Abstract
Free full text

Stage-Specific Changes in GDNF Expression by Rat Sertoli Cells: A Possible Regulator of the Replication and Differentiation of Stem Spermatogonia1
Abstract
In the adult testis, the precise control of the self-renewing replication and differentiation of stem spermatogonia is fundamental to male fertility. Previous studies have shown that the replication of A single (As) spermatogonia, a population that includes the stem cells, is maximal at stage I of the cycle of the rat seminiferous epithelium and minimal at stage VII, while the ratio of A-paired spermatogonia to As spermatogonia increases from stages I to VII. It has been hypothesized that these changes in As spermatogonia replication and differentiation result from changes in the expression of glial cell-line derived neurotrophic factor (GDNF) by Sertoli cells. To directly test this hypothesis, we used immunocytochemistry and confocal microscopy to demonstrate that within intact seminiferous tubules, GDNF is detectable only in Sertoli cells and that its amount and its location within these cells changes with progression of the stages of the cycle. The identification of Sertoli cells as the primary source of GDNF was confirmed by RT-PCR analysis of RNA isolated from purified populations of Sertoli cells, pachytene spermatocytes, and round spermatids. Stage-specific changes in GDNF expression were confirmed by quantifying GDNF mRNA in seminiferous tubules at defined stages of the cycle. Expression of this transcript was maximal at stage I, fell 14-fold by stage VIIc,d, and then increased 12-fold by stages XIII–XIV. This pattern of expression was the opposite of the control, cathepsin L mRNA. Taken together, these data support the hypothesis that cyclical changes in GDNF expression by Sertoli cells are responsible for the stage-specific replication and differentiation of stem spermatogonia, the foundational cells of spermatogenesis.
INTRODUCTION
Although the rat testis contains only about 800,000 stem spermatogonia, these cells are the ultimate source of the 107 spermatozoa that are produced daily by a normal, mature testis [1–3]. Therefore, maintaining the proper balance between the self-renewing replication and differentiation of these cells is fundamental to continued male fertility. In a series of meticulous studies, Huckins [1, 4, 5] demonstrated that both the replication and differentiation of rat A single (As) spermatogonia, a population that includes the stem cells, varied as the differentiated spermatogenic cells with which they were associated progressed through the stages of the cycle of the seminiferous epithelium. The number of As spermatogonia undergoing replicative DNA synthesis was maximal at stage I and decreased 10-fold by stage VII [4]. Huckins also enumerated A-paired (Apr) spermatogonia, the differentiating progeny of the As cells, and demonstrated that the ratio of Apr to As spermatogonia increased 1.8-fold from stage I to VII [1]. This result suggests that there is also stage-specific control of the differentiation of As spermatogonia.
As yet, there is uncertainty about how stem spermatogonial replication and differentiation is regulated in the normal adult testis, as most of what has been deduced about this regulation has come from studies of knockout and transgenic mice or from in vitro studies. De Rooij proposed that the stage-specific changes in As spermatogonia replication and differentiation are regulated by a Sertoli cell product, glial cell-line derived neutrotrophic factor (GDNF) [6]. This proposal was based in part on the observation that GDNF overexpression resulted in the accumulation of masses of As spermatogonia within the seminiferous tubules, while the reduced expression of GDNF resulted in the depletion of the stem spermatogonial pool [7]. Later, GDNF was shown to stimulate the replication of stem spermatogonia and to suppress their differentiation in vitro [8]. Though it is tempting to speculate from these studies that stage-specific changes in GDNF production by Sertoli cells are responsible for the reported changes in replication and differentiation of As spermatogonia, it is premature to do so, for there are studies in the literature that argue against the Sertoli cell as the sole source of GDNF and against its stage-specific expression [9, 10].
In the present studies, we analyzed both the cellular source of GDNF expression within the seminiferous epithelium of the adult rat testis and its potential stage-specific expression. To do so, we used an experimental approach that both preserved the antigenicity of GDNF and allowed a high-resolution analysis of the distribution of this growth factor within an intact seminiferous epithelium. We demonstrate here that the Sertoli cells are, indeed, the principal source of GDNF within the rat seminiferous epithelium and show definitively that GDNF expression is maximal at stages XIII–I and that expression of this growth factor varies with progression of the stages of the cycle. We interpret these data in the context of Huckins's analyses of the replication and differentiation of As spermatogonia and conclude that changes in GDNF production by Sertoli cells are the likely cause for the stage-specific changes in the replication and differentiation of As spermatogonia [1, 4].
MATERIALS AND METHODS
Animals
Cells and seminiferous tubules were isolated from mature Sprague-Dawley rats. The use of animals for the studies reported herein was approved by the Johns Hopkins Institutional Animal Care and Use Committee.
