Abstract
Free full text

Integration of Regulatory Networks by NKX3-1 Promotes Androgen-Dependent Prostate Cancer Survival
Associated Data
Abstract
The NKX3-1 gene is a homeobox gene required for prostate tumor progression, but how it functions is unclear. Here, using chromatin immunoprecipitation coupled to massively parallel sequencing (ChIP-seq) we showed that NKX3-1 colocalizes with the androgen receptor (AR) across the prostate cancer genome. We uncovered two distinct mechanisms by which NKX3-1 controls the AR transcriptional network in prostate cancer. First, NKX3-1 and AR directly regulate each other in a feed-forward regulatory loop. Second, NKX3-1 collaborates with AR and FoxA1 to mediate genes in advanced and recurrent prostate carcinoma. NKX3-1- and AR-coregulated genes include those found in the “protein trafficking” process, which integrates oncogenic signaling pathways. Moreover, we demonstrate that NKX3-1, AR, and FoxA1 promote prostate cancer cell survival by directly upregulating RAB3B, a member of the RAB GTPase family. Finally, we show that RAB3B is overexpressed in prostate cancer patients, suggesting that RAB3B together with AR, FoxA1, and NKX3-1 are important regulators of prostate cancer progression. Collectively, our work highlights a novel hierarchical transcriptional regulatory network between NKX3-1, AR, and the RAB GTPase signaling pathway that is critical for the genetic-molecular-phenotypic paradigm in androgen-dependent prostate cancer.
INTRODUCTION
Androgens such as testosterone and 5α-dihydrotestosterone (DHT) are steroid hormones that are required for key physiological events ranging from the acquisition and development of male characteristics during embryogenesis to the proper maturation and maintenance of male sexual reproductive organs such as the prostate and epididymis (20, 38). In addition to their roles in normal physiological processes, androgens are also key players in the initiation, development, and growth of prostate cancer (PCa) (13, 25, 42, 62), which is the most commonly diagnosed cancer and the second leading cause of cancer death among European and American males (36). Although initial androgen deprivation causes regression of androgen-dependent prostate tumors, prognosis is frequently poor, as they will eventually acquire an androgen-independent phenotype with disease progression that currently has no cure (19, 24).
The effects of androgens are mediated via the androgen receptor (AR), a member of the nuclear hormone receptor superfamily (54). Upon ligand binding, AR undergoes a conformational change, dissociates from heat shock proteins (HSPs) in the cytoplasm, homodimerizes, and translocates to the nucleus, where it binds to the palindromic androgen response element (ARE), which consists of two hexameric half sites (5′-AGAACA-3′) arranged as an inverted repeat separated by a 3-bp spacer (12, 17, 28). AR then recruits a combination of factors, including components of the general transcriptional machinery, chromatin-remodeling complexes, and specific transcriptional coregulators, in a cell- and gene-specific manner for the modulation of downstream transcriptional activities (4, 33, 34, 49).
The spatial and temporal expression program of a given gene is usually dictated by the unique combination of transcription factors recruited to the regulatory DNA regions that function together to either activate or repress transcription. Although much effort toward the description of coactivators (e.g., SRCs, p300/CBP, and mediators) and corepressors (e.g., NCoR and SMRT) has been made in the past, the understanding of collaborative DNA binding transcription factors that contribute to AR-dependent transcription is considerably less established. Furthermore, there remains insufficient evidence to clearly distinguish direct targets from the indirect gene targets despite the generation of whole-genome transcriptional profiles of ligand-regulated genes.
