Abstract
Free full text

Cell cycle–regulated phosphorylation of p220NPAT by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription
Abstract
Cyclin E/Cdk2 acts at the G1/S-phase transition to promote the E2F transcriptional program and the initiation of DNA synthesis. To explore further how cyclin E/Cdk2 controls S-phase events, we examined the subcellular localization of the cyclin E/Cdk2 interacting protein p220NPAT and its regulation by phosphorylation. p220 is localized to discrete nuclear foci. Diploid fibroblasts in Go and G1 contain two p220 foci, whereas S- and G2-phase cells contain primarily four p220 foci. Cells in metaphase and telophase have no detectable focus. p220 foci contain cyclin E and are coincident with Cajal bodies (CBs), subnuclear organelles that associate with histone gene clusters on chromosomes 1 and 6. Interestingly, p220 foci associate with chromosome 6 throughout the cell cycle and with chromosome 1 during S phase. Five cyclin E/Cdk2 phosphorylation sites in p220 were identified. Phospho-specific antibodies against two of these sites react with p220 within CBs in a cell cycle–specific manner. The timing of p220 phosphorylation correlates with the appearance of cyclin E in CBs at the G1/S boundary, and this phosphorylation is maintained until prophase. Expression of p220 activates transcription of the histone H2B promoter. Importantly, mutation of Cdk2 phosphorylation sites to alanine abrogates the ability of p220 to activate the histone H2B promoter. Collectively, these results strongly suggest that p220NPAT links cyclical cyclin E/Cdk2 kinase activity to replication-dependent histone gene transcription.
Cyclin E, an essential regulatory subunit of Cdk2 (Dulic et al. 1992; Koff et al. 1992), plays a central role in coordinating both the onset of S phase and centrosome duplication in multicellular eukaryotes (Sherr 1996; Reed 1997). Cyclin E/Cdk2 complexes have two major roles in promoting S phase. First, cyclin E/Cdk2 participates, together with cyclin D/Cdk4, in the control of transcriptional processes that are critical to cell cycle progression. The best understood example is control of the E2F/DP transcription factor via phosphorylation of a family of transcriptional repressors (Rb, p130, and p107) (for review, see Reed 1997; Dyson 1998; Nevins 1998). E2F complexes regulate the S-phase-dependent expression of a number of proteins required for the synthesis of nucleic acids as well as proteins such as Cdc2 and cyclin A that promote subsequent cell cycle transitions. Second, cyclin E/Cdk2 can function in an E2F-independent manner to activate DNA replication. Accumulation of cyclin E is required for S-phase entry, and ectopic cyclin E expression can bypass the requirement for Rb inactivation and E2F activation for S-phase entry (Ohtsubo et al. 1995; Leng et al. 1997; Lukas et al. 1997).
An understanding of the role of cyclin E/Cdk2 in promoting S phase requires knowledge of its essential substrates. Insight into Cdk targets has been advanced by the finding that several Cdk substrates bind tightly to the cyclin subunit. In some cases, this interaction involves a motif in the substrate, the RXL motif, and a conserved pocket in the cyclin box (Zhu et al. 1995; Adams et al. 1996; Russo et al. 1996; Schulman et al. 1998; Brown et al. 1999; Ma et al. 1999). We and others have exploited this property of cyclins to identify relevant cyclin E/Cdk2 substrates with the use of expression cloning (Zhao et al. 1998; Ma et al. 1999). One of these, p220NPAT, interacts with cyclin E/Cdk2 in extracts from tissue culture cells and accelerates S-phase entry when overexpressed (Zhao et al. 1998). Moreover, retroviral insertion into the mouse p220NPAT gene leads to embryonic lethality at the eight-cell stage, indicating an essential role for p220 in cell division or development (Di Fruscio et al. 1997). However, the precise function of p220 and the role of cyclin E/Cdk2 in its action remain unknown.
Emerging data (Zhao et al. 2000, this paper) suggest that p220 is involved in S-phase-specific histone gene transcription. Histones, components of nucleosomes, have to be supplied on demand during DNA replication. This regulation is attributed to both transcriptional and posttranscriptional control mechanisms (Harris et al. 1991; Heintz 1991), mediated in part by the activation of histone gene–specific transcription factors (Oct-1 in the case of the H2B promoter and H1TF2 in the case of the H1 promoter) through an unknown mechanism (Fletcher et al. 1987; Segil et al. 1991). Once generated, histone messages are stabilized and processed preferentially in S phase. Implicated in histone gene transcription are Cajal bodies (CBs; sometimes referred to as coiled bodies). CBs were initially described as small nuclear organelles (Cajal 1903), but their function has remained obscure for the better part of the twentieth century. Recent work has led to the hypothesis that CBs are sites of assembly of transcription and splicing complexes (Gall et al. 1999). The link to histone transcription comes from the finding that a subset of CBs is physically associated with histone gene clusters on chromosomes 1 (1q21) and 6 (6p21) (Frey and Matera 1995) and with histone gene loci in Xenopus lampbrush chromosomes (Abbott et al. 1999). Moreover, CBs also contain a component of the histone mRNA 3′-end processing machinery SLBP1 (Abbott et al. 1999).
Here, we report that p220NPAT is localized to discrete foci that are coincident with a subset of CBs in normal diploid fibroblasts. The number of p220 foci increased from two in Go and G1 cells in association with chromosome 6 to four in S and G2 phases in association with both chromosomes 6 and 1. Foci are lost during mitosis. Consistent with these observations, Zhao et al. (2000) have found that p220 is associated directly with histone gene clusters and that overexpression of p220 can activate histone 2B and histone 4 transcription. We also demonstrate that cyclin E is contained in p220 foci and that p220 within CBs is phosphorylated on Cdk sites in a cell cycle–dependent manner. Moreover, mutation of cyclin E/Cdk2 phosphorylation sites in p220 reduces its ability to activate expression from histone H2B reporter constructs in transiently transfected cells. These data, together with those of Zhao et al. (2000), suggest that cyclin E/Cdk2 functions in conjunction with p220 to coordinate S-phase-dependent histone gene transcription; they also demonstrate a role for CBs in cell cycle–regulated transcriptional control.
Results
p220 is localized in cell cycle–regulated nuclear foci
foci
We previously identified a C-terminal fragment of NPAT (residues 1054–1397) in a cyclin E/Cdk3 interaction screen (Ma et al. 1999). Affinity-purified anti-NPAT antibodies generated against this C-terminal fragment recognize a closely spaced protein doublet of 220 kD in molecular mass in nuclear extracts from HeLa and 293 cells, as determined by either immunoblotting or immunoprecipitation (Fig. (Fig.1a,1a, lane 2; data not shown). The identity of the p220 protein obtained by immunoprecipitation was confirmed by mass spectral analysis of tryptic peptides (see below).
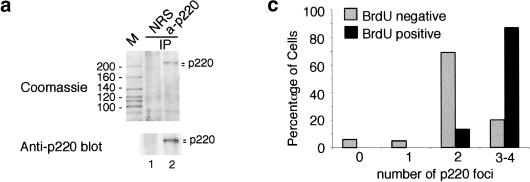

p220 is located in cell cycle–regulated nuclear foci. (a) Affinity-purified polyclonal antibodies against p220 immunoprecipitate a closely spaced doublet of proteins 220 kD in molecular mass from tissue culture cells. For a large-scale immunoprecipitation, nuclear extracts from 293T cells (44 mg in 9 mL) were immunoprecipitated with 20 μg of anti-p220 antibodies or pre-immune IgG bound to 80 μL of protein A–Sepharose. Washed immunoprecipitates were separated using SDS–PAGE, and the gel was stained with Coomassie blue (top). A small fraction of this immune complex was immunoblotted with anti-p220 antibodies (bottom). (M) Molecular mass markers with masses indicated at left; (NRS) normal rabbit sera; (IP) immunoprecipitate. (b) p220 is localized in discrete nuclear foci. WI38 fibroblasts were subjected to indirect immunofluorescence using anti-p220 antibodies in the presence (right) or absence (left) of 0.5 μg of antigen. (red) p220; (blue) nuclei stained with 4′,6-diamidino-2-phenylindole (DAPI). (c) Cells with four p220 foci accumulate during S phase. Asynchronous bjTERT fibroblasts were pulse-labeled with BrdU for 60 min and then stained for p220 and BrdU. The number of p220 foci in BrdU-positive and BrdU-negative cells was determined from a minimum of 100 cells. (d) An example of BrdU-positive (green) cells displaying three or four p220 foci (red), whereas a BrdU-negative cell had two p220 foci. DAPI staining of nuclei is in blue.
To examine the subcellular localization of p220, we performed immunofluorescence by using normal diploid fibroblasts (Fig. (Fig.1b).1b). The majority of cells (>80%) in an asynchronous culture contained either two or four nuclear foci staining for p220, whereas the remaining cells contained one or three obvious p220 foci. This immunoreactivity was blocked by competition with antigen (Fig. (Fig.1b).1b). The variation of p220 staining patterns suggested that p220 localization might be cell cycle regulated. To test this possibility, we examined p220 localization in several asynchronous growing fibroblast lines (normal dermal fibroblasts, WI38 fibroblasts, and bjTERT fibroblasts) labeled with BrdU to mark S-phase cells. Similar results were observed, and the data for bjTERT cells are shown in Figure Figure1c,d.1c,d. The vast majority (70%) of cells lacking BrdU staining contained two p220 foci, whereas 87% of BrdU-positive cells (in green) contained four p220 foci (in red) (Fig. (Fig.11d).