Isolation and Processing of Cells and of Seminiferous Tubules at Defined Stages of the Cycle of the Seminiferous Epithelium
Highly purified pachytene spermatocytes, round spermatids, and Sertoli cells were isolated by elutriation of enzymatically dispersed testes of 90- to 120-day-old rats as previously described [11, 12]. The purity of the cell preparations, which were 85% or greater, was determined by examination of cells by phase contrast microscopy. Seminiferous tubules at defined stages of the cycle of the rat seminiferous epithelium were isolated by transillumination-assisted microdissection [11, 13]. To ensure accurate identification of the stages, we collected small segments of tubules, extruded their germ cells, and used phase contrast microscopy to identify the step in spermiogenesis of the spermatids. Cells and tubules that were used for RNA isolation were immediately solubilized in Passive Lysis Buffer (Qiagen Corp., Valencia, CA) and frozen in liquid nitrogen. RNA was subsequently isolated using the RNeasy Mini Kit, and during this isolation, samples were digested with RNase-free DNAse (Qiagen). Seminiferous tubules at stages XIII–I, II–V, VII, and XI–XII were isolated for immunocytochemical localization of GDNF and were fixed for 2 h in 4% PFA-PBS at 4°C. The tubules were then washed for 2 h with multiple changes of PBS.
Immunocytochemical Localization of GDNF
Fixed seminiferous tubules were incubated at 4°C for 2 h in blocking solution: 2% goat serum, 1% BSA, 0.4% Triton X-100 in Tris-saline. Tubules then were incubated overnight at 4°C either in 200 ng/ml rabbit anti-GDNF (D-20; SC-328; Santa Cruz Biotechnology, Santa Cruz, CA) diluted in blocking solution, in 200 ng/ml of primary antibody that had been preadsorbed with the antigen for this antiserum (SC-328P; Santa Cruz Biotechnology), or in preimmune rabbit IgG. Subsequently, tubules were washed for 2 h with multiple changes of PBS and then incubated overnight in 1 μg/ml of Alexa-fluor 488 goat anti-rabbit IgG (Invitrogen Corp., Carlsbad, CA) diluted in blocking solution. Tubules were then washed for a total of 4 h and 20 min with multiple changes of PBS and mounted in Vectashield Mounting Medium with DAPI (Vector Laboratories, Burlingame, CA). A Zeiss 510 meta confocal microscope equipped with Argon 488-nm and 405-nm diode lasers was used to capture overlapping optical sections through the middle of a seminiferous tubule. To localize GDNF to individual cell types, we used a 100× Planapo oil lens and captured 320-nm-thick overlapping optical sections. To determine if the distribution of GDNF within the seminiferous epithelium changed with progression of the stages of the cycle, we used a 40× PlanNeofluar oil immersion lens and captured overlapping 600-nm overlapping optical sections. During image acquisition, the gain on the detector was set at 850 to 930. Images were processed using Zeiss LSM Image software. Multiple tubule segments at each stage or group of stages were examined.
Because there were differences between tubules in overall fluorescence intensities, for images that were selected for presentation, we used Photoshop (Adobe Systems Incorporated, San Jose, CA) to equalize the intensities of the blue fluorescence produced by the binding of DAP1 to DNA. For tubules incubated with antigen-adsorbed anti-GDNF, the brightness of the image was increased further so that the light green background fluorescence could be seen. This manipulation was not performed for the images that were used to quantify the relative amounts of GDNF in Sertoli cells at different stages of the cycle or within different domains of the Sertoli cell.
To test if there were stage-specific changes in the concentration of GDNF within Sertoli cells, we isolated stage XIII–I and stage VII tubules from five different testes, processed the tubules for GDNF immunocytochemistry, and then optically sectioned the tubules by confocal microscopy. Identical conditions were used to generate all optical sections, and the intensities of all images were within the linear range of the instrument. Images as TIFF files were imported into iVision (Biovision Technologies, Inc., Exton, PA), and fluorescence intensities of the Sertoli cells in the optical section were quantified. Fluorescence in areas outside of the Sertoli cells was also recorded, and this fluorescence was subtracted as background from the fluorescence measured in the Sertoli cells. Corrected fluorescence resulting from immunostaining of GDNF in Sertoli cells is presented as mean pixel intensities from the fluorescence emitted by the immunostained Sertoli cells. Data were obtained from Sertoli cells within 7–10 separate optical sections per stage for each set of tubules.
To test if there were significant stage-specific changes in GDNF concentration within Sertoli cells, we measured the relative concentration of GDNF in Sertoli cells within stage XIII–I, II–V, VII, and XI–XII tubules. All the tubules were obtained from a single animal. and 7–10 different optical sections per stage were analyzed. Images of Sertoli cells within the plane of the optical section were divided into a basal domain, a middle domain. and an apical domain. and fluorescence intensities in each domain were measured as described above.