Recent advances in genomic technologies such as microarray-based chromatin immunoprecipitation (ChIP-on-chip) and chromatin immunoprecipitation coupled to massively parallel sequencing (ChIP-seq) are beginning to provide to us with a better understanding of the transcriptional role of AR collaborative factors in prostate cancer cells (37, 48, 59, 60, 67, 73). For example, the pioneer transcription factor, FoxA1, which is overexpressed in prostate tumors, was shown to bind at AR binding sites (ARBS) prior to androgen signaling (67). Furthermore, FoxA1 was recently shown to possess a lineage-specific transcription cistrome as defined by the distribution of mono- and dimethylated H3K4 as well as dimethylated H3K9 histone marks in both prostate and breast cancers (46). Several groups have subsequently identified additional AR collaborative factors such as GATA2 (67), ETS1 (48), and ERG (73). Given that transcriptional regulation is a complex process involving the delicate coordination between multiple transcription factors, it is therefore important to identify and characterize additional players that are part of the AR cistrome in androgen-dependent prostate cancer.
Molecular and phenotypic differences between normal and cancerous prostate cells are frequently attributed to altered gene expression and activities which lead to modifications of regulatory pathways that eventually result in aberrant cellular events, including abnormal cell growth and proliferation, disturbed cell cycle, and enhanced cell viability, as well as altered cellular adhesion and cohesion. Expression of AR in the AR-null prostate cancer cell line PC3 under different doses of androgen stimulation has been shown to result in differential gene expression, with approximately 5.7% of these genes involved in cell survival/apoptosis pathways (43). Such phenotypic effects observed upon androgen signaling generally occur through regulation of critical cell survival pathways, such as the insulin-like growth factor 1 (IGF-1), epidermal growth factor (EGF), and mitogen-activated protein kinase (MAPK) signaling pathways, as well as cell death pathways, such as the transforming growth factor β1 (TGF-β1), p53, or death receptor-mediated, caspase-dependent apoptotic pathway (21, 77). Therefore, the identification and characterization of primary AR target genes are required to better understand the overview of cross talk between AR signaling and multiple biological signaling pathways. In our present study, we combined genome-wide, molecular, and cell-based approaches to identify and functionally characterize a novel collaborative factor of AR. Our results suggest that the NKX3-1 gene, a homeobox gene involved in prostate cancer biology, regulates AR transcriptional activity via two distinct mechanisms to drive the AR transcriptional network toward a program that favors prostate cancer cell survival.
MATERIALS AND METHODS
Reagents and antibodies.
Dihydrotestosterone (DHT) was purchased from Tokyo Chemical Industry. The following antibodies were used for ChIP and Western blot analyses: anti-AR (sc-816 and sc-815x), anti-NKX3-1 (sc-15022), anti-Rab3B (sc-81911), normal rabbit IgG (sc-2027), normal goat IgG (sc-2028), and donkey anti-goat IgG–horseradish peroxidase (HRP) (sc-2033) from Santa Cruz; anti-FoxA1 (ab5089), anti-Pol II (ab5131), and anti-alpha-tubulin (ab4074) from Abcam; and donkey anti-rabbit IgG–HRP (NA934V) and sheep anti-mouse IgG–HRP (NA931V) from Amersham.
Cell culture and transient-transfection reporter assays.
LNCaP cells (ATCC) were maintained in RPMI medium 1640 (RPMI) supplemented with 10% fetal bovine serum (FBS), 1 mM sodium pyruvate, 100 units/ml penicillin, and 100 μg/ml streptomycin as well as 30 μg/ml gentamicin. VCaP cells (ATCC) were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% FBS, 1 mM sodium pyruvate, 0.08% sodium bicarbonate, 100 units/ml penicillin, and 100 μg/ml streptomycin. Prior to ethanol (EtOH) or DHT treatment, LNCaP cells were deprived of hormones for at least 3 days in phenol red-free RPMI containing 5% charcoal-dextran-treated FBS (HyClone), while VCaP cells were grown for at least a day in phenol red-free DMEM containing 10% charcoal-dextran treated FBS.
