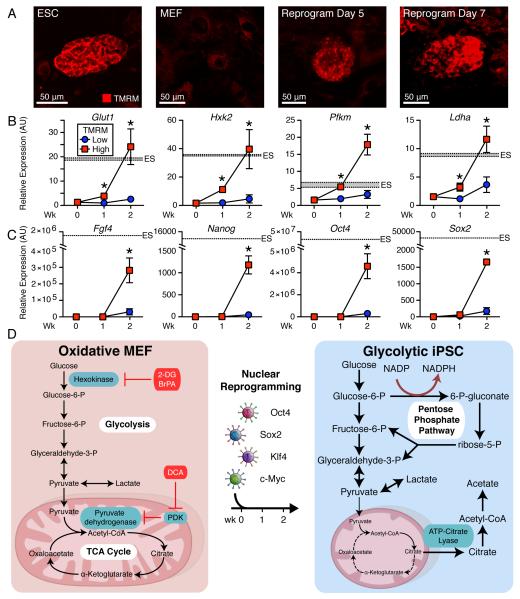
Similar to ESC and distinct from MEF, live cell imaging of mitochondrial membrane potential identified nascent compact cell clusters with high TMRM fluorescence within 5-7 days of nuclear reprogramming (A). Compared to the low TMRM fluorescence population, high fluorescence cells had significantly elevated glycolytic gene expression (Glut1, Hxk2, Pfkm and Ldha) within 1-week of reprogramming, which by 2-weeks of reprogramming equaled ESC glycolytic gene expression (B). Of note, at week one of reprogramming pluripotent gene expression (Fgf4, Nanog, Oct4 and Sox2) remained low in high TMRM fluorescence cells, similar to the starting MEF, with pluripotent gene induction apparent during the second week (C). Shaded region represents mean ± SEM for ESC gene expression. Values are mean ± SEM, n=3. * P<0.05 versus low TMRM population. Nuclear reprogramming switches oxidative MEF into glycolytic iPSC (D). See also Figure S3.
Reprogramming-Induced Metabolic Remodeling Independent of c-Myc Induction
As c-Myc gene targets control rates of glycolysis and mitochondrial biogenesis (Dang, 2007), a cell line was derived without c-Myc (3F iPSC). Similar to 4F iPSC, NMR metabolomic footprinting and fingerprinting segregated 3F iPSC away from parental MEF (Figures 4A-B), based upon greater intracellular and extracellular accumulation of glycolytic end products, acetate (intracellular: 30.4 ± 0.8 versus 21.3 ± 0.5 pmol/μg protein; extracellular: 0.41 ± 0.03 versus 0.035 ± 0.001 nmol/μg protein/h, n=3, p<0.05, Figure 4C) and lactate (intracellular: 188 ± 11 versus 95 ± 5 pmol/μg protein; extracellular: 4.1 ± 0.1 versus 3.0 ± 0.1 nmol/μg protein/h n=3, p<0.05, Figure 4D). Consistent with accelerated glycolysis, 3F iPSC displayed reduced energy turnover compared to MEF (0.039 ± 0.005 versus 0.068 ± 0.006, n=3, p<0.05, Figure 4E). Both 3F and 4F iPSC demonstrated limited oxidative capacity and elevated lactate production (Figure 4F). Thus, nuclear reprogramming-induced metabolic remodeling is characteristic of the iPS phenotype, independent of c-Myc transduction.