Generation and Cloning of a Partial cDNA for Rat GDNF
We used 150 ng of RNA from Sertoli cells, pachytene spermatocytes, and round spermatids to synthesize cDNA by use of the Superscript First Stand Synthesis System with random hexamers employed as primers (Invitrogen). Negative controls were treated in the same manner with the exception that reverse transcriptase was omitted from the reaction. One microliter of cDNA or of the negative control reaction product was used as template when primers were used to amplify a partial cDNA for rat Gdnf mRNA. The cDNA and negative control RT reaction products were diluted 1:50 when primers for 18S rRNA, the positive control, were used in the PCR reaction. The reaction was conducted with 50 pmol of forward and reverse primers (see Table 1 for sequences) and buffer and deoxynucleotides provided in Optimized Buffer B (Invitrogen). The PCR conditions were 30 cycles of 95°C, 62°C, and 72°C; the reactants were incubated for 1 min at each temperature. PCR products were fractionated on 1% agarose gels and stained with ethidium bromide. To clone the Gdnf PCR product, we used cDNA from Sertoli cells as the template for seven replicate PCR reactions. The products of these seven reactions were pooled, and the Gdnf cDNA was isolated by agarose gel electrophoresis and column chromatography (MinElute PCR purification kit; Qiagen). The partial Gdnf cDNA was then digested with Hind III and XhoI, ligated into pGL2 basic (Promega Corp., Madison WI), and cloned. The DNA sequences of three independent clones were determined (Johns Hopkins University Genetic Resources Core Facility, Baltimore, MD).
TABLE 1.
Sequences of primers.

Evaluation of the Stage-Specific Changes in Expression of Gdnf mRNA and Its Comparison to Cathepsin L (Ctsl) mRNA
RNA was isolated and cDNA synthesized from rat seminiferous tubules at defined stages of the cycle as described above. Gdnf and Ctsl cDNAs were quantified by use of Lux and Taqman primers, respectively, and data were normalized to the amount of 18S rRNA in each sample as previously described [14]. 18S RNA concentration was measured using Lux primers. Table 1 provides the sequences of the Lux primers for Gdnf. Lux primers were purchased from Invitrogen, and Taqman primers were purchased from Applied Biosystems (Foster City, CA). Data were obtained from three independent sets of samples and were expressed as the ratio of expression of Gdnf mRNA and Ctsl mRNA at each stage compared to the expression of those two transcripts in the whole testis.
Statistical Analysis
Differences between experimental groups within an experiment were established by analysis of variance and the Fisher PLSD test by use of StatView (SAS Institute Inc., Cary, NC).
RESULTS
Cellular Distribution of GDNF Within Intact Seminiferous Tubules at Different Stages of the Cycle of the Seminiferous Epithelium
We initially attempted to evaluate the distribution of GDNF in the seminiferous epithelium by studying sections of rat testes that had been fixed in Bouin solution or in paraformaldehyde and then embedded in paraffin, an approach that has been taken by others [9]. (The methods we used for those preliminary studies are described as Supplemental Methods, available online at www.biolreprod.org.) These studies were unproductive, however, because when 5-μm sections were incubated with anti-GDNF from two different sources, results were identical to those we obtained when testes sections were incubated with an equivalent concentration of nonimmune IgG (data not shown). These results suggested that the organic solvents or heat that we used to process the tissue were denaturing antigenic epitopes of GDNF. To circumvent these potential problems, we examined the distribution of GDNF within intact seminiferous tubules that had been permeabilized with Triton X-100. These tubules were incubated with anti-GDNF followed by a fluorescently labeled secondary antibody, and consecutive optical sections of the tubule segments were examined by confocal microscopy. Figure 1 shows representative results from these studies. GDNF protein was present in columns that converged on Sertoli cell nuclei, which were recognizable by their prominent nucleolus (Fig. 1A). Both the amount of GDNF and its distribution within Sertoli cells appeared to change with progression of the surrounding spermatogenic cells through the stages of the cycle of the seminiferous epithelium. Differences in total GDNF expression per Sertoli cell are most apparent when Sertoli cells within stage XIII–I tubules are compared with Sertoli cells within stage VII tubules (compare Fig. 1B with Fig. 1D).

Confocal microscopic analysis of the distribution of GDNF within the rat seminiferous epithelium. Fixed seminiferous tubule segments were incubated with anti-GDNF IgG, the bound antibody was detected by use of a Alexaflour 488 (green)-goat anti-rabbit IgG, and the tubules were counterstained with DAPI (blue). All images are optical sections taken through the middle of a segment of a seminiferous tubule. The bar in the lower right corner of each image in this figure is equal to 10 μm in the tissue. Optical sections are either 320 nm thick (A) or 600 nm thick (B–F). The basement membrane of a tubule is located toward the upper left-hand corner of the image (A) or toward the top of the image (B–F). A) A high-resolution optical section showing the distribution of GDNF in a Sertoli cell within the basal half of a stage VII seminiferous tubule. The arrow points to a Sertoli cell nucleus and its distinctive nucleolus. B) An optical section through a stage XIII–I seminiferous tubule. C) An optical section through a stage II–V seminiferous tubule. D) An optical section through a stage VII seminiferous tubule. E) An optical section through a stage XI–XII seminiferous tubule. F) An optical section through a stage XI–XII tubule that had been incubated with anti-GDNF IgG that was preadsorbed with its antigen.
At stages XIII–I, the amount of GDNF per Sertoli cell appeared to be greater. Changes in the distribution of GDNF within Sertoli cells was most apparent when Sertoli cells within stages XIII–I are compared to those at stages XI–XII. At stages XIII–I, GDNF was present throughout the entirety of the Sertoli cell (Fig. 1B). At stages XI–XII, GDNF concentration in the basal domain of the Sertoli cell appeared to be greater than in its apical domain (Fig. 1E). Control experiments verified that the immunostaining we obtained was specific for GDNF. We detected only very light background green fluorescence when the primary antibody was adsorbed against the peptide used as antigen (Fig. 1F) or when the primary antibody was replaced with preimmune rabbit IgG (data now shown).