Consistent with the hypothesis of cell cycle–regulated foci formation, quantitative analysis of relative 4′,6-diamidino-2-phenylindole (DAPI) signal intensity of 200 cells indicated that nuclear DNA content of normal human dermal fibroblasts with two or fewer foci was generally lower than that of cells with three or four foci (Fig. (Fig.2a).2a). Thus, the majority of cells containing three or four foci appeared to have undergone at least partial DNA replication and were in S or G2 phases. To substantiate this conclusion, we examined the status of p220 and DNA replication in cells stimulated to re-enter the cell cycle from quiescence (Fig. (Fig.2c).2c). Normal dermal fibroblasts were pulsed-labeled with BrdU before harvest to mark S-phase cells. After 72 h of growth arrest (0 h), only one out of 100 cells scored was BrdU positive, and this cell had four p220 foci. In contrast, 81% of BrdU-negative cells had two foci, 15% had one or no focus, and 3% had three or four foci. At 12 h after release, 11% of the cells had entered S phase and were BrdU positive. Among these cells, 78% contained four p220 foci and 20% had three foci. Only 2% had two foci. In contrast, 90% of the BrdU-negative cells had two foci and 4% had one focus at this time point, whereas the balance had three or four foci. A similar pattern was observed for the 18- and 24-h time points, when 66% and 80% of cells were in S phase, respectively. At 48 h after release, 35% of the cells entered the second G1 phase and became BrdU negative. Of these cells, 83% again contained only two p220 foci, and 7% had one or no focus, with the remainder containing three or four foci. We paid special attention to the small population of mitotic cells. p220 foci persisted in prophase (Fig. (Fig.3b,3b, left panel, and top cell in the right panel), but they were absent by metaphase (Fig. (Fig.3b,3b, middle panel) or telophase (Fig. (Fig.3b,3b, right panel, bottom cell). The loss of p220 foci during this short period would explain the small number of cells with one or no p220 focus (Fig. (Fig.2a).2a). Taken together, these data demonstrate that the number of p220 foci is cell cycle regulated and that S phase is accompanied by the generation of two additional p220 foci not seen in G1 or Go cells.
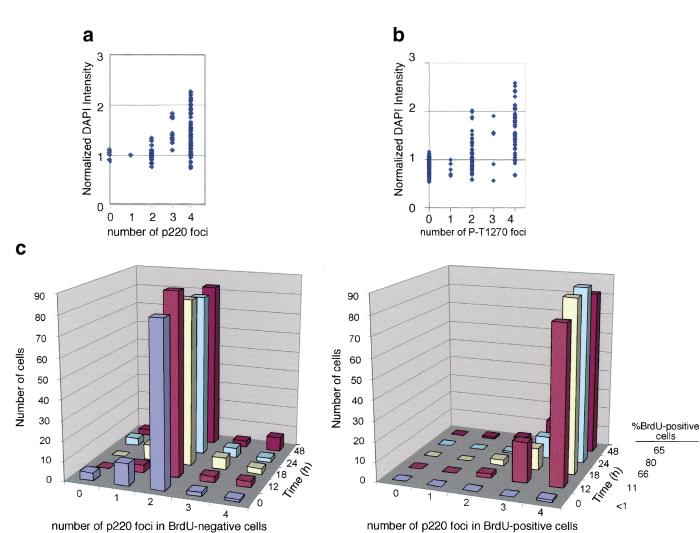
S-phase entry from quiescence is accompanied by the generation of four p220 foci in normal diploid fibroblasts. Quantification of nuclear DNA contents in human dermal fibroblasts containing different numbers of foci as detected with antibodies against p220 (a) or with an antibody against the phosphopeptide-spanning T1270 (b). Two hundred cells were analyzed for each experiment. (c) Quiescent normal dermal fibroblasts were stimulated to enter the cell cycle by serum addition. Cells were pulsed-labeled with BrdU at the indicated times before immunofluorescence to detect p220 and BrdU. The number of p220 foci was determined in 200 cells per time point, 100 each for BrdU-positive and BrdU-negative cells. (DAPI) 4′,6-diamidino-2-phenylindole.
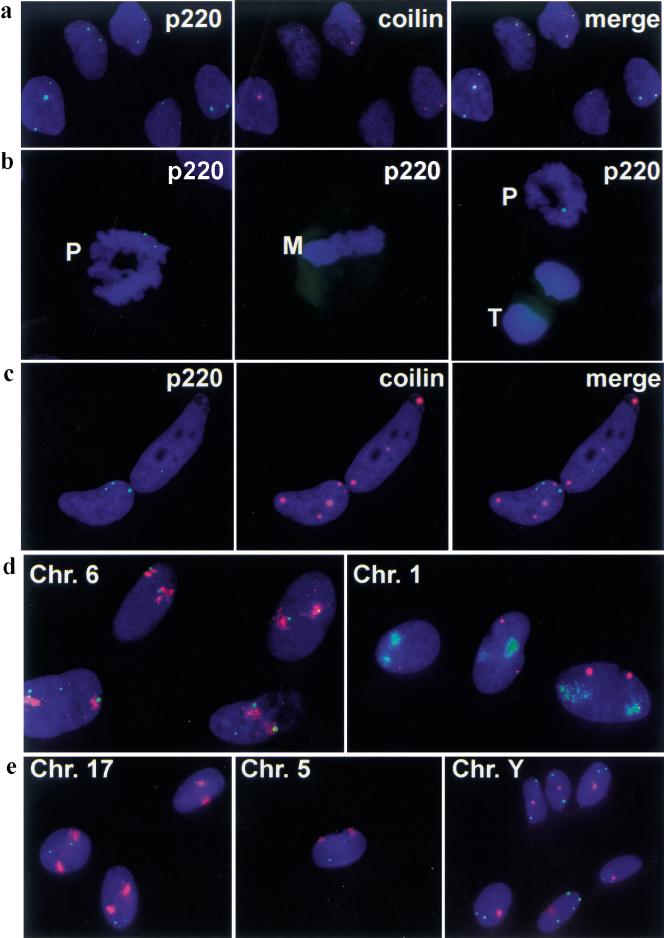
Association of p220 foci with Cajal bodies (CBs) and with domains of chromosomes 1 and 6. In all panels, 4′,6-diamidino-2-phenylindole (DAPI) stained nuclear DNA blue. (a) Normal dermal fibroblasts were subjected to immunofluorescence by using anti-p220 (green) and anti-p80coilin monoclonal antibodies (red) known to stain CBs. Co-localization is demonstrated in the merged image. (b) p220 foci are present in prophase (left; right, cell on top), but are no longer detectable in metaphase (middle) and telophase (right, cell at bottom). Prophase cells with four p220 foci were also observed (not shown). (c) HeLa cells contain CBs devoid of p220 foci. In a–c, p220 is green and DAPI is blue; in a and c, coilin is red. (d) and (e) p220 foci are associated with chromosomes 1 and 6 but not with other chromosomes. Normal dermal fibroblasts were stained for p220 and for the indicated chromosomal domains by using chromosome paints. For chromosomes 6, 17, 5, and Y, p220 is green and chromosome paint is red. For chromosome 1, p220 is red and chromosome paint green. (Chr.) Chromosome; (M) metaphase; (P) prophase; (T) telophase.
p220 foci are associated with CBs and with chromosomes 1 and 6
6
The size and number of p220 foci observed in S-phase cells are reminiscent of those displayed by CBs, as detected by antibodies against a component p80coilin. CBs are present in variable numbers in tissue culture cell lines (three to eight CBs/cell) (Frey and Matera 1995; Almeida et al. 1998). Because CBs typically are difficult to detect in nontransformed cells, we used antigen retrieval to examine whether p220 might be associated with CBs in normal human dermal fibroblasts. As shown in Figure Figure3a,3a, p220 foci coincide with coilin-containing CBs. In contrast to the colocalization of p220 and coilin throughout most of the cell cycle observed with three lines of fibroblasts (diploid dermal fibroblasts, WI38 lung fibroblasts, and bjTERT fibroblasts), transformed cells, including HeLa, Caski, SiHa, and MCF7 cells, displayed a larger and more variable number of p220 foci, ranging from three to >12 (data not shown). In HeLa cells, most if not all of the p220 foci are associated with CBs, but only a subset of CBs is associated with p220 foci (Fig. (Fig.33c).
Because CBs have previously been demonstrated to associate with histone gene clusters on chromosomes 1 and 6, it follows that one or more p220 foci may be expected to associate with these chromosomal domains. By using interphase chromosome painting in normal dermal fibroblasts, we found that chromosome 6 signals were closely associated with both p220 foci in 100% of cells containing two p220 foci. In 87% of cells containing four foci, the signals were associated with two foci, whereas the remaining 13% had more than two associated foci (Fig. (Fig.3d;3d; Table Table1).1). In contrast, chromosome 1 signals typically were not associated with p220 foci in cells containing two foci, but 93% of cells with four foci had two associated foci (Fig. (Fig.3e;3e; Table Table1).1). In our experience, false positive association occurs at a frequency of ~10% or lower. Because a subset of CBs is physically associated with an snRNA U2 gene loci at 17q21 (Frey and Matera 1995), we painted chromosome 17 as well as chromosomes 5 and Y as additional controls. These chromosomal domains displayed only rare association with p220 foci (Fig. (Fig.3e).3e). For example, out of 400 cells, two were found to have one p220 focus associated with chromosome Y. Taken together, these data indicate that p220 foci are intimately linked with chromosome 1– and chromosome 6–associated CBs and that the chromosome 6 domain is associated with p220 foci throughout the cell cycle, whereas association with chromosome 1 occurs during S phase and coincides with the increase in p220 foci from two to four. These data also imply the existence of mechanisms that restrict association of p220 with particular chromosomes to particular points in the cycle. The increased number of p220 foci observed in some transformed cells (data not shown) likely reflects at least in part an increased ploidy in chromosomes 1 and 6.
Table 1
Association of p220 foci with chromosomes 6 and 1
1
|
| No. of foci associated
| Chromosome 6 (%)
| Chromosome 1 (%)
|
|---|---|---|---|
| Cells with 2 foci | 0 | 0 | 91 |
| 1 | 0 | 9 | |
| 2 | 100 | 0 | |
| Cells with 4 foci | 0 | 0 | 0 |
| 1 | 0 | 1 | |
| 2 | 87 | 93 | |
| 3 | 11 | 6 | |
| 4 | 2 | 0 |
One hundred cells with 2 and 4 foci, respectively, were counted for association with the indicated chromosomes. With chromosomes Y, 5, and 17, association was rare; for example, 2 of 400 cells displayed one p220 foci associated with chromosome Y.
p220 is specifically phosphorylated by cyclin E/Cdk2 on sites near the cyclin E interaction domain
domain
Cyclin E and p220 co-immunoprecipitate from cell extracts, and cyclin E/Cdk2 can phosphorylate associated p220 (Zhao et al. 1998). To elucidate the role of cyclin E/Cdk2 in p220 regulation, we sought to determine the specificity of phosphorylation. Initially we examined the ability of p220 to bind to various cyclin/Cdk complexes. Flag-tagged p220 was expressed in insect cells and cell lysates used in binding assays with immobilized cyclin/Cdk complexes (Fig. (Fig.4).4). Although p220 associated efficiently with the cyclin E/Cdk2 complex (lane 16), it did not associate with the cyclin D1/Cdk4, cyclin A/Cdk1, or cyclin B/Cdk1 complex (lanes 4, 10, and 13, respectively) and bound only weakly with cyclin A/Cdk2 (lane 7). Thus, p220 displays specificity for cyclin E/Cdk2.