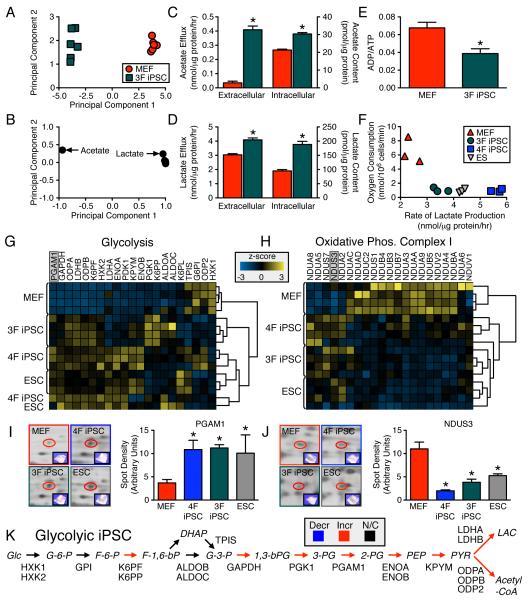
1H NMR cellular metabolomic fingerprints (n=6) of three stemness factor induced iPSC (3F iPSC) segregated, away from the somatic, the acquired pattern (first principal component accounts for 97% and second for 1% of total variance) (A). Glycolytic end products, acetate and lactate, were key metabolites responsible for segregation (B). Intracellular content and efflux of acetate and lactate were significantly elevated in the 3F iPSC compared to MEF (C and D) and associated with reduced energy turnover (n=6) (E). Compared to MEF, 3F iPSC had lower maximal oxidative capacity and higher lactate production similar to that of ESC, albeit not fully overlapping with 4F iPSC (n=3) (F). Proteome-wide label-free quantification segregated iPSC away from MEF towards an ESC pattern based on agglomerative clustering of z-score transformed data (n=4) due to predominant glycolytic enzyme upregulation (G). Electron transport chain complex I subunits were predominantly downregulated in pluripotent cytotypes, which clustered away from MEF (H). 2-D gel quantification and MS/MS identification (n=3) independently confirmed glycolytic upregulation (I) and complex I downregulation (J). iPSC proteomic upregulation was mapped across the glycolytic pathway (K). Values are mean ± SEM. *P<0.05 versus MEF. Proteins are abbreviated by Swiss-Prot gene name. See also Figure S4 and Tables S1 and S2.
iPS Metabotype Arises through Proteome Restructuring
Metaboproteome dissection delineated a transformed molecular signature in iPSC, distinct from parental MEF, yet synonymous to the metabolic protein profile of ESC (Figures 4G-J and S4, Table S1 and S2). A label-free, quantitative proteomics approach revealed the identities of glycolytic enzymes consistently upregulated in pluripotent cytotypes relative to MEF (Figures (Figures4G4G and S4A), confirmed independently by 2-D gel based analysis (Figure 4I). Concomitantly, 65% of detected complex 1 subunits were downregulated in iPSC extracts (Figures (Figures4H4H and S4B, Table S1) as validated by 2-D gel based analysis (Figure 4J), underscoring departure from MEF patterns and acquisition of a glycolytic-dependent profile (Figure 4K). In addition, reprogramming was associated with selective downregulation of the reducing equivalent entry points (complex I and II) (Figure S4C, Table S1). Thus, the resolved iPSC metaboproteome unmasks a bioenergetic switch in energy utilization infrastructure underlying metabolic remodeling in nuclear reprogramming.
DISCUSSION
Nuclear reprogramming sets in motion dedifferentiation processes leading to acquisition of pluripotency. This study resolved the metabolic attributes underpinning iPSC generation. Metaboproteome restructuring was demonstrated to underlie conversion from somatic oxidative bioenergetics to glycolysis of pluripotent progeny. Promotion of a glycolytic flux-dependent, mitochondria-independent metabotype fostered nuclear reprogramming.
Regression of the parental somatic network of abundant tubular and cristae-rich mitochondria into sparse spherical and cristae-poor structures characterized iPSC progeny. A mitochondria-poor capacity would enable iPSC to revert reactive oxygen species-dependent telomere shortening associated with cellular aging (Passos et al., 2007; Marion et al., 2009). In fact, mitochondrial regression and reduction of mtDNA as a result of nuclear reprogramming is consistent with the undeveloped mitochondrial morphology of ESC (Kondoh et al., 2007; Armstrong et al., 2010; Prigione et al., 2010; Zeuschner et al., 2010). Induced reduction in mitochondria number and maturity suggests metamorphosis of the bioenergetic infrastructure underlying dedifferentiation.