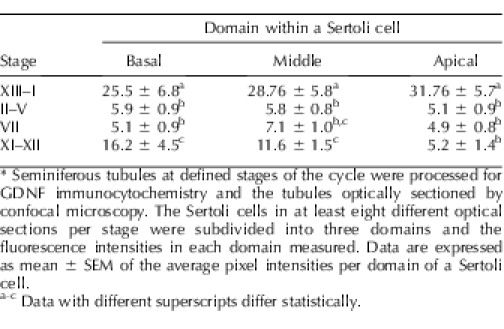
To confirm that the concentration of GDNF within Sertoli cells was greater at stages XIII–I than at stage VII, we used image analysis to quantify the amount of this growth factor in Sertoli cells within five independent sets of tubules at both sets of stages. Data (Fig. 2) demonstrate that GDNF concentration within Sertoli cells of stage XIII–I tubules is significantly greater than in Sertoli cells of stage VII tubules. To determine if there are significant stage-specific changes in GDNF concentration within Sertoli cells, we quantified the expression of this growth factor in the basal, middle, and apical domains of Sertoli cells within tubules at stages XIII–I, II–V, VII, and XI–XII. Data obtained from tubules of a single animal are presented in Table 1. Consistent with the data shown in Figure 2, data in Table 2 show that the concentration of GDNF in Sertoli cells within stage XIII–I tubules is greater than in Sertoli cells within tubules at all other stages. Furthermore, Table 2 demonstrates that at stages XI–XII, GDNF concentration in the apical domain of Sertoli cells is significantly lower than its concentration in the middle and basal domains of the same cell. This reduced GDNF concentration in the apical domain of Sertoli cells was not observed at any other stage.

Quantitative analysis of GDNF concentration in Sertoli cells within XIII–I and VII tubules. The relative concentration of GDNF in Sertoli cells at both stages was assayed by immunocytochemistry, confocal microscopy, and image analysis. Results were obtained from five independent sets of tubules. The relative concentrations (mean ± SEM) of GDNF in Sertoli cells within tubules at the two stages are expressed as average pixel intensities per Sertoli cell.
TABLE 2.
Relative concentrations of GDNF in the basal, middle, and apical domains of Sertoli cells within tubules at different stages of the cycle.*
Analysis of Gdnf mRNA Expression by Mature Sertoli Cells, Pachytene Spermatocytes, and Round Spermatids
Figure 1A supports the hypothesis that in the seminiferous epithelium, Sertoli cells are the primary source of GDNF. As these data are not consistent with results of some other studies reporting GDNF expression both by Sertoli cells and by pachytene spermatocytes and found spermatids [9, 10], we used a second, independent method to identify the cell type(s) that express GDNF. To this end, we purified total RNA from Sertoli cells, pachytene spermatocytes, and round spermatids that had been isolated from mature rat testes, and we used RT-PCR and DNA sequencing to identify cells that expressed Gdnf mRNA. Figure 3 shows typical results obtained from three independent isolates of these three cell types. A PCR product of the anticipated size, approximately 240 base pairs (bp), was obtained from Sertoli cells, while no PCR product was obtained from pachytene spermatocytes or round spermatids. In contrast, a PCR product for 18S rRNA was obtained from all three cell types, indicating that for all cell types, the RNA used for the RT reaction was intact (Fig. 3). The PCR product obtained from Sertoli cell RNA was cloned and sequenced and determined to be 100% identical to the comparable DNA sequence of rat Gdnf encoded in the Celera assembly of the rat genome (NW_001084796.1). These data, in combination with the results in Figure 1, indicate clearly that Sertoli cells are the principal source of GDNF within the seminiferous epithelium.

RT-PCR analysis of the expression of Gdnf mRNA by pachytene spermatocytes, round spermatids, and Sertoli cells; 150 ng of RNA from each cell type was used as a template in an RT reaction or in a negative control reaction, which lacked RT. One microliter, or the equivalent of 20 nanoliters of the RT or negative control reaction, was used as template for PCR reactions for Gdnf cDNA or 18S rRNA, respectively. The products of the PCR reactions were fractionated on 1% agarose gels along with a 100-bp DNA ladder (right-hand lane), and the DNA was stained with ethidium bromide. These results are representative of results obtained from three independent isolates of pachytene spermatocytes, round spermatids, and Sertoli cells.
Stage-Specific Changes in Expression of Gdnf mRNA
We used real-time PCR to quantify Gdnf mRNA in seminiferous tubules at defined stages of the cycle. As a control, we also quantified Ctsl mRNA, and levels of both transcripts were normalized to 18S rRNA. Figure 4 shows that Gdnf mRNA expression by Sertoli cells is maximal at stage I, is reduced 14-fold by stage VIIc,d, and then is increased 8- and 12-fold by stage XII and stages XIII–XIV, respectively. This pattern of expression is nearly the opposite to the pattern of expression of Ctsl mRNA, which is maximal at stages VIIa,b and minimal at stages XIII–I. Thus, Gdnf mRNA expression changes with progression of the stages of the cycle and its pattern of expression differs from that of a transcript encoding another Sertoli cell product, cathepsin L.