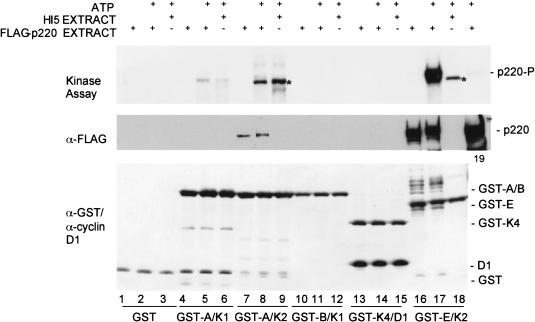
p220 preferentially associates with and is phosphorylated by cyclin E/Cdk2 in vitro. Immobilized cyclin/Cdk complexes were incubated with control insect cell lysates or insect cell lysates containing Flag-p220, as described in Materials and Methods. Complexes were washed with lysis buffer followed by 10 mM MgCl2 and 20 mM Tris-HCl. Some samples were supplemented with γ-[32P]ATP for 20 min before SDS–PAGE and visualization of proteins by immunoblotting or autoradiography. Flag-p220 was detected by anti-flag antibodies. The quantities of GST–cyclin/Cdk complexes were similar, as determined by immunoblotting with GST antibodies. Cdk2 complexes associated with an insect cell protein migrating slightly faster than p220 that was also a substrate for the kinase (indicated by an asterisk). An anti-flag immunoprecipitate of Flag-p220 (lane 19) was included as a control.
As expected, p220 was readily phosphorylated by the associated cyclin E/Cdk2 complex, and this phosphorylation was accompanied by reduced mobility of p220 (Fig. (Fig.4,4, lanes 16 and 17). Although p220 bound weakly to cyclin A/Cdk2, the associated protein underwent a similar mobility shift in the presence of ATP (lanes 7 and 8), suggesting that cyclin A/Cdk2 can also phosphorylate p220 when bound. We note that control reactions employing control insect cell lysates revealed the presence of a cyclin E/Cdk2–associated substrate (indicated by the asterisk) that migrated slightly faster than did p220 (Fig. (Fig.4,4, lanes 9 and 18). It can be distinguished from the human p220, because it was also phosphorylated to similar levels by cyclin A/Cdk2.
We next sought to determine the sites of p220 phosphorylation in vitro by using mass spectrometry (Zhang et al. 1998). p220 contains 18 potential Cdk phosphorylation sites (Thr/Ser followed by Pro). Four tryptic p220 phosphopeptides containing five phosphorylation sites were identified in recombinant p220 phosphorylated by associated cyclin E/Cdk2 in vitro (Fig. (Fig.5a,b;5a,b; Table Table2).2). Three singly phosphorylated peptides were sequenced by liquid chromatography/mass spectrometry/mass spectrometry (LC/MS/MS) to identify the phosphorylation sites as S1100 (site 3), T1270 (site 4), and T1350 (site 5), respectively (Table (Table2).2). The sequence of a doubly phosphorylated peptide encompassing residues 742–788 could not be determined because of its large size (m/z 5077.5, average mass; see Fig. Fig.5a).5a). However, this peptide contains two consensus Cdk substrates at S775 and S779 (sites 1 and 2) (Table (Table2),2), allowing a tentative assignment as sites of modification by cyclin E/Cdk2. Indeed, we found that p220 mutated in both of these serine residues was resistant to a cyclin E/Cdk2–induced shift in mobility (Fig. (Fig.5d).5d). In contrast, p220 proteins mutated in one or more of the other identified phosphorylation sites still underwent a mobility shift in response to cyclin E/Cdk2 treatment (Fig. (Fig.5d).5d). These data are consistent with the assignment of S775 and S779 as in vitro substrates and indicate that they are primarily responsible for the mobility shift observed upon phosphorylation by cyclin E/Cdk2.
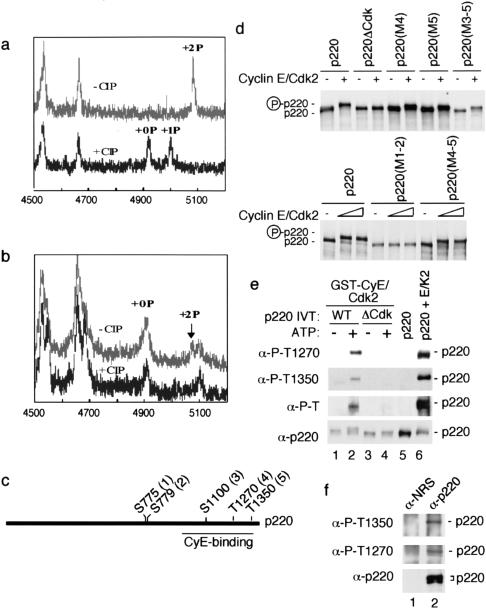
Identification of phosphorylation sites in p220. (a,b) A portion of the matrix-assisted laser desorption/ionization mass spectrometry (MALDI/TOF) mass spectra before (upper spectra) and after (lower spectra) treatment with calf intestinal phosphatase (CIP), showing the doubly phosphorylated peptide encompassing the sequence of 742–788 from recombinant (a) and endogenous (b) p220. To generate cyclin E/Cdk2–phosphorylated p220, 2 μg of Flag-p220 immobilized on anti-flag agarose was allowed to associate with 1 μg of cyclin E/Cdk2 and washed complexes incubated with 1 mM ATP (50 min at 25°C). (c) Schematic diagram of p220 phosphorylation sites and the cyclin E binding domain inferred by expression cloning (Zhao et al. 1998; Ma et al. 1999). (d) Altered mobility of p220 in response to cyclin E/Cdk2–mediated phosphorylation requires S775 and S779. In vitro translation products were incubated in the presence or absence of 20 nM cyclin E/Cdk2 for 20 min at 30°C (top) or with 20 and 50 nM cyclin E/Cdk2 for 20 min at 30°C (bottom) before electrophoresis and autoradiography. (e) Specificity of phosphopeptide-specific antibodies. In vitro–translated p220 or p220ΔCdk (50 μL) was incubated with 1 μg of GST–cyclin E/Cdk2 immobilized on GSH-Sepharose, and washed complexes were incubated in the presence or absence of 1 mM ATP (lanes 1–4). Proteins were separated by SDS–PAGE and immunoblotted. The anti-phosphoThr antibody (New England Biolabs), which reacts with a large number of different phosphoThr-Pro-containing sequences, did not recognize p220ΔCdk. As a control, Flag-p220 from insect cells (~100 ng) was incubated with or without 50 nM cyclin E/Cdk2 and 1 mM ATP before immunoblotting. (WT) Wild type. (f) Anti-p220 immune complexes from 293T cells were separated by SDS–PAGE and immunoblotted with the indicated antibodies. (NRS) Normal rabbit serum.
Table 2
Mass spectral identification of p220 phosphorylation sites
sites
| Sites
| Peptides
| Molecular mass (measured/calculated)
| No. of PO3 group
| |||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| In vitro | ||||||||||||||||||||||||||||||||||||||||||||||||||
 S775,S779 S775,S779 | 742 | V | I | I | S | D | D | P | F | V | S | S | D | T | E | L | T | S | A | V | S | S | I | N | G | E | N | L | P | T | I | I | L | S | S | P | T | K | S | P | T | K | N | A | E | L | V | K788 | 4918/4916.5 | 2 |
 S1100 S1100 | 1091 | N | A | V | S | F | P | N | L | D | S | P | N | V | S | S | T | L | K | P | P | S | N | N | A | I | K | 1116 | 2712/2713.0 | 1 | ||||||||||||||||||||
 T1270 T1270 | 1259 | L | A | D | S | S | D | L | P | V | P | R | T | P | G | S | G | A | G | E | K | 1278 | 1708/1707.9 | 1 | ||||||||||||||||||||||||||
 T1350 T1350 | 1346 | T | T | S | A | T | P | L | K | D | N | T | Q | Q | F | R | 1360 | 1954/1954.1 | 1 | |||||||||||||||||||||||||||||||
| In vivo | ||||||||||||||||||||||||||||||||||||||||||||||||||
 S775,S779 S775,S779 | 742 | V | I | I | S | D | D | P | F | V | S | S | D | T | E | L | T | S | A | V | S | S | I | N | G | E | N | L | P | T | I | I | L | S | S | P | T | K | S | P | T | K | N | A | E | L | V | K788 | 4918/4916.5 | 2 |
Average molecular masses of dephosphorylated peptides are shown.