Metabolomic footprinting and fingerprinting indeed demonstrated distinct features of iPSC metabolism. The key metabolic rates contributing to the iPSC phenotype were consistent among multiple clones, and included elevated utilization of glucose and accumulation of intracellular and extracellular lactate (via lactate dehydrogenase) and acetate (via acetyl-CoA synthase or ATP-citrate lyase), suggesting that accelerated glycolysis results in accumulation of metabolic byproducts within the cell, followed by metabolite exportation. iPSC demonstrated diminished basal oxygen consumption and uncoupled oxidative capacity, indicating departure from oxidative metabolism. ESC also rely on glycolytic ATP generation (Cho et al., 2006; Facucho-Oliveira et al., 2007), with pluripotency maintenance sustained under hypoxic conditions (Ezashi et al., 2005). Acquisition of an embryonic-like metabolic state is concomitant with global transcriptional and epigenetic reorganization that includes demethylation and activation of the promoter region of pluripotency inductors (Bock et al., 2011; Plath and Lowry, 2011). A glycolytic burst could promote linkage of energy metabolism with nuclear reprogramming-induced epigenetic resetting (Huangfu et al., 2008; Friis et al., 2009; Wellen et al., 2009; Teperino et al., 2010), priming the cell for pluripotent induction. Indeed, expression of glycolytic genes preceded pluripotent gene expression, implicating metabolic remodeling as a facilitator of nuclear reprogramming.
Complete oxidation of glucose limits generation of substrates for biosynthetic pathways. Transition to glycolytic metabolism would fuel specific cellular processes, such as ATP consumption in the cell nucleus and provide the necessary balance between ATP generation and production of metabolic precursors to meet anabolic requirements of dedifferentiation and associated proliferation (DeBerardinis et al., 2008; Chung et al., 2010). Strategies that in part stimulate glycolysis, including induction of hypoxia or inhibition of the p53 pathway, increase nuclear reprogramming efficiency (Kruse and Gu, 2006; Krizhanovsky and Lowe, 2009; Yoshida et al., 2009). Consistent with the ability of small molecules with known metabolic targets to modulate the appearance of pluripotent colonies (Zhu et al., 2010), here stimulation of glycolytic flux potentiated reprogramming whereas oxidative metabolism promotion preserved the somatic cytotype.
c-Myc, through its regulation of both glycolysis and mitochondrial metabolism, is a key contributor to the metabolic Warburg effect, reflecting the tendency of highly proliferative cell types to convert glucose to lactate despite sufficient oxygen for mitochondrial oxidative metabolism (Dang, 2007). Here 3F iPSC displayed equivalent metabolic profiles to 4F iPSC counterparts, verifying that nuclear reprogramming-induced metabolic remodeling did not require c-Myc transduction. Proteomic dissection resolved a metabolic infrastructure that distinguished 3F and 4F iPSC from fibroblasts. Specifically, reprogramming induced reciprocal changes in glycolytic enzymes and electron transport chain components, indicating a molecular substrate contributing to the switch from oxidative to glycolytic metabolism. The observed decreased expression of complex I and II subunits would uncouple oxidative metabolism and ATP generation, favoring glycolytic dependent ATP production.
In summary, dedifferentiation of parental somatic cells induced mitochondrial regression, electron transport chain downregulation, and glycolysis enzyme upregulation, revealing a pluripotent glycolytic signature with limited dependence on mitochondrial metabolism. Induction of the glycolytic metabotype support anabolic and catabolic requirements for bona fide pluripotency to regulate cell fate.
EXPERIMENTAL PROCEDURES
Transduction
Mouse embryonic fibroblasts were transduced with in house HIV-based viral vectors encoding OCT3/4, SOX2, and KLF4 in the presence or absence of c-MYC to produce iPSC clones that met pluripotent criteria including expression of stem cell markers, embryoid body differentiation, teratoma formation, diploid aggregation and contribution to organogenesis (Martinez-Fernandez et al., 2009; Nelson et al., 2009a; Nelson et al., 2009b). Alternatively iPSC were derived with a viPS (Open Biosystems) kit. iPSC and mouse ESC (R1) were induced and maintained in DMEM supplemented with 15% FBS, 25 mM glucose, 2 mM Glutamax (Invitrogen) and 1 mM sodium pyruvate. Reprogramming efficiency was quantified using an alkaline phosphatase staining kit (Stemgent) or FACS analysis of SSEA1 expression (Millipore) on a LSR II flow cytometer.