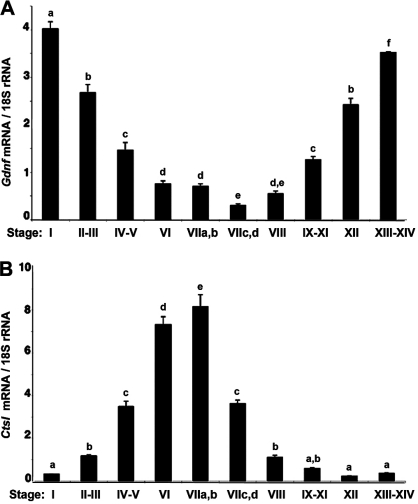
Stage-specific changes in expression of Gdnf mRNA and cathepsin L (Ctsl) mRNA. Seminiferous tubules at defined stages of the cycle were isolated by transillumination-assisted microdissection, and RNA was purified and used as template for cDNA synthesis. Expression of both Gdnf and Ctsl mRNAs were normalized for 18S rRNA. Data (mean ± SEM) are obtained from three independent samples of tubules and are expressed as the ratio of expression of Gdnf and Ctsl mRNA at each stage compared to the expression of those two transcripts in the whole testis. Means marked with different superscripts are statistically different.
DISCUSSION
Sertoli Cells Are the Primary Source of GDNF Within the Rat Seminiferous Epithelium
We have used two independent approaches—immunocytochemical examination of intact seminiferous tubules and RT-PCR analysis of RNA isolated from purified Sertoli cells and from spermatogenic cells—to demonstrate clearly that Sertoli cells are the principal source of GDNF within the rat seminiferous epithelium. Our data agree with previous reports that Sertoli cells produce GDNF [9, 10, 15]. However, in striking contrast to some other studies, we did not detect GDNF expression by spermatogenic cells. What are possible causes for the differences between the results from ours and previous studies? To date, only Caires et al. [9] have examined the distribution of GDNF within an intact murine seminiferous epithelium. In that study, sections of fixed, paraffin-embedded testes were incubated with anti-GDNF, and immunostaining was observed throughout the entire seminiferous epithelium. When the primary antibody was omitted, no staining was observed. As noted in Results, we used a comparable experimental approach in our preliminary studies, but when we included another control, incubation of testis sections with nonimmune rabbit IgG (rabbits were used to generate the anti-GDNF employed in our studies), the entire seminiferous epithelium was also stained. Thus, we concluded that the antibodies to GDNF that we used were unable to detect GDNF within histological sections of fixed and paraffin-embedded testes. These results lead us to initiate studies of the distribution of GDNF within intact seminiferous tubules.
Our identification of Sertoli cells as the principal source of Gdnf mRNA in the testis disagrees with previous reports. Fochecourt et al. [9] examined purified populations of Sertoli cells, pachytene spermatocytes, and round spermatids for the presence of Gdnf mRNA and its protein product. Similar to our experiments, they generated RT-PCR products for Gdnf mRNA and sequenced the cloned cDNA to confirm its identity. However, in contrast to the data presented here, Fochecourt et al. [9] conclude that pachytene spermatocytes and Sertoli cells contain similar levels of Gdnf mRNA. One possible explanation for the differences between our data and that of Fochecourt et al. [9] lies in the methods that were used to isolate Sertoli cells. Fochecourt et al. [9] exposed the Sertoli cells to repeated sheer stress to kill germ cells, a procedure that we have found damages Sertoli cells and alters their gene expression. In contrast, the rapid method we use for isolation of mature Sertoli cells does not expose the Sertoli cells to sheer stress. and array analysis indicates that high levels of Sertoli cell-specific gene expression are maintained in the isolated cells [11]. Thus, the more stressful method used by Fochecourt et al. [9] to isolate Sertoli cells may have led to a significant loss of Gdnf mRNA and its protein product, reducing the levels found in the isolated cells. Although we did not detect GDNF in spermatogenic cells, our results do not rule out the possibility that germ cells within an intact seminiferous epithelium express low levels of this growth factor [9, 10]. However, the data in Figures 1 and and22 lead us to conclude that Sertoli cells express substantially more Gdnf mRNA and its protein product than spermatogenic cells and therefore that these somatic cells are the principal source of GDNF within the rat seminiferous epithelium.
Stage-Specific Changes in Expression of GDNF
Our data demonstrate clearly that the expression of GDNF mRNA and protein by Sertoli cells changes as the surrounding germ cells progress through the stages of the cycle. Expression of the protein and the transcript are at a maximum at stages XIII–I and reach their nadir at stage VII. This pattern of expression is almost the opposite of cathepsin L described herein as well as previously [16–18]. The difference in the pattern of expression of these two genes is consistent with our demonstration that stage-specific gene expression is a fundamental characteristic of Sertoli cells and that different genes are maximally expressed by Sertoli cells at different stages of the cycle [11].