To examine phosphorylation in vivo, we purified p220 from a 293T cell nuclear lysate by using immunoprecipitation (Fig. (Fig.1a)1a) and subjected it to mass spectral analysis. p220 is present at low levels in this cell line, and from 44 mg of nuclear extract, we obtained 200 ng of p220. We were able to identify one peptide with the mass expected for a doubly phosphorylated peptide spanning Val742–Lys788 whose quantity was consistent with the level of protein available for analysis (Fig. (Fig.5b;5b; Table Table2).2). Importantly, this peptide is absent from spectra after treatment with a phosphatase (Fig. (Fig.5b),5b), indicating that S775 and S779 are phosphorylated in vivo. Signals for the three singly phosphorylated peptides observed in vitro were not evident, possibly because of the small amounts of material available for analysis.
p220 phosphopeptide antibodies recognize phosphorylated p220 in vitro and in vivo
vivo
To investigate T1270 and T1350 phosphorylation in vivo, we generated phospho-specific antibodies against these sites. These antibodies recognize Flag-p220 purified from insect cells only after phosphorylation with cyclin E/Cdk2 (Fig. (Fig.5e,5e, lanes 5 and 6). To examine the specificity of these antibodies for reaction with p220, we bound either wild-type p220 or p220ΔCdk (containing Ala substitutions at S775, S779, S1100, T1270, and T1350) to immobilized cyclin E/Cdk2 and incubated these complexes in the presence or absence of ATP (Fig. (Fig.5e).5e). Both antibodies recognized phosphorylated p220 (lane 2) but did not recognize p220ΔCdk. The two proteins were present at comparable levels based on anti-p220 immunoblotting (lanes 3 and 4). The low levels of reactivity toward the in vitro–translated p220 in the absence of added ATP likely reflect phosphorylation that occurred during immunoprecipitation from ATP-containing reticulocyte lysates, because larger amounts of recombinant p220 purified from insect cells did not react with the phosphopeptide-specific antibodies in the absence of phosphorylation by cyclin E/Cdk2 (lanes 5 and 6). We also note that a general phosphothreonine-proline antibody gave similar results, indicating that other TP sequences in p220ΔCdk are not phosphorylated by bound cyclin E/Cdk2 in vitro. We found that both phosphopeptide antibodies reacted with p220 immunoprecipitated from cycling 293T cells (Fig. (Fig.5f),5f), indicating that these sites are indeed phosphorylated in vivo. On the basis of the comparison with p220 in 293T cells examined in parallel (Fig. (Fig.5f),5f), the more slowly migrating p220 protein was preferentially detected by the phosphopeptide antibodies.
CB-associated p220 co-localizes with cyclin E and is phosphorylated on Cdk sites in a cell cycle–dependent manner
manner
The data described thus far suggest that p220 is targeted to CBs and is a substrate of cyclin E/Cdk2. However, it is unclear whether p220 is phosphorylated while in CBs. To examine whether CB-associated p220 is phosphorylated on Cdk2 sites, we performed immunofluorescence by using antibodies against phospho-T1270 and phospho-T1350. Both recognized nuclear foci similar to the antibody against p220 (Fig. (Fig.6a;6a; data not shown). To demonstrate that the foci coincided with those found with anti-p220, we performed double immunofluorescence staining with antibodies against phospho-T1270 and coilin in cycling fibroblasts (Fig. (Fig.6).6). From 500 cells scored, 30.2% displayed primarily two foci reactive toward both antibodies, whereas 40.4% had primarily four co-localized foci (Fig. (Fig.6a),6a), including cells in prophase (Fig. (Fig.6b).6b). The remaining 29.4% of the cells lacked staining with the phospho-T1270 antibody. This is in marked contrast to staining with p220 antibodies in which cells lacking antibody reactivity were very rare except for those in metaphase and telophase (Figs. (Figs.2a2a and and3b).3b). In cells that were negative for reactivity with anti-phospho-T1270, coilin reactivity was still observed (Fig. (Fig.6a).6a). Quantification of relative DAPI intensity of 200 additional cells demonstrated that cells nonreactive with the phosphopeptide antibodies had a lower DNA content than did reactive cells, which is consistent with these cells being in the G1 phase, whereas phospho-T1270 antibody-positive cells had a higher DNA content consistent with S- or G2-phase cells (Fig. (Fig.2b).2b). In this separate experiment, a somewhat higher percentage of cells had no detectable focus (data not shown). In agreement with the results with p220 antibodies, the few cells in metaphase and telophase were negative for staining with the phosphopeptide antibody, whereas coilin signals appeared dispersed (Fig. (Fig.6d,e).6d,e).
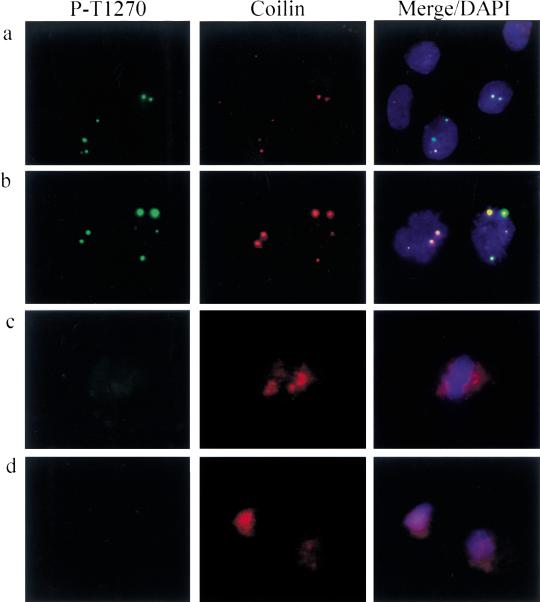
p220 in Cajal bodies (CBs) is phosphorylated on Cdk sites in a cell cycle–specific manner. Dual detection with anti-phospho-T1270 antibodies and p80coilin in dermal fibroblasts is shown. Focal co-localization was observed in S phase (a) and prophase (b), but antiphospho-T1270 did not detect any foci in metaphase (c) or telophase (d). a also contains three cells that display anti-coilin reactive foci but not anti-phospho-T1270 reactive foci. The DNA content of these cells is consistent with G1 phase (Fig. (Fig.2b).2b). Only diffused coilin signals were observed in c and d. (Green) anti-phospho-T1270; (red) anti-coilin; (blue) 4′,6-diamidino-2-phenylindole (DAPI).
If cyclin E is responsible for phosphorylation of p220 within CBs, one would predict that p220 would co-localize with cyclin E and that the timing of p220 phosphorylation would be coincident with co-localization. To examine this issue directly, we initially performed co-localization experiments using anti–cyclin E and anti-p220 antibodies. As shown in Figure Figure7a,7a, cyclin E was concentrated in foci that are coincident with p220 foci. At longer exposure, cyclin E was evident as faint dust throughout the nucleus as well. These results are consistent with the recent report that cyclin E is concentrated in CBs in S phase (Lui et al. 2000).
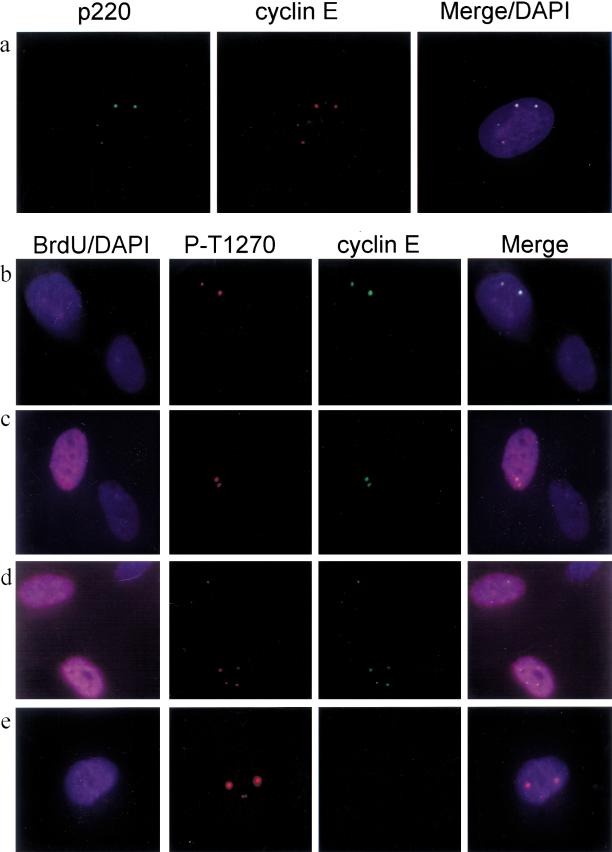
Co-localization of cyclin E with anti-phospho-T1270 reactive foci in a cell cycle–dependent manner. (a) Cyclin E (red) co-localizes with p220 (green) in a cell containing four p220 foci. Nuclei are in blue. (b–e) Growing diploid fibroblasts were labeled with BrdU for 1 h and subjected to immunofluorescence using anti-phospho-T1270 (red), anti-cyclin E (green), anti-BrdU (magenta), and 4′,6-diamidino-2-phenylindole (DAPI; blue) to visualize nuclei. (b) A BrdU-negative cell containing two phospho-T1270 foci that co-localize with cyclin E. This field also contains a BrdU-negative cell that is negative for both anti–cyclin E and anti-phospho-T1270 antibodies. (c) A BrdU-positive cell containing two phospho-T1270 foci that co-localize with cyclin E adjacent to a BrdU-negative cell that is negative for both anti–cyclin E and anti-phospho-T1270 antibodies. (d) BrdU-positive cells containing two or four phospho-T1270 foci that co-localize with cyclin E. (e) A BrdU-negative cell containing four phospho-T1270 foci that lacks cyclin E staining.
To examine whether phosphorylation of p220 correlates with co-localization with cyclin E, we pulse-labeled asynchronous diploid fibroblasts with BrdU and determined the presence of phospho-T1270, cyclin E, BrdU, and DAPI-stained nuclei (Fig. (Fig.7b–e).7b–e). Among cells containing two phospho-T1270 foci, both BrdU-positive and BrdU-negative cells were observed; in both cases, however, these foci contained cyclin E (Fig. (Fig.7b–d).7b–d). In contrast, among cells containing four anti-phospho-T1270 foci, those that were BrdU positive most frequently displayed co-localized cyclin E, whereas those that were BrdU negative typically lacked cyclin E staining (Fig. (Fig.7d,e).7d,e). Given the data presented previously, we believe the latter class of cells to be G2 cells that have lost cyclin E expression but maintain p220 in a phosphorylated form.