Ultrastructure
Mitochondrial density and morphology was evaluated in 1% glutaraldehyde and 4% formaldehyde fixed cells, and examined as ultramicrotome sections on a JEOL 1200 EXII electron microscope (Perez-Terzic et al., 2007).
Metabolomic Footprinting and Fingerprinting
For footprinting of extracellular metabolites (see Supplemental Experimental Procedures), 540 μL of media collected following 24 h of culture was added to 60 μL of D2O (Sigma) containing 5 mM sodium 3-(trimethylsilyl)propionate-2,2,3,3-d4 (TSP) (Sigma) for chemical shift reference and 81.84 mM formate (Sigma) for peak quantification reference (Turner et al., 2008). For intracellular metabolite fingerprinting, neutralized perchloric acid extracts were concentrated with a SpeedVac and suspended in 600 μl of 100 mM phosphate buffer (pH 7.0) in D2O (Sigma) containing 0.5 mM TSP (Beckonert et al., 2007). Identities of 1H NMR spectra peaks obtained on a Bruker Ultrashield 700 MHz spectrometer with a zgpr water suppression pre-saturation pulse were assigned by comparison to reference values for chemical shift and multiplicity, and confirmed by comparison to spectra of pure compounds in the Human Metabolome database (Govindaraju et al., 2000; Wishart et al., 2009).
Metabolites and Oxygen Consumption
Lactate efflux rate was assessed in extracellular media using a lactate assay (SUNY). Nucleotide concentrations were determined in neutralized perchloric acid extracts by high performance liquid chromatography, using a 0.1 M phosphate (pH 6.5), 0.01 M tetrabutylammoniumhydrogensulfate, and 40% methanol elution buffer (Chung et al., 2007). Oxygen consumption was assessed using an electrode system (Hansatech) on 5 million trypsinized cells suspended in DMEM. Maximal rate of uncoupled oxygen consumption was assessed by serial additions of 2,4-dinitrophenol (Sigma).
TMRM Fluorescence Sorting and Gene Expression
Mitochondrial membrane potential was assessed at day 4-14 of reprogramming by incubating with 20 nM TMRM (Anaspec) for 30 min at 37°C, and imaged with a LSM 510 Axiovert laser confocal microscope. By 1 and 2-weeks of reprogramming, single cell suspensions were incubated in TMRM and separated by a FACS Aria Cell Sorter. Gene expression was examined on an Eco RT-PCR system (Illumina).
Proteomics
Protein extracts were resolved by 2-D gel electrophoresis and 4-15% SDS-PAGE (100 and 30 μg, respectively) and silver stained (Zlatkovic-Lindor et al., 2010). For comparative analysis, SDS-PAGE lanes were excised, destained, and prepared for LC-MS/MS, as were protein species from 2-D gels identified as significantly altered by PDQuest analysis (Zlatkovic-Lindor et al., 2010). Isolated tryptic peptides were analyzed and identified by LTQ-Orbitrap mass spectrometry. Label-free quantitative comparison of SDS-PAGE protein and peptide abundance was carried out on acquired MS spectra using Rosetta Elucidator’s differential workflow, with annotation performed using PeptideTeller and ProteinTeller (Neubert et al., 2008; Lomenick et al., 2009). See Supplemental Experimental Procedures.
Statistical Analysis
Data are presented as mean ± SEM. Metabolic footprinting and fingerprinting were analyzed using principle component analysis and JMP. Student t-test was used to evaluate two group comparisons, and ANOVA with a Bonferroni post-hoc correction for three group comparisons. A value of P<0.05 was considered significant.