Our results raise the issue of how the stage-specific changes in expression of the Gdnf gene is regulated. Published data indicate that Gdnf mRNA expression by Sertoli cells is increased in testes that are severely depleted of germ cells [19]. Thus, spermatogenic cells may repress transcription of the Gdnf gene. This possibility is important because we have shown that stage-specific expression of the Ctsl gene is also controlled by suppressive signals from spermatids within tubules at stages I–IV and stages IX–XIV [20]. A combination of transgenic and transient transfection analyses of the Ctsl promoter have generated the first model of how stage-specific changes in gene expression by Sertoli cells may be regulated [21, 22]. It is possible that while the patterns of Gdnf and Ctsl genes expression differ, the ways in which repressive signals from spermatogenic cells are interpreted and integrated by their respective promoters may be similar. Thus, a brief review of a model that potentially explains stage-specific Ctsl gene expression is a starting point for considering how the transcriptional expression of the Gdnf gene by Sertoli cells may be regulated.
Stage-specific expression of the Ctsl gene by Sertoli cells involves the functional integration of cis-acting elements within three regulatory domains of a 2-kb promoter [21, 22]. Inhibitory signals from spermatids act via elements that reside 935 bp and 2 kb upstream from the transcription start site [22]. Activation of these elements prevents more downstream elements residing between −450 and −935 from recruiting histone modifying enzymes to the 100-bp proximal promoter. Thus, suppressive signals from spermatids lead to compaction of the chromatin over the proximal promoter, rendering it inaccessible to key transcription factors and/or to RNA polymerase II. However, at stages V–VIII, loss of the suppressive signals from the spermatids allows for the recruitment of enzymes that make transcriptionally permissive posttranslational modifications to the histones on the proximal promoter. These modifications cause relaxation of the chromatin structure, rendering the proximal promoter accessible to essential transcription factors and/or to RNA polymerase II. If, as proposed above, transcriptional regulation of the Gdnf gene in Sertoli cells shares characteristics with the regulation of the Ctsl gene, then stage-specific changes in expression of the Gdnf gene also may be controlled by regulating its chromain structure and thereby the access of essential transcription factors or RNA polymerase II to its promoter.
Relationship of Stage-Specific Changes in Expression of GDNF and Distribution of GDNF in Sertoli Cells
We have demonstrated a strong positive relationship between the expression of Gdnf mRNA and protein and the distribution of GDNF protein within Sertoli cells. At stages XII–I, where Gdnf mRNA expression is maximal, GDNF is distributed throughout the entire cell. However, at stages XI–XII, when GDNF expression is increasing after its nadir at stage VII, this growth factor is concentrated in the basal and middle domains of the cells. This relationship between changes in transcript expression and protein distribution in Sertoli cells is reminiscent of what we have observed in our studies of cathepsin L. For example, at stages IV–V, when the stage-specific increase in Ctsl mRNA expression is initiated (Fig. 3), the protein is concentrated in the base of the Sertoli cell [23]. However, at stage VIIa,b, the peak of Ctsl mRNA expression, cathepsin L protein, is distributed throughout the entire cell [23, 24]. These data lead us to propose that the distribution of a secreted protein in a Sertoli cell is dependent on the amount of protein that is being synthesized and not specifically on the stage of the cycle. Additionally, we suggest that the stage-specific increase in transcriptional expression of any Sertoli cell product initially leads to an increase in its secretion into the basal compartment of the seminiferous epithelium.
Potential Implications of the Stage-Specific Changes in Expression of GDNF of the Replication and Differentiation of Stem Spermatogonia
The stage-specific changes in expression of GDNF protein and transcript and the distribution of GDNF protein within Sertoli cells correlate well with stage-specific changes in the replication of As spermatogonia, a population of undifferentiated spermatogonia that contains the stem cells [6]. Huckins [4, 5] indicated that replicative DNA synthesis by rat As spermatogonia is minimal at stage VII, is increased at stages X–XII, and is maximal at stage I. Thus, replication of As spermatogonia increases coincidently with the rise in Gdnf mRNA expression by Sertoli cells and the increased concentration of the growth factor in these somatic cells. Furthermore, when this increase in replication is initiated, GDNF is concentrated in the base of the Sertoli cells the cellular domain nearest to the stem cells. Huckins also reports that there is a significant decrease in replication of As spermatogonia after stage II [4]. This decrease is associated with a substantial decrease in GDNF expression by Sertoli cells; at stage VIIc,d, Gdnf mRNA expression is only 7% of its level of expression at stage I. At this stage the percentage of As spermatogonia that have undergone replicative DNA synthesis is less than 3% of the percentage observed at stage I [4]. The decrease from stage II to stage VII in replicative DNA synthesis by As spermatogonia may be accompanied by an increase in frequency in the differentiation of these cells, for the ratio of Apr to As spermatogonia increases by 80% from stage I to stage VII [1].