The in situ results with antibodies to p220, phospho-T1270, phospho-T1350, coilin, and cyclin E (Figs. (Figs.11–3, ,6;6; Liu et al. 2000) indicate the following: (1) p220 is an in vivo Cdk2 substrate and can be phosphorylated on cyclin E/Cdk2 sites while present in CBs. There is a tight correlation between the appearance of cyclin E in foci and the occurrence of p220 phosphorylation such that (2) cells that are nonreactive with antibody to phospho-T1270 are in the early G1 phase before cyclin E/Cdk2 is present to phosphorylate p220 in CBs. Consistent with this, cells that lacked anti-phospho-T1270 foci also lacked detectable cyclin E. (3) Cells containing two anti-phospho-T1270 reactive foci that co-localize with coilin and cyclin E are in late G1 or early S phases when cyclin E/Cdk2 levels peak. (4) Cells that have four co-localized foci are well into S phase, and p220 remains phosphorylated (Figs. (Figs.11 and and2).2). Although the pattern of p220 phosphorylation persists into prophase, cyclin E staining is lost at some point in late S phase or G2 phase, as exemplified by the presence of BrdU-negative cells containing four phospho-p220 foci but lacking cyclin E co-localization. (5) Around the time of metaphase and later, p220 foci are not detected with either p220 or phospho-T1270 antibodies.
Mutation of cyclin E/Cdk2 sites in p220 reduces p220-mediated histone H2B promoter activation
activation
Our data suggest that cyclin E/Cdk2 may regulate p220 during the G1/S-phase transition. To examine this question, we took advantage of the recent finding that p220 expression in tissue culture cells leads to increased expression of histone 2B (H2B) promoter– and histone 4 (H4) promoter–luciferase reporter constructs, independent of its effects on S-phase acceleration (Zhao et al. 2000). Histone gene expression is complex, involving both message stabilization (approximately sevenfold) and transcriptional activation (approximtely fivefold), which together account for an ~35-fold increase in histone mRNA levels during S phase (Harris et al. 1991; Heintz 1991). But the process by which the histone transcriptional apparatus senses cell cycle position is unknown.
We compared the ability of a vector expressing p220 (pCMV-p220) or p220ΔCdk (pCMV-p220ΔCdk) to activate luciferase expression from an H2B (−200/0) promoter–luciferase reporter plasmid in transiently transfected 293T cells (Fig. (Fig.8a).8a). The level of induction by p220 relative to control transfections ranged from two- to 10-fold, depending on the quantity of p220 plasmid used and the level of p220 expression achieved, as determined by immunoblotting (Fig. (Fig.8b).8b). In contrast, p220ΔCdk displayed a substantially reduced ability to activate the H2B reporter construct when expressed at comparable levels (Fig. (Fig.8a–e).8a–e). Similar results were obtained with a minimal H2B promoter (−127/−27) (data not shown). At low levels of expression, p220ΔCdk displayed levels of H2B reporter activation comparable to control transfected cells (Fig. (Fig.8a,b);8a,b); however, at higher levels of expression, a twofold increase in reporter activity over control transfected cells was typically observed (Fig. (Fig.8c,d).8c,d). Transfected cells displayed p220 foci as well as diffuse signals throughout the nucleoplasm because of elevated levels of expression from the transfected plasmids (Fig. (Fig.8e).8e). Taken together, these results suggest that phosphorylation at one or more cyclin E/Cdk2 sites contributes substantially to this aspect of p220 function. However, it is possible that elevated levels of p220 can partially bypass a requirement for phosphorylation at these sites. In these experiments, p220 appeared to be primarily in a more slowly migrating phosphorylated form, while p220ΔCdk remained in a more rapidly migrating form (Fig. (Fig.8b,d).8b,d). Thus, it appeared that sufficient cyclin E/Cdk2 existed in these cells to phosphorylate fully the transiently expressed p220. This may explain why co-expression of cyclin E/Cdk2 had little effect on the levels of H2B reporter activity in 293T cells (data not shown).
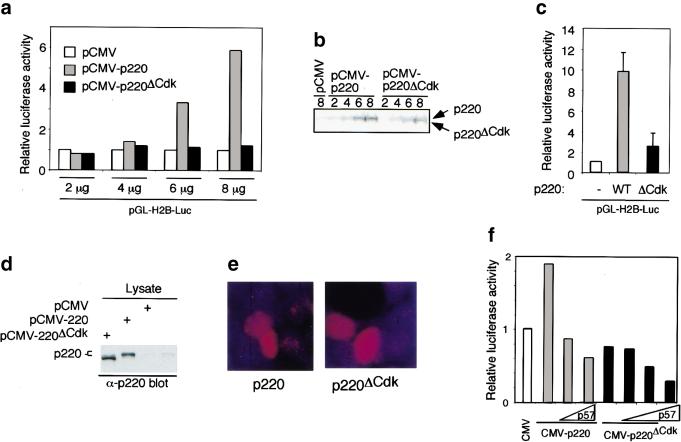
Cyclin E/Cdk2–mediated phosphorylation of p220 is important for optimal p220-induced histone H2B transcriptional activation. (a) pCMV, pCMV-p220, or pCMV-p220ΔCdk was transfected into 293T cells (3.5-cm dish), along with 50 ng of pCMV-β-galactosidase and 50 ng of pGL-H2B-luciferase reporter plasmid, and subsequently processes for luciferase and β-galactosidase activity as described in Materials and Methods. Luciferase activities were normalized relative to β-galactosidase activities. (b) A portion of extracts used in a was immunoblotted using anti-p220 antibodies. The wild-type p220 protein comigrates with the endogenous protein found at low levels, whereas p220ΔCdk migrates slightly faster as a result of the absence of phosphorylation. (c) The results of two independent experiments each performed using triplicate independent calcium phosphate precipitates derived from 10 μg of pCMV, pCMV-p220, or pCMV-p220ΔCdk are shown. (−) Lacking p220; (WT) wild type. (d) and (e) Expression levels for p220 and p220ΔCdk for experiments shown in c were similar as determined by immunoblotting (d) or immunofluorescence (e). (f) Expression of p57KIP2 blocks activation of the H2B promoter by p220 overexpression. 293T cells were transfected with limiting amounts of pCMV-p220 or p200ΔCdk expressing plasmids (5 μg) in the presence or absence of either 1 or 2 μg of pCMV-p57KIP2. At this level of expression, p220 leads to a twofold increase in reporter activity. p57KIP2 expression leads to a dramatic reduction in the level of p220-induced reporter activity in the presence or absence of p220 expression. The averages of duplicate independent transfections are shown.
Consistent with a role for cyclin E/Cdk2–mediated phosphorylation in p220 function, we found that co-expression of p57KIP2, which can inhibit Cdk2 activity and block cells at the G1/S-phase transition (Matsuoka et al. 1995), led to a reduction in the ability of p220 to activate H2B promoter activity in transiently transfected cells (Fig. (Fig.8f).8f). In this experiment, the levels of p220 expression plasmid used were such that a twofold increase in H2B promoter activity was observed, but p57KIP2 reduced luciferase levels below that obtained with control transfected cells. As expected, expression of p57KIP2 alone also reduced the levels of promoter activity in the absence of p220 expression (Fig. (Fig.8f).8f). This repression likely reflects the fact that cells are blocked in G1 with low cyclin E/Cdk2 activity. Immunoblotting demonstrated comparable levels of expression of p220 and p220ΔCdk and verified the expression of p57KIP2 (data not shown).
S-phase entry in quiescent fibroblasts by cyclin E/Cdk2 expression is associated with the accumulation of four p220 foci
foci
Expression of cyclin E/Cdk in quiescent fibroblasts leads to S-phase entry (Connell-Crowley et al. 1998; Leone et al. 1998). Because the appearance of four p220 foci is associated with S phase in asynchronous and serum-stimulated cells (Figs. (Figs.11 and and2),2), we wondered whether S-phase entry by an alternative mechanism would also lead to the appearance of four p220 foci. To this end, quiescent WI38 cells were stimulated to enter the cell cycle by infection with adenoviruses (Ad) expressing cyclin E and Cdk2. At 24 h after infection, cells were pulse-labeled with BrdU for 1 h before analysis of p220 by immunofluorescence. BrdU-negative cells in control cultures maintained in low serum contained predominantly two p220 foci (Fig. (Fig.9a,b).9a,b). In contrast, a large fraction of BrdU-positive cells in the cyclin E/Cdk2–treated culture contained four foci, whereas BrdU-negative cells in the culture contained predominantly two foci. These results suggest that cyclin E/Cdk2 can function upstream of the pathway responsible for the establishment of four p220 foci during S phase.
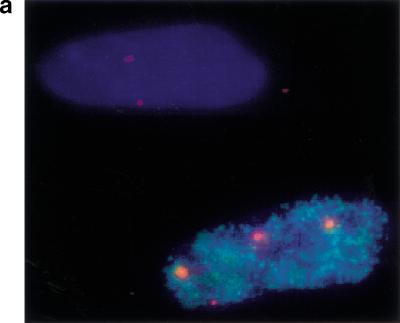
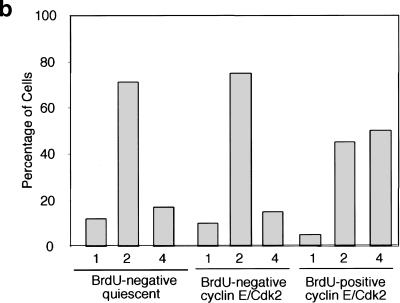
S-phase entry in quiescent fibroblasts by cyclin E/Cdk2 expression is associated with the appearance of four p220 foci. WI38 fibroblasts were subjected to serum deprivation for 72 h before infection with Ad-cyclin E/Cdk2 and maintained in 0.1% serum. Twenty-four h after infection, cells were pulse-labeled with BrdU for 1 h before analysis of p220 staining by immunofluorescence. (a) An example of an S-phase cell from a cyclin E/Cdk2 infection containing four p220 foci adjacent to a non-S-phase cell containing two p220 foci. (b) Quantitation of p220 foci. Thirty to 100 nuclei of each class were counted.
Discussion
Cell cycle transitions are driven in part by transcriptional programs that generate proteins needed for subsequent processes. Although it is clear that these transcriptional programs are ultimately linked to the basic cell cycle machinery, how this linkage is accomplished is largely unknown. The best understood connection between transcription and the basic cell cycle machinery in mammalian cells is the activation of E2F by Cdk-mediated phosphorylation of Rb family members (Dyson 1998; Nevins 1998). In this article, we provide evidence, at the cellular and molecular level, that cyclin E/Cdk2 directly regulates the activity of p220NPAT, which in turn controls S-phase-specific activation of histone gene transcription. Thus, cyclin E/Cdk2 not only regulates the production of DNA synthesis machinery through E2F but also supports the production of nucleosome components required for completion of DNA replication.