For transient transfection, LNCaP cells were grown in phenol red-free medium (without antibiotics) in 24-well culture plates for 4 days prior to transfection. Luciferase reporter and Renilla (Promega) constructs were cotransfected into the cells using Lipofectamine 2000 reagent (Invitrogen). After 18 to 24 h, cells were treated with EtOH or 100 nM DHT for another 24 h before harvesting for luciferase reporter assay. Firefly and Renilla luciferase activities were assayed using the Dual Luciferase System kit (Promega) and a Centro LB960 luminometer (Berthold Technologies) according to the manufacturers' protocols. The relative reporter gene activity was obtained after normalization of the firefly luciferase activity with Renilla luciferase activity. Each experiment was repeated at least three times to ensure reproducibility. All primers used for cloning and mutagenesis of reporter and overexpression constructs can be found in Table S3 in the supplemental material.
ChIP and re-ChIP.
ChIP and sequential ChIP (re-ChIP) assays were performed as described previously (61). Androgen-deprived LNCaP or VCaP cells were treated with EtOH or 100 nM DHT for 2 h before harvesting. For re-ChIP, dimethyl pimelimidate (DMP) (Pierce) was included as a protein cross-linker. Real-time quantitative PCR (qPCR) primer sequences for ChIP and re-ChIP assays are listed in Table S3 in the supplemental material. All experiments were performed at least three times.
Solexa sequencing and binding site determination.
ChIP-enriched DNA was quantified with the Quant-iT Pico Green double-stranded DNA (dsDNA) assay kit (Invitrogen, Molecular Probes). Five to 15 nanograms of DNA was used for library preparation using the ChIP-seq DNA Sample Prep kit from Illumina with minor modifications. The Illumina adaptor-ligated DNA was amplified with Phusion DNA polymerase (for AR and Pol II libraries) or Platinum Pfx DNA polymerase (for NKX3-1 and FoxA1 libraries) (Invitrogen) for 15 cycles. Amplified DNA products of 200 to 300 bp were gel excised and purified. After cluster amplification and sequencing on the Solexa GAIIx, ChIP-seq reads were aligned to the reference human genome (UCSC, hg18) using our in-house program, BATMAN, and binding peaks with a cutoff false-discovery rate (FDR) of 0.05 were determined using CCAT (71) with input DNA as a control.
siRNA studies.
LNCaP or VCaP cells suspended in phenol red-free medium were transfected with 100 nM small interfering RNA (siRNA) (Dharmacon or 1stBase Pte Ltd.) using Lipofectamine RNAiMAX transfection reagent (Invitrogen) according to the manufacturer's protocol. After 48 h of incubation, the cells were transfected again in a similar manner with 50 nM siRNA for LNCaP cells or 100 nM siRNA for VCaP cells. At 48 h after the second round of transfection, cells were treated with EtOH or 10 nM DHT for another 8 h before harvesting for real-time reverse transcription-qPCR (RT-qPCR) and Western blot analyses. Each target siRNA was paired with the control nontargeting siRNA (siCtrl) from the same company. siRNA and real-time qPCR primer sequences are listed in Table S3 in the supplemental material. Gene expression profiles for knockdown studies were obtained from at least three independent experiments. For flow cytometry and migration assay, 100 nM siRNA was used for both rounds of transfection.
Co-IP.
LNCaP cells were treated for 24 h with either EtOH or 100 nM DHT, trypsinized, and lysed to obtain whole-cell lysate. An aliquot of the cell lysate was kept for input in Western blot analysis. For coimmunoprecipitation (co-IP) of FoxA1- or NKX3-1-bound complexes with AR, whole-cell lysate was first precleared with protein A/G-agarose beads (Roche Applied Science) at 4°C for 4 h before the precleared supernatant was incubated with anti-FoxA1 or anti-NKX3-1 antibody overnight. The next day, protein A/G-agarose beads were added to the mixture and incubated at 4°C for 1.5 h. The beads were then pelleted and washed four times with Tris-buffered saline (TBS) before boiling and eluting with SDS loading buffer, followed by Western blot analysis. However, for co-IP of FoxA1-bound complexes with NKX3-1, the whole-cell lysate was first incubated with Preclearing Matrix D (sc-45055) at 4°C for 2 h before the precleared lysate was immunoprecipitated with anti-NKX3-1 and IP matrix D complex (sc-45041) according to the manufacturer's protocol.