In summary, our data, when viewed from the context of the pioneering work of Huckins and deRooij [1, 4–6], lead to the hypothesis that differentiated spermatogonia, spermatocytes, and/or spermatids regulate the expression of GDNF by Sertoli cells and thereby indirectly control the replication and differentiation of stem spermatogonia. As GDNF stimulates the growth of cultures enriched in stem spermatogonia [8] and as GDNF suppresses the differentiation of stem spermatogonia in vivo [7], we propose that the increase from stage VIIc,d to stage XIV in GDNF expression by Sertoli cells and the concentration of this protein around the stem spermatogonia stimulates stem and other As spermatogonia to enter the cell cycle. This stimulation begins to wane by stage III, when expression of the growth factor is substantially decreased. This decrease may allow for some of the replicating cells to begin to differentiate and thus form Apr spermatogonia, which are the first step in the formation of A1 spermatogonia, cells that are committed to the orderly process of spermatogenesis.
ACKNOWLEDGMENTS
We thank Barry Zirkin and Terry Brown for constructive critiques of this manuscript. We also thank Amy Albin for her expert assistance with confocal microscopy.
Footnotes
1Supported by the Eunice Kennedy Shriver NICHD/NIH through cooperative agreement U54055740 as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research.
REFERENCES
- Huckins C. The spermatogonial stem cell population in adult rats. I. Their morphology, proliferation and maturation. Anat Rec 1971; 169: 533 557 [Abstract] [Google Scholar]
- Orwig KE, Shinohara T, Avarbock MR, Brinster RL. Functional analysis of stem cells in the adult rat testis. Biol Reprod 2002; 66: 944 949 [Abstract] [Google Scholar]
- Wing TY, Christensen AK. Morphometric studies on rat seminiferous tubules. Am J Anat 1982; 165: 13 25 [Abstract] [Google Scholar]
- Huckins C. The spermatogonial stem cell population in adult rats. II. A radioautographic analysis of their cell cycle properties. Cell Tissue Kinet 1971; 4: 313 334 [Abstract] [Google Scholar]
- Huckins C. The spermatogonial stem cell population in adult rats. 3. Evidence for a long-cycling population. Cell Tissue Kinet 1971; 4: 335 349 [Abstract] [Google Scholar]
- de Rooij DG. Proliferation and differentiation of spermatogonial stem cells. Reproduction 2001; 121: 347 354 [Abstract] [Google Scholar]
- Meng X, Lindahl M, Hyvonen ME, Parvinen M, de Rooij DG, Hess MW, Raatikainen-Ahokas A, Sainio K, Rauvala H, Lakso M, Pichel JG, Westphal H, et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science 2000; 287: 1489 1493 [Abstract] [Google Scholar]
- Oatley JM, Avarbock MR, Brinster RL. Glial cell line-derived neurotrophic factor regulation of genes essential for self-renewal of mouse spermatogonial stem cells is dependent on Src family kinase signaling. J Biol Chem 2007; 282: 25842 25851 [Europe PMC free article] [Abstract] [Google Scholar]
- Fouchecourt S, Godet M, Sabido O, Durand P. Glial cell-line-derived neurotropic factor and its receptors are expressed by germinal and somatic cells of the rat testis. J Endocrinol 2006; 190: 59 71 [Abstract] [Google Scholar]
- Caires K, Broady J, McLean D. Maintaining the male germline: regulation of spermatogonial stem cells. J Endocrinol 2010; 205: 133 145 [Abstract] [Google Scholar]
- Johnston DS, Wright WW, Dicandeloro P, Wilson E, Kopf GS, Jelinsky SA. Stage-specific gene expression is a fundamental characteristic of rat spermatogenic cells and Sertoli cells. Proc Natl Acad Sci U S A 2008; 105: 8315 8320 [Europe PMC free article] [Abstract] [Google Scholar]
- Shaper NL, Wright WW, Shaper JH. Murine beta 1,4-galactosyltransferase: both the amounts and structure of the mRNA are regulated during spermatogenesis. Proc Natl Acad Sci U S A 1990; 87: 791 795 [Europe PMC free article] [Abstract] [Google Scholar]
- Kotaja N, Kimmins S, Brancorsini S, Hentsch D, Vonesch JL, Davidson I, Parvinen M, Sassone-Corsi P. Preparation, isolation and characterization of stage-specific spermatogenic cells for cellular and molecular analysis. Nat Methods 2004; 1: 249 254 [Abstract] [Google Scholar]
- Johnston DS, Jelinsky SA, Bang HJ, Dicandeloro P, Wilson E, Kopf GS, Turner TT. The mouse epididymal transcriptome: transcriptional profiling of segmental gene expression in the epididymis. Biol Reprod 2005; 73: 404 413 [Abstract] [Google Scholar]
- Yue F, Cui L, Johkura K, Ogiwara N, Sasaki K. Induction of midbrain dopaminergic neurons from primate embryonic stem cells by coculture with Sertoli cells. Stem Cells 2006; 24: 1695 1706 [Abstract] [Google Scholar]