In normal fibroblasts, p220 is localized to discrete foci in the nucleus, and the number of these foci change during the cell cycle (Zhao et al. 2000; this work). Cells in G1 contain primarily two p220 foci, whereas cells in S phase contain four foci. Anti-p220-reactive foci are only absent during the short span of metaphase and telophase (Figs. (Figs.2,2, ,3,3, and and6).6). These p220 foci coincide with small nuclear organelles, the CBs (Fig. (Fig.3).3). CBs contain a bewildering number of transcriptional and splicing/polyadenylation proteins, and recent studies have led to the hypothesis that CBs function as sites of assembly of pol II transcriptosomes, complexes of pol II transcription factors and splicing machinery (Gall et al. 1999). CBs also contain specialized proteins such as histone stem–loop binding proteins that function in the processing of histone 3′ ends (Abbott et al. 1999). In addition, cyclin E/Cdk2 is present in CBs during S phase (Liu et al. 2000). Although it was suggested that this localization of cyclin E/Cdk2 reflects the presence of the Cdk-activating kinase cyclin H/Cdk7 in coiled bodies, our results suggest that cyclin E/Cdk2 plays an S-phase-promoting role within CBs, at least in part by phosphorylating p220.
An interesting aspect of CBs is their physical attachment to discrete gene loci. In HeLa cells, a subset of CBs co-localize with replication-dependent histone gene clusters on chromosomes 1 and 6 (1q21 and 6p21, respectively) and with snRNA genes located elsewhere in the genome (Frey and Matera 1995). Our results indicate that p220-containing CBs associate with the chromosome 6 domain during G1 and S/G2 and additionally with the chromosome 1 domain during S phase and G2, but we found little evidence for association with the domains of chromosomes 5, 17, or Y (Fig. (Fig.3).3). Typically, the p220 foci linked with chromosome 6 appeared to be larger than those associated with chromosome 1 (Fig. (Fig.3),3), potentially reflecting the larger numbers of histone genes located in the chromosome 6 cluster. The cell cycle–dependent association of p220 with CBs on chromosomes 6 and 1 then accounts for the oscillation in p220 foci numbers. Zhao et al. (2000) have also demonstrated that p220 is localized with chromosomes 1 and 6 and have shown that p220 is physically linked to histone gene loci.
On a biochemical level, p220 preferentially binds to cyclin E/Cdk2 over other Cdk complexes and is phosphorylated by cyclin E/Cdk2 in vitro and in vivo (Figs. (Figs.44 and and5).5). Indeed p220 foci also contain cyclin E (Fig. (Fig.7).7). Moreover, antibodies specific to p220 (Figs. (Figs.11–3) and to specific phosphopeptides of p220 (Figs. (Figs.66 and and7)7) demonstrated that p220 is present in CBs in both Cdk2 phosphorylated form and unphosphoryated forms and that these two forms alternate during the cell cycle. Three major classes of staining patterns were observed with phospho-specific antibodies against p220. Thirty percent of cells in an asynchronous population lacked phospho-T1270 antibody reactivity and displayed predominantly a G1 DNA content while maintaining detectable CBs. Because the vast majority of G1 cells contain two p220 foci co-localized with CBs (Figs. (Figs.11 and and2),2), we conclude that p220 in a distinct population of G1 cells is not phosphorylated on Cdk2 sites. This population of cells also lacked cyclin E staining, which is consistent with these cells being in early G1. Cells in the second class (30%) contain two phospho-T1270 antibody reactive foci that are co-localized with cyclin E. Our analysis indicates that these cells are in either late G1 or early S phase and suggests that chromosome 6–associated p220 can be phosphorylated on Cdk2 sites in advance of the formation of four obvious foci. In asynchronous cells, a small fraction of BrdU-positive cells have two clear p220 foci (Fig. (Fig.1c),1c), suggesting that S phase can be initiated before the accumulation of p220 in chromosome 1–associated CBs. This idea is substantiated by the finding that some cells containing two anti-phospho-T1270 and anti–cyclin E reactive foci display partial replication, as determined by quantitation of DNA content or BrdU incorporation (Figs. (Figs.2b2b and and7).7). However, we cannot rule out the possibility that p220 is already present in chromosome 1–associated CBs but is present at levels below detection. Cells in a third class (40%) each contained four phospho-antibody reactive foci, and a substantial fraction of these cells contained co-localized cyclin E. These cells have a higher DNA content, consistent with p220 being phosphorylated in S and G2. p220 phosphorylation is maintained in prophase, with some cells displaying two foci and some displaying four foci, but p220 foci are absent in metaphase and telophase. Thus, p220 foci appear to be lost sequentially during the prophase-to-metaphase transition. The fate of p220 during mitosis is unclear at present. It could be dispersed and therefore beyond our means to detect at this stage in the cycle. Alternatively, p220 could be degraded. Consistent with the latter possibility is the finding that 293T cell extracts from mitotic cells (obtained by mitotic shake-off) have no detectable p220 by immunoprecipitation/immunoblotting analysis (data not shown). Thus, if p220 is degraded in mitosis, new p220 must be synthesized early in G1 phase and be incorporated into CBs in the unphosphorylated form before the activation of cyclin E/Cdk2 at late G1 and early S phase. In HeLa cells released from mitosis, p220 foci and coilin foci reappear 2–3 h after release, consistent with the formation of foci in early G1 phase (data not shown).
The link between p220, a cyclin E/Cdk2 interacting protein (Zhao et al. 1998; Ma et al. 1999), and the transcription of histone 2B and histone 4 genes (Zhao et al. 2000) led us to address whether cyclin E/Cdk2 directly regulates this aspect of p220 function. In principle, cyclin E/Cdk2 could function to relay cell cycle positional information to p220, thereby playing a role in controlling the timing of S-phase-specific histone gene transcription. We found that p220 lacking five Cdk2 phosphorylation sites, four of which were phosphorylated in vivo, displayed a reduced ability to activate transcription from an H2B reporter construct (Fig. (Fig.8),8), consistent with a role for cyclin E/Cdk2 in p220 activation and H2B transcription. p220-dependent activation of the H2B promoter requires the Oct-1 element (data not shown) known to be involved in S-phase-specific induction of H2B expression (Fletcher et al. 1987; Segil et al. 1991). Using a U2OS-based tissue culture system, Zhao et al. (2000) found that the ability of p220 to activate H4 transcription was stimulated by co-expression of cyclin E, again pointing to a role for cyclin E in this process. Although we did not observe a stimulatory effect of cyclin E/Cdk2 co-expression in our system, we found that the vast majority of ectopic p220 in 293T cells is in the slower mobility phosphorylated form (Fig. (Fig.8),8), suggesting that cyclin E is not a limiting component in these cells at the levels produced with our p220 expression plasmid. Apparently, at the levels of expression achieved in U2OS cells, cyclin E/Cdk2 is limiting. Regardless of these differences, both studies indicate that the cyclin E/Cdk2–mediated, cell cycle–dependent activation of p220 is an important component of the S-phase-specific histone transcriptional program.
Although cyclin E/Cdk2 activation seems to be central to p220 function, several issues remain to be addressed. First, what is the significance of p220 localization in CBs? Although p220 is clearly localized in these organelles and these organelles are physically linked to target genes, it is conceivable that the localization of p220 reflects its accumulation before release from CBs in a form that then activates histone gene transcription. Many transcription factors accumulate in inactive pools and are not present in detectable levels at target gene loci. Nevertheless, the co-localization of p220 with cyclin E, CBs, and histone gene clusters, the cell cycle–dependent phosphorylation of p220, the persistent association of phosphorylated p220 with CBs throughout S phase when histone genes are transcribed, as well as the activation of histone gene transcription by Cdk2-dependent p220 phosphorylation are most striking and strongly point to a functional role for p220 and cyclin E localization in CBs, as summarized in Figure Figure10.10. Second, what is the basis of the appearance of p220 foci on chromosome 1 during S phase, and how is this regulated? Presumably, the association of p220 foci with chromosome 1 reflects a role in S-phase-dependent histone transcription, but why then do p220 foci exist on chromosome 6 throughout most of the cell cycle? In this regard, it is important to determine whether transcription from endogenous histone genes is linked to accumulation of p220 at histone gene clusters. Third, the histone gene cluster on chromosome 6 contains >50 copies of the four classes of core histones as well as the linker histone H1 (Ahn and Gruen 1999). Does p220 coordinately regulate the transcription of all classes of histone genes in the locus, and if so, how is this achieved? One possibility is that p220 could generate a chromosomal context that allows transcriptional activation of the whole region during S phase. If this is the case, p220 might also regulate the expression of nearby genes during S phase. Analysis of the 6p21 region reveals a large number of nonhistone genes, and it is possible that one or more of these genes are under the control of a p220-dependent S-phase transcriptional program. Alternately, p220 might function within the CB to assemble histone-specific transcription complexes, in keeping with the proposed function of CBs (Gall et al. 1999). Fourth, although p220 overexpression increases the S-phase population in transiently transfected cells (Zhao et al. 1998), it remains to be determined whether this activity is related to histone transcription and whether the phosphorylation events that we have identified are relevant to this activity. Finally, the finding that cyclin E is concentrated in CBs suggests the possibility that these organelles are important control centers linking the cell cycle machinery to S-phase-specific processes. It will therefore be important to determine whether other relevant cyclin E substrates gain access to the kinase through localization in CBs.
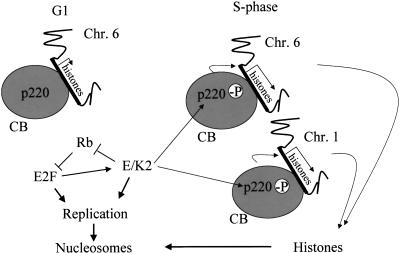
Summary and proposed model of cell cycle–dependent transcriptional activation of histone genes on chromosomes 1 and 6 in association with Cajal bodies (CBs) as mediated by cyclin E/Cdk2 phosphorylation of p220. p220 foci are no longer detectable by metaphase and telophase but reappear in G1 phase. However, phosphorylation of p220 in CBs does not occur until cyclin E/Cdk2 is activated during late G1 phase/S phase.