Supplementary Material
01
02
03
ACKNOWLEDGMENTS
Authors thank Mayo Clinic Nuclear Magnetic Resonance Core and Flow Cytometry/Optical Morphology Shared Resource for assistance. This work was supported by National Institutes of Health (R01HL083439, T32HL007111, R01HL085208, R56AI074363), Canadian Institutes of Health Research, Marriott Program, and Mayo Clinic.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Armstrong L, Tilgner K, Saretzki G, Atkinson SP, Stojkovic M, Moreno R, Przyborski S, Lako M. Human induced pluripotent stem cell lines show similar stress defence mechanisms and mitochondrial regulation to human embryonic stem cells. Stem Cells. 2010;28:661–673. [Abstract] [Google Scholar]
- Beckonert O, Keun HC, Ebbels TM, Bundy J, Holmes E, Lindon JC, Nicholson JK. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat Protoc. 2007;2:2692–2703. [Abstract] [Google Scholar]
- Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, Ziller M, Croft GF, Amoroso MW, Oakley DH, et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. [Europe PMC free article] [Abstract] [Google Scholar]
- Cho YM, Kwon S, Pak YK, Seol HW, Choi YM, Park do J, Park KS, Lee HK. Dynamic changes in mitochondrial biogenesis and antioxidant enzymes during the spontaneous differentiation of human embryonic stem cells. Biochem. Biophys. Res. Commun. 2006;348:1472–1478. [Abstract] [Google Scholar]
- Chung S, Arrell DK, Faustino RS, Terzic A, Dzeja PP. Glycolytic network restructuring integral to the energetics of embryonic stem cell cardiac differentiation. J. Mol. Cell. Cardiol. 2010;48:725–734. [Europe PMC free article] [Abstract] [Google Scholar]
- Chung S, Dzeja PP, Faustino RS, Perez-Terzic C, Behfar A, Terzic A. Mitochondrial oxidative metabolism is required for the cardiac differentiation of stem cells. Nat. Clin. Pract. Cardiovasc. Med. 2007;4(Suppl 1):S60–67. [Europe PMC free article] [Abstract] [Google Scholar]
- Chung S, Dzeja PP, Faustino RS, Terzic A. Developmental restructuring of the creatine kinase system integrates mitochondrial energetics with stem cell cardiogenesis. Ann. N. Y. Acad. Sci. 2008;1147:254–263. [Europe PMC free article] [Abstract] [Google Scholar]
- Dang CV. The interplay between MYC and HIF in the Warburg effect. Ernst Schering Found Symp Proc. 2007:35–53. [Abstract] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. [Abstract] [Google Scholar]
- Dzeja PP, Chung S, Faustino RS, Behfar A, Terzic A. Developmental Enhancement of Adenylate Kinase-AMPK Metabolic Signaling Axis Supports Stem Cell Cardiac Differentiation. PLoS One. 2011;6:e19300. [Europe PMC free article] [Abstract] [Google Scholar]
- Ezashi T, Das P, Roberts RM. Low O2 tensions and the prevention of differentiation of hES cells. Proc Natl Acad Sci USA. 2005;102:4783–4788. [Abstract] [Google Scholar]
- Facucho-Oliveira JM, Alderson J, Spikings EC, Egginton S, St John JC. Mitochondrial DNA replication during differentiation of murine embryonic stem cells. J. Cell Sci. 2007;120:4025–4034. [Abstract] [Google Scholar]
- Friis RM, Wu BP, Reinke SN, Hockman DJ, Sykes BD, Schultz MC. A glycolytic burst drives glucose induction of global histone acetylation by picNuA4 and SAGA. Nucleic Acids Res. 2009;37:3969–3980. [Europe PMC free article] [Abstract] [Google Scholar]
- Govindaraju V, Young K, Maudsley AA. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR Biomed. 2000;13:129–153. [Abstract] [Google Scholar]
- Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat. Biotechnol. 2008;26:795–797. [Abstract] [Google Scholar]
- Ko YH, Pedersen PL, Geschwind JF. Glucose catabolism in the rabbit VX2 tumor model for liver cancer: characterization and targeting hexokinase. Cancer Lett. 2001;173:83–91. [Abstract] [Google Scholar]