- Erickson-Lawrence M, Zabludoff SD, Wright WW. Cyclic protein-2, a secretory product of rat Sertoli cells, is the proenzyme form of cathepsin L. Mol Endocrinol 1991; 5: 1789 1798 [Abstract] [Google Scholar]
- Kim GH, Wright WW. A comparison of the effects of testicular maturation and aging on the stage-specific expression of CP-2/cathepsin L messenger ribonucleic acid by Sertoli cells of the Brown Norway rat. Biol Reprod 1997; 57: 1467 1477 [Abstract] [Google Scholar]
- Zabludoff SD, Charron M, DeCerbo JN, Simukova N, Wright WW. Male germ cells regulate transcription of the cathepsin l gene by rat Sertoli cells. Endocrinology 2001; 142: 2318 2327 [Abstract] [Google Scholar]
- Tadokoro Y, Yomogida K, Ohta H, Tohda A, Nishimune Y. Homeostatic regulation of germinal stem cell proliferation by the GDNF/FSH pathway. Mech Dev 2002; 113: 29 39 [Abstract] [Google Scholar]
- Wright W, Zabludoff SD, Penttilä T-L, Parvinen M. Germ cell-Sertoli cell interactions: regulation by germ cells of the stage-specific expression of CP-2/Cathepsin l mRNA by Sertoli cells. Dev Genet 1995; 16: 104 113 [Abstract] [Google Scholar]
- Charron M, Folmer JS, Wright WW. A. 3-kilobase region derived from the rat cathepsin L gene directs in vivo expression of a reporter gene in Sertoli cells in a manner comparable to that of the endogenous gene. Biol Reprod 2003; 68: 1641 1648 [Abstract] [Google Scholar]
- Visone T, Charron M, Wright WW. Activation and repression domains within the promoter of the rat cathepsin L gene orchestrate Sertoli cell-specific and stage-specific gene transcription in transgenic mice. Biol Reprod 2009; 81: 571 579 [Europe PMC free article] [Abstract] [Google Scholar]
- Zabludoff S, Erickson-Lawrence M, Wright W. Sertoli cells, proximal convoluted tubules in the kidney and neurons in the brain contain cyclic protein-2. Biol Reprod 1990; 43: 15 24 [Abstract] [Google Scholar]
- Anway MD, Wright WW, Zirkin BR, Korah N, Mort JS, Hermo L. Expression and localization of cathepsin k in adult rat Sertoli cells. Biol Reprod 2004; 70: 562 569 [Abstract] [Google Scholar]
Articles from Biology of Reproduction are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1095/biolreprod.110.087676
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/biolreprod/article-pdf/85/4/763/10578195/biolreprod0763.pdf
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1095/biolreprod.110.087676
Article citations
Omega-3 polyunsaturated fatty acids and its metabolite 12-HEPE rescue busulfan disrupted spermatogenesis via target to GPR120.
Cell Prolif, 57(2):e13551, 24 Sep 2023
Cited by: 4 articles | PMID: 37743695 | PMCID: PMC10849791
Effects of Follicle-Stimulating Hormone on Human Sperm Motility In Vitro.
Int J Mol Sci, 24(7):6536, 31 Mar 2023
Cited by: 0 articles | PMID: 37047508 | PMCID: PMC10095528
Sertoli Cell-Germ Cell Interactions Within the Niche: Paracrine and Juxtacrine Molecular Communications.
Front Endocrinol (Lausanne), 13:897062, 10 Jun 2022
Cited by: 9 articles | PMID: 35757413 | PMCID: PMC9226676
Review Free full text in Europe PMC
Impact of germ cell ablation on the activation of the brain-pituitary-gonadal axis in precocious Atlantic salmon (Salmo salar L.) males.
Mol Reprod Dev, 89(10):471-484, 13 Jul 2022
Cited by: 2 articles | PMID: 35830347 | PMCID: PMC9796531
The Regulation of Spermatogonial Stem Cells in an Adult Testis by Glial Cell Line-Derived Neurotrophic Factor.
Front Endocrinol (Lausanne), 13:896390, 03 Jun 2022
Cited by: 4 articles | PMID: 35721702 | PMCID: PMC9203831
Review Free full text in Europe PMC
Go to all (50) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
RefSeq - NCBI Reference Sequence Database
- (1 citation) RefSeq - NW_001084796.1
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Glial cell-line-derived neurotropic factor and its receptors are expressed by germinal and somatic cells of the rat testis.
J Endocrinol, 190(1):59-71, 01 Jul 2006
Cited by: 31 articles | PMID: 16837611
Stage-specific gene expression is a fundamental characteristic of rat spermatogenic cells and Sertoli cells.
Proc Natl Acad Sci U S A, 105(24):8315-8320, 10 Jun 2008
Cited by: 91 articles | PMID: 18544648 | PMCID: PMC2448834
The production of glial cell line-derived neurotrophic factor by human sertoli cells is substantially reduced in sertoli cell-only testes.
Hum Reprod, 32(5):1108-1117, 01 May 2017
Cited by: 20 articles | PMID: 28369535 | PMCID: PMC6075567
Regulation of GDNF expression in Sertoli cells.
Reproduction, 157(3):R95-R107, 01 Mar 2019
Cited by: 23 articles | PMID: 30620720 | PMCID: PMC6602878
Review Free full text in Europe PMC