Materials and methods
Cell culture
Normal diploid fibroblasts (dermal fibroblasts, WI38, and bjTERT), 293T, and HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). To generate quiescent fibroblasts, cells were plated at ~50% confluence before culture for 72 h without serum. Cells were released into DMEM containing 10% FBS for various lengths of time. In some experiments, the cells were exposed to BrdU (10–50 μg/mL) for 1 h before harvest to reveal cells in S phase. To examine cell cycle entry via cyclin E/Cdk2 expression, WI38 cells were maintained for 72 h in 0.1% FBS and infected with adenoviruses expressing cyclin E and Cdk2 (generously provided by J. Nevins, Duke University, Durham, NC) (Leone et al. 1998) at a multiplicity of infection of 100.
Antibodies and immunofluorescence assays
assays
Bacterial GST-p220 (residues 1054–1397) was used to generate antibodies in rabbits. Antibodies were depleted of reactivity to the GST protein and affinity-purified using immobilized GST-p220. Anti-coilin monoclonal antibodies (π-isotype) were provided by M. Carmo-Fonseca (University of Lisbon, Portugal; Almeida et al. 1998). Anti-cyclin E (HE12) came from Pharmingen. Antibodies against Thr-1270 (Asp-Leu-Pro-Val-Pro-Arg-phosphoThr-Pro-Gly-Ser-Gly-Ala-Gly-Cys) and Thr-1350 (Ser-Arg-Thr-Thr-Ser-Ala-phosphoThr-Pro-Leu-Lys-Asp-Asn-Thr-Cys) were generated in rabbits after coupling to keyhole limpet hemocyanin. For immunofluorescence, cells were fixed in either ethanol or formalin and permeabilized with 0.1% Triton X-100. Detection of rabbit antibodies was accomplished using secondary antibodies labeled with Cy3 or Alexa 488 (Molecular Probes, Eugene, OR) or with anti-rabbit horseradish peroxidase (HRP) (Roche, Indianapolis, IN) and tyramides (NEN, Boston, MA) (see below). BrdU was detected using fluorescein isothiocyanate (FITC)–conjugated anti-BrdU antibody. Nuclear DNA was revealed with DAPI staining. Microscopic analysis was performed using either an Olympus BX-60 fitted with an Optronics CCD camera (Baylor College of Medicine) or an AX-70 Olympus microscope and an Olymix digital camera (University of Alabama). Images were captured with a 100× objective lens by using either multiband pass filters or single pass filters with merging using Adobe Photoshop.
Correlation of p220 foci or phospho-T1270 foci with DNA content
content
Asynchronous human dermal fibroblasts were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100 in phosphate-buffered saline (PBS), and then treated with 3% hydrogen peroxide. Slides were then incubated with a 1:100 dilution of anti-p220 or anti-phosphopeptide for 2 h at 37°C and washed three times in PBS containing 0.1% Tween 20. After incubation with anti-rabbit HRP (1:100), cells were washed three times, and the signals were developed using a 1:100 dilution of cyanine-3-tyramide (NEN). To detect BrdU, the cells were fixed once more with 4% paraformaldehyde, treated with 3 N HCl for 15 min, incubated with an anti-BrdU FITC antibody for 2 h at 37°C, DAPI (Sigma) stained, and mounted with antifade.
Relative nuclear DNA content was determined as follows: After staining with various antibodies, we took images of DAPI and of the p220 or phosphopeptide foci separately, but all images came from the same slide to avoid any variability in DAPI staining. Using Image Pro Plus software (Media Cybernetics, Silver Spring, MD), we identified nuclei to be measured and determined the average DAPI density individually. The average background for each image was then determined and subtracted, giving the corrected average nuclear DAPI density for each cell. Cells were then individually correlated to the foci number. For graphic representation (Fig. (Fig.2a,b),2a,b), the average DAPI density of all two-foci nuclei (the majority of which were BrdU negative; see Figs. Figs.11 and and2c)2c) was used to divide the density of each individual cell. This normalization procedure yielded a number between 1 (G1) and 2 (G2) and somewhere in between (S).
Interphase chromosome painting
Asynchronous primary human dermal fibroblasts were fixed and stained with antibodies to p220, and signals were developed with tyramide as described above. Slides were fixed again with 4% paraformaldehyde and treated with RNase A (100 μg/mL) in 2 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate) for 1 h at 37°C. They were then ethanol-dehydrated and denatured in 70% formamide, 2 × SSC, pH 7.0, for 2 min at 72°C, dehydrated in ethanol, and hybridized individually with chromosome paints overnight at 37°C. The chromosome 1 and chromosome Y paints were a direct FITC conjugate or cyanine 3 conjugate, respectively, from Vysis (Downers Grove, IL). Signals were detected according to the manufacturer's protocol. Paints for chromosome 5, 6, or 17 were digoxigenin probes from Oncor (Gaithersburg, MD). They were detected using an anti-dig Texas Red antibody. The slides were DAPI stained before imaging.
Immunoprecipitation and phosphorylation
For nuclear extracts, 293T cells were lysed in 50 mM Tris-HCl, pH 7.5, containing 1 mM EDTA, 100 mM NaCl, 0.3% Nonidet P-40, and protease/phosphatase inhibitors, and nuclei pelleted by centrifugation. Nuclear proteins were solubilized using RIPA buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 0.5% deoxycholate, 1% Nonidet P-40, 0.1% SDS, and protease/phosphatase inhibitors) before centrifugation. Extracts were precleared with protein A–bound normal rabbit IgG before incubation with affinity-purified anti-p220 antibodies bound to protein A–Sepharose. Beads were washed with RIPA buffer. Proteins were separated by SDS–PAGE before staining with Coomassie Brilliant Blue R 250 or transferred to nitrocellulose filters for immunoblotting with anti-p220 antibodies. Detection was accomplished using enhanced chemiluminescence (Amersham).
For expression of p220 in insect cells, the coding sequence (Imai et al. 1996) was cloned into pUNI-50, and in vitro recombination was then used to generate a pVL1392-based plasmid with p220 fused at its N terminus to the Flag tag as described (Liu et al. 1998). Viruses were made using Baculogold (Pharmingen). To examine interaction with Cdks, cyclin/Cdk complexes were purified with glutathione-Sepharose beads (Ma et al. 1999). Immobilized complexes were then incubated with insect cell extracts with or without Flag-p220. Complexes were washed with lysis buffer and with 20 mM Tris-HCL, pH 7.5, and 10 mM MgCl2. A portion of each mixture was used for kinase assays employing 50 μM γ-[32P]ATP. Samples were separated by SDS–PAGE and transferred to nitrocellulose before immunoblotting and autoradiography.
Matrix-assisted laser desorption/ionization mass spectrometry (MALDI/TOF) with delayed extraction (Voyager-DE, Perseptive Biosystems, Framingham, MA) was used for the identification of phosphopeptides, as described by Zhang et al. (1998). An electrospray ion trap mass spectrometer (LCQ, Finnigan, San Jose, CA) coupled on-line with a capillary high-pressure liquid chromatograph (Magic 2002, Auburn, CA) was used for identification of phosphorylation sites. A MAGICMS C18 column (5 μm particle diameter, 150 Å pore size, 0.1 × 50 mm dimension) was used for the LC/MS/MS analysis.
Plasmids and reporter assays
Mutations were generated using a Gene Editor kit (Promega). Mutated and wild-type p220 coding sequences were cloned into pcDNA3.1 for expression. To generate histone gene reporter plasmids, H2B regulatory sequences (−200/0 or −127/−27) were cloned into the luciferase reporter pGL3 (Promega). To examine histone transcription, pCMV-p220, pGL3 reporter plasmids, and pCMV-β-galactosidase plasmids were co-transfected into 293T cells at 50%–80% confluence by using calcium phosphate. Twenty-four h after precipitate removal, extracts were generated and normalized for β-galactosidase activity before measurement of luciferase activity.
Acknowledgments
We thank J. Gall for discussions, J. Zhao, A.G. Matera, and E. Harlow for communicating results before publication, M. Carmo-Fonseca for anti-coilin antibodies, and Richard Atkinson, Brian Streib, and Heather Benedict-Hamilton for assistance. J.W.H. was supported by U.S. Public Health Service (USPHS) grant GM54137 and by the Welch Foundation. L.T.C. was supported by USPHS grants CA36200 and DE/CA11910. B.A.V.T was partially supported by the University of Alabama Medical Scientist Training Program. The Digital Imaging Microscopy Facility at the University of Alabama was supported by the University of Alabama Health Services Foundation and by grant DE/CA11901.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ude.cmt.mcb@reprahj; FAX (713) 796-9438.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.829500.