- Kondoh H, Lleonart ME, Nakashima Y, Yokode M, Tanaka M, Bernard D, Gil J, Beach D. A high glycolytic flux supports the proliferative potential of murine embryonic stem cells. Antioxid. Redox. Signal. 2007;9:293–299. [Abstract] [Google Scholar]
- Krizhanovsky V, Lowe SW. Stem cells: The promises and perils of p53. Nature. 2009;460:1085–1086. [Europe PMC free article] [Abstract] [Google Scholar]
- Kruse JP, Gu W. p53 aerobics: the major tumor suppressor fuels your workout. Cell Metab. 2006;4:1–3. [Abstract] [Google Scholar]
- Lomenick B, Hao R, Jonai N, Chin RM, Aghajan M, Warburton S, Wang J, Wu RP, Gomez F, Loo JA, et al. Target identification using drug affinity responsive target stability (DARTS) Proc Natl Acad Sci USA. 2009;106:21984–21989. [Abstract] [Google Scholar]
- Marion RM, Strati K, Li H, Tejera A, Schoeftner S, Ortega S, Serrano M, Blasco MA. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell. 2009;4:141–154. [Abstract] [Google Scholar]
- Martinez-Fernandez A, Nelson TJ, Yamada S, Reyes S, Alekseev AE, Perez-Terzic C, Ikeda Y, Terzic A. iPS programmed without c-MYC yield proficient cardiogenesis for functional heart chimerism. Circ. Res. 2009;105:648–656. [Europe PMC free article] [Abstract] [Google Scholar]
- Meissner A, Wernig M, Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat. Biotechnol. 2007;25:1177–1181. [Abstract] [Google Scholar]
- Nelson TJ, Martinez-Fernandez A, Terzic A. Induced pluripotent stem cells: developmental biology to regenerative medicine. Nat Rev Cardiol. 2010;7:700–710. [Abstract] [Google Scholar]
- Nelson TJ, Martinez-Fernandez A, Yamada S, Mael AA, Terzic A, Ikeda Y. Induced pluripotent reprogramming from promiscuous human stemness-related factors. Clin Transl Sci. 2009a;2:118–126. [Europe PMC free article] [Abstract] [Google Scholar]
- Nelson TJ, Martinez-Fernandez A, Yamada S, Perez-Terzic C, Ikeda Y, Terzic A. Repair of acute myocardial infarction by human stemness factors induced pluripotent stem cells. Circulation. 2009b;120:408–416. [Europe PMC free article] [Abstract] [Google Scholar]
- Neubert H, Bonnert TP, Rumpel K, Hunt BT, Henle ES, James IT. Label-free detection of differential protein expression by LC/MALDI mass spectrometry. J. Proteome Res. 2008;7:2270–2279. [Abstract] [Google Scholar]
- Passos JF, Saretzki G, Ahmed S, Nelson G, Richter T, Peters H, Wappler I, Birket MJ, Harold G, Schaeuble K, et al. Mitochondrial dysfunction accounts for the stochastic heterogeneity in telomere-dependent senescence. PLoS Biol. 2007;5:e110. [Abstract] [Google Scholar]
- Perez-Terzic C, Faustino RS, Boorsma BJ, Arrell DK, Niederlander NJ, Behfar A, Terzic A. Stem cells transform into a cardiac phenotype with remodeling of the nuclear transport machinery. Nat. Clin. Pract. Cardiovasc. Med. 2007;4(Suppl 1):S68–76. [Abstract] [Google Scholar]
- Plath K, Lowry WE. Progress in understanding reprogramming to the induced pluripotent state. Nat. Rev. Genet. 2011;12:253–265. [Europe PMC free article] [Abstract] [Google Scholar]
- Prigione A, Fauler B, Lurz R, Lehrach H, Adjaye J. The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cells. 2010;28:721–733. [Abstract] [Google Scholar]
- St John JC, Ramalho-Santos J, Gray HL, Petrosko P, Rawe VY, Navara CS, Simerly CR, Schatten GP. The expression of mitochondrial DNA transcription factors during early cardiomyocyte in vitro differentiation from human embryonic stem cells. Cloning Stem Cells. 2005;7:141–153. [Abstract] [Google Scholar]
- Stacpoole PW. The pharmacology of dichloroacetate. Metabolism. 1989;38:1124–1144. [Abstract] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. [Abstract] [Google Scholar]
- Teperino R, Schoonjans K, Auwerx J. Histone methyl transferases and demethylases; can they link metabolism and transcription? Cell Metab. 2010;12:321–327. [Europe PMC free article] [Abstract] [Google Scholar]