References
- Abbott J, Marzluff WF, Gall JG. The stem–loop binding protein (SLBP1) is present in coiled bodies of the Xenopus germinal vesicle. Mol Biol Cell. 1999;10:487–499. [Europe PMC free article] [Abstract] [Google Scholar]
- Adams PD, Sellers WR, Sharma SK, Wu AD, Nalin CM, Kaelin WG., Jr Identification of a cyclin-cdk2 recognition motif present in substrates and p21-like cyclin-dependent kinase inhibitors. Mol Cell Biol. 1996;16:6623–6633. [Europe PMC free article] [Abstract] [Google Scholar]
- Ahn J, Gruen JR. The genomic organization of the histone clusters on human 6p21.3. Mamm Genome. 1999;10:768–770. [Abstract] [Google Scholar]
- Almeida F, Saffrich R, Ansorge W, Carmo-Fonseca M. Microinjection of anti-coilin antibodies affects the structure of coiled bodies. J Cell Biol. 1998;142:899–912. [Europe PMC free article] [Abstract] [Google Scholar]
- Brown NR, Noble ME, Endicott JA, Johnson LN. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat Cell Biol. 1999;1:438–443. [Abstract] [Google Scholar]
- Cajal SRY. Un sencillo metodo de coloracion seletiva del reticulo protoplasmatico y sus efectos en los diversos organos nerviosos de vertebrados e invertebrados. Trab Lab Invest Biol. 1903;2:129–221. [Google Scholar]
- Connell-Crowley L, Elledge SJ, Harper JW. G1 cyclin-dependent kinases are sufficient to initiate DNA synthesis in quiescent human fibroblasts. Curr Biol. 1998;8:65–68. [Abstract] [Google Scholar]
- Di Fruscio M, Weiher H, Vanderhyden BC, Imai T, Shiomi T, Hori TA, Jaenisch R, Gray DA. Proviral inactivation of the Npat gene of Mpv 20 mice results in early embryonic arrest. Mol Cell Biol. 1997;17:4080–4086. [Europe PMC free article] [Abstract] [Google Scholar]
- Dulic V, Lees E, Reed SL. Association of human cyclin E with a periodic G1-S phase protein kinase. Science. 1992;257:1958–1961. [Abstract] [Google Scholar]
- Dyson N. The regulation of E2F by pRb-family proteins. Genes & Dev. 1998;12:2245–2262. [Abstract] [Google Scholar]
- Fletcher C, Heintz N, Roeder RG. Purification and characterization of OTF-1, a transcription factor regulating cell cycle expression of a human histone H2b gene. Cell. 1987;51:773–781. [Abstract] [Google Scholar]
- Frey MR, Matera AG. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc Natl Acad Sci. 1995;92:5915–5919. [Europe PMC free article] [Abstract] [Google Scholar]
- Gall JG, Bellini M, Wu Z, Murphy C. Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol Biol Cell. 1999;10:4385–4402. [Europe PMC free article] [Abstract] [Google Scholar]
- Harris ME, Bohni R, Schneiderman MH, Ramamurthy L, Schumperli D, Marzluff WF. Regulation of histone mRNA in the unperturbed cell cycle: Evidence suggesting control at two posttranscriptional steps. Mol Cell Biol. 1991;11:2416–2424. [Europe PMC free article] [Abstract] [Google Scholar]
- Heintz N. The regulation of histone gene expression during the cell cycle. Biochim Biophys Acta. 1991;1088:327–339. [Abstract] [Google Scholar]
- Imai T, Yamauchi M, Seki N, Sugawara T, Saito T, Matsuda Y, Ito H, Nagase T, Nomura N, Hori T. Identification and characterization of a new gene physically linked to the ATM gene. Genome Res. 1996;6:439–447. [Abstract] [Google Scholar]
- Koff A, Giordano A, Desai D, Yamashita K, Harper JW, Elledge SJ, Nishimoto T, Morgan DO, Franza BR, Roberts JM. Formation and activation of a cyclin E–cdk2 complex during the G1 phase of the human cell cycle. Science. 1992;257:1689–1694. [Abstract] [Google Scholar]
- Leng X, Connell-Crowley L, Goodrich D, Harper JW. S-phase entry upon ectopic expression of G1 cyclin-dependent kinases in the absence of retinoblastoma protein phosphorylation. Curr Biol. 1997;7:709–712. [Abstract] [Google Scholar]
- Leone G, DeGregori J, Jakoi L, Cook JG, Nevins JR. Collaborative role of E2F transcriptional activity and G1 cyclindependent kinase activity in the induction of S phase. Proc Natl Acad Sci. 1998;96:6626–6631. [Europe PMC free article] [Abstract] [Google Scholar]
- Liu J, Hebert MD, Ye Y, Templeton DJ, Kung H, Matera AG. Cell cycle–dependent localization of the CDK2–cyclin E complex in Cajal (coiled) bodies. J Cell Sci. 2000;113:1543–1552. [Abstract] [Google Scholar]
- Liu Q, Li MZ, Leibham D, Cortez D, Elledge SJ. The univector plasmid-fusion system, a method for rapid construction of recombinant DNA without restriction enzymes. Curr Biol. 1998;8:1300–1309. [Abstract] [Google Scholar]
- Lukas J, Herzinger T, Hansen K, Moroni MC, Resnitzky D, Helin I, Reed SI, Bartek J. Cyclin E–induced S phase without activation of the Rb/E2F pathway. Genes & Dev. 1997;11:1479–1492. [Abstract] [Google Scholar]
- Ma T, Zou N, Lin BY, Chow LT, Harper JW. Interaction between cyclin-dependent kinases and human papillomavirus replication-initiation protein E1 is required for efficient viral replication. Proc Natl Acad Sci. 1999;96:382–387. [Europe PMC free article] [Abstract] [Google Scholar]
- Matsuoka S, Edwards MC, Bai C, Parker S, Zhang P, Baldini A, Harper JW, Elledge SJ. p57KIP2, a structurally distinct member of the p21CIP1 Cdk inhibitor family, is a candidate tumor suppressor gene. Genes & Dev. 1995;9:650–662. [Abstract] [Google Scholar]
- Nevins JR. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ. 1998;9:585–593. [Abstract] [Google Scholar]
- Ohtsubo M, Theodoras AM, Schumacher J, Roberts JM, Pagano M. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol Cell Biol. 1995;15:2612–2624. [Europe PMC free article] [Abstract] [Google Scholar]
- Reed SI. Control of the G1/S transition. Cancer Surv. 1997;29:7–23. [Abstract] [Google Scholar]
- Russo AA, Jeffrey PD, Patten AK, Massague J, Pavletich NP. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A–Cdk2 complex. Nature. 1996;382:325–331. [Abstract] [Google Scholar]
- Schulman BA, Lindstrom DL, Harlow E. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc Natl Acad Sci. 1998;95:10453–10458. [Europe PMC free article] [Abstract] [Google Scholar]
- Segil N, Roberts SB, Heintz N. Mitotic phosphorylation of the Oct-1 homeodomain and regulation of Oct-1 DNA binding activity. Science. 1991;254:1814–1816. [Abstract] [Google Scholar]
- Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–1677. [Abstract] [Google Scholar]
- Zhang X, Herring CJ, Romano PR, Szczepanowska J, Brezeska H, Hinnebusch AG, Qin J. Identification of phosphorylation sites in proteins separated by polyacrylamide gel electrophoresis. Anal Chem. 1998;70:2050–2059. [Abstract] [Google Scholar]
- Zhao J, Dynlacht BD, Imai T, Hori T, Harlow E. Expression of NPAT, a novel substrate of cyclin E–CDK2, promotes S-phase entry. Genes & Dev. 1998;12:456–461. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhao J, Kennedy BK, Lawrence BD, Barbie D, Matera AG, Fletcher JA, Harlow E. NPAT links cyclin E–cdk2 to the regulation of replication-dependent histone gene transcription. Genes & Dev. 2000;14:2283–2297. (this issue). [Europe PMC free article] [Abstract] [Google Scholar]
- Zhu L, Harlow E, Dynlacht BD. p107 uses a p21CIP1-related domain to bind cyclin/cdk2 and regulate interactions with E2F. Genes & Dev. 1995;9:1740–1752. [Abstract] [Google Scholar]
Articles from Genes & Development are provided here courtesy of Cold Spring Harbor Laboratory Press
Full text links
Read article at publisher's site: https://doi.org/10.1101/gad.829500
Read article for free, from open access legal sources, via Unpaywall:
http://genesdev.cshlp.org/content/14/18/2298.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Transcriptional inhibition after irradiation occurs preferentially at highly expressed genes in a manner dependent on cell cycle progression.
Elife, 13:RP94001, 11 Oct 2024
Cited by: 1 article | PMID: 39392398 | PMCID: PMC11469672
Role of Neurocellular Endoplasmic Reticulum Stress Response in Alzheimer's Disease and Related Dementias Risk.
Genes (Basel), 15(5):569, 28 Apr 2024
Cited by: 0 articles | PMID: 38790197 | PMCID: PMC11121587
Recent Advances in Structural Optimization of Quinazoline-Based Protein Kinase Inhibitors for Cancer Therapy (2021-Present).
Molecules, 29(4):875, 16 Feb 2024
Cited by: 3 articles | PMID: 38398626 | PMCID: PMC10892255
Review Free full text in Europe PMC
Histone locus bodies: a paradigm for how nuclear biomolecular condensates control cell cycle regulated gene expression.
Nucleus, 14(1):2293604, 14 Dec 2023
Cited by: 1 article | PMID: 38095604 | PMCID: PMC10730174
Review Free full text in Europe PMC
EMP3 sustains oncogenic EGFR/CDK2 signaling by restricting receptor degradation in glioblastoma.
Acta Neuropathol Commun, 11(1):177, 07 Nov 2023
Cited by: 0 articles | PMID: 37936247 | PMCID: PMC10629159
Go to all (218) article citations
Other citations
Wikipedia
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The cyclin E/Cdk2 substrate and Cajal body component p220(NPAT) activates histone transcription through a novel LisH-like domain.
Mol Cell Biol, 23(10):3669-3680, 01 May 2003
Cited by: 64 articles | PMID: 12724424 | PMCID: PMC164767
The cyclin E/Cdk2 substrate p220(NPAT) is required for S-phase entry, histone gene expression, and Cajal body maintenance in human somatic cells.
Mol Cell Biol, 23(23):8586-8600, 01 Dec 2003
Cited by: 86 articles | PMID: 14612403 | PMCID: PMC262656
HiNF-P directly links the cyclin E/CDK2/p220NPAT pathway to histone H4 gene regulation at the G1/S phase cell cycle transition.
Mol Cell Biol, 25(14):6140-6153, 01 Jul 2005
Cited by: 73 articles | PMID: 15988025 | PMCID: PMC1168814
Cell cycle dependent phosphorylation and subnuclear organization of the histone gene regulator p220(NPAT) in human embryonic stem cells.
J Cell Physiol, 213(1):9-17, 01 Oct 2007
Cited by: 47 articles | PMID: 17520687
Review
Funding
Funders who supported this work.
NCI NIH HHS (2)
Grant ID: CA36200
Grant ID: R01 CA036200
NIDCR NIH HHS (1)
Grant ID: DE/CA11910
NIGMS NIH HHS (2)
Grant ID: GM54137
Grant ID: R01 GM054137