- Turner WS, Seagle C, Galanko JA, Favorov O, Prestwich GD, Macdonald JM, Reid LM. Nuclear magnetic resonance metabolomic footprinting of human hepatic stem cells and hepatoblasts cultured in hyaluronan-matrix hydrogels. Stem Cells. 2008;26:1547–1555. [Abstract] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. [Europe PMC free article] [Abstract] [Google Scholar]
- Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37:D603–610. [Europe PMC free article] [Abstract] [Google Scholar]
- Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell. 2009;5:237–241. [Abstract] [Google Scholar]
- Zeuschner D, Mildner K, Zaehres H, Scholer HR. Induced pluripotent stem cells at nanoscale. Stem Cells Dev. 2010;19:615–620. [Abstract] [Google Scholar]
- Zhu S, Li W, Zhou H, Wei W, Ambasudhan R, Lin T, Kim J, Zhang K, Ding S. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell. 2010;7:651–655. [Europe PMC free article] [Abstract] [Google Scholar]
- Zlatkovic-Lindor J, Arrell DK, Yamada S, Nelson TJ, Terzic A. ATP-sensitive K+ channel-deficient dilated cardiomyopathy proteome remodeled by embryonic stem cell therapy. Stem Cells. 2010;28:1355–1367. [Europe PMC free article] [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.cmet.2011.06.011
Read article for free, from open access legal sources, via Unpaywall:
https://www.cell.com/article/S1550413111002609/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1016/j.cmet.2011.06.011
Article citations
Lactylation of Hdac1 regulated by Ldh prevents the pluripotent-to-2C state conversion.
Stem Cell Res Ther, 15(1):415, 13 Nov 2024
Cited by: 0 articles | PMID: 39533309 | PMCID: PMC11559218
Understanding the autophagic functions in cancer stem cell maintenance and therapy resistance.
Expert Rev Mol Med, 26:e23, 08 Oct 2024
Cited by: 0 articles | PMID: 39375840 | PMCID: PMC11488345
Review Free full text in Europe PMC
piRNAs are regulators of metabolic reprogramming in stem cells.
Nat Commun, 15(1):8405, 27 Sep 2024
Cited by: 0 articles | PMID: 39333531 | PMCID: PMC11437085
Regulation of glucose metabolism: Effects on oocyte, preimplantation embryo, assisted reproductive technology and embryonic stem cell.
Heliyon, 10(19):e38551, 28 Sep 2024
Cited by: 0 articles | PMID: 39403464 | PMCID: PMC11471579
Review Free full text in Europe PMC
Blocking tumor-intrinsic MNK1 kinase restricts metabolic adaptation and diminishes liver metastasis.
Sci Adv, 10(37):eadi7673, 13 Sep 2024
Cited by: 0 articles | PMID: 39270021 | PMCID: PMC11397505
Go to all (632) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Interference with the mitochondrial bioenergetics fuels reprogramming to pluripotency via facilitation of the glycolytic transition.
Int J Biochem Cell Biol, 45(11):2512-2518, 09 Aug 2013
Cited by: 23 articles | PMID: 23939289
Nuclear reprogramming with c-Myc potentiates glycolytic capacity of derived induced pluripotent stem cells.
J Cardiovasc Transl Res, 6(1):10-21, 18 Dec 2012
Cited by: 37 articles | PMID: 23247633 | PMCID: PMC3750736
Energy metabolism in nuclear reprogramming.
Biomark Med, 5(6):715-729, 01 Dec 2011
Cited by: 38 articles | PMID: 22103608 | PMCID: PMC3477517
Review Free full text in Europe PMC
Metabolome and metaboproteome remodeling in nuclear reprogramming.
Cell Cycle, 12(15):2355-2365, 08 Jul 2013
Cited by: 22 articles | PMID: 23839047 | PMCID: PMC3841314
Funding
Funders who supported this work.
CIHR
NHLBI NIH HHS (11)
Grant ID: R01 HL083439-04
Grant ID: R01 HL085744-05
Grant ID: R01HL083439
Grant ID: R01 HL085744
Grant ID: T32HL007111
Grant ID: R01 HL083439
Grant ID: R01 HL083439-05
Grant ID: T32 HL007111
Grant ID: R01 HL085208
Grant ID: R01 HL085744-04
Grant ID: R01HL085208
NIAID NIH HHS (2)
Grant ID: R56 AI074363
Grant ID: R56AI074363