Cell cycle flow cytometry analysis.
Transfected LNCaP cells were grown in serum or androgen-deprived medium (without antibiotics) before being treated with EtOH or 100 nM DHT for 72 h. Similarly, transfected androgen-deprived VCaP cells were treated with EtOH or DHT for 48 h. Cells were then harvested by trypsinization, washed with cold phosphate-buffered saline (PBS), and fixed with 70% EtOH for 45 min at 4°C. After another wash with cold PBS, the cell pellet was incubated with 100 μg/ml RNase (Sigma) at room temperature for 5 min before incubation with 50 μg/ml propidium iodide (PI) (Sigma) for an hour in the dark, followed by flow cytometry analysis of DNA content using a FACSCalibur (Becton-Dickson) with the CellQuest analysis software. A total of 1 × 104 cells were analyzed for each sample, and the percentage of cells in the sub-G1 phase was obtained from the DNA histogram. The cell cycle profile was obtained from at least three independent experiments.
Caspase assay.
Similar to the cell cycle flow cytometry analysis, androgen-depleted LNCaP or VCaP cells were treated with EtOH or DHT for 72 or 48 h, respectively, before harvesting by trypsinization, followed by washing with cold PBS. The cell pellet was fixed and permeabilized with the BD Cytofix/Cytoperm solution and washed with 1× BD Perm/Wash buffer (BD Cytofix/Cytoperm fixation/permeabilization kit). The cell pellet was then incubated with fluorescein isothiocyanate (FITC)–rabbit anti-active caspase-3 (BD Pharmingen) for an hour in the dark before washing with 1 × BD Perm/Wash buffer again. Caspase-3 activity was measured using a FACSCalibur (Becton-Dickson) with the CellQuest analysis software. The caspase activity was determined from at least three independent experiments.
Microarray analysis.
Microarray analysis was performed as described previously (61).
GO analysis.
Gene ontology (GO) analysis was performed using the Panther web tool on genes with a ≥1.5-fold or ≤0.67-fold change in transcript level relative to that with EtOH treatment upon either 3, 6, 12, or 24 h of DHT treatment (referred to as androgen-responsive genes) and which contained at least one transcription factor binding site within 50 kb of the respective gene transcription start site (TSS).
Oncomine molecular concept map (MCM).
To identify the enrichment network between our genes of interest and gene signatures from the different molecular concepts within the Oncomine database of tumor microarrays (https://www.oncomine.org/), we performed analysis of the association of our androgen-responsive genes cooccupied by AR and NKX3-1 within 50 kb of the TSS with the cancer transcriptome profiles. The odds ratio was set at 1.6 and above to categorize genes considered to be differentially expressed within the context of each concept. A node represents a molecular concept, and the node size is made proportional to the number of genes within each gene cluster. An edge represents statistically significant overlap (P value of <0.01) between the gene sets in two interconnected nodes.
Accession numbers.
The ChIP-seq and microarray gene expression data have been deposited at the NCBI GEO repository under accession numbers GSE28264 and GSE28596.
RESULTS
Genome-wide profile of AR binding events in prostate cancer cells.
To begin examining the transcriptional network of AR in prostate cancer cells, we used chromatin immunoprecipitation coupled to massively parallel sequencing (ChIP-seq) to map the in vivo binding sites of AR in the androgen-dependent prostate cancer cell line LNCaP. We identified 18,117 and 75,296 AR binding sites (ARBS) before and after DHT treatment, respectively (see Table S1 in the supplemental material). We validated the AR ChIP-seq data set by ChIP-qPCR and observed a good correlation (r = 0.75) between ChIP-seq peak intensities and ChIP-qPCR (see Fig. S1 in the supplemental material). A comparison of AR binding before and after DHT treatment revealed a significant overlap in ARBS between the two conditions (Fig. 1A). Moreover, DHT stimulation increased the overall intensity of AR binding (as measured by ChIP-seq tag density) by ≥2.8-fold (Fig. 1B and C).
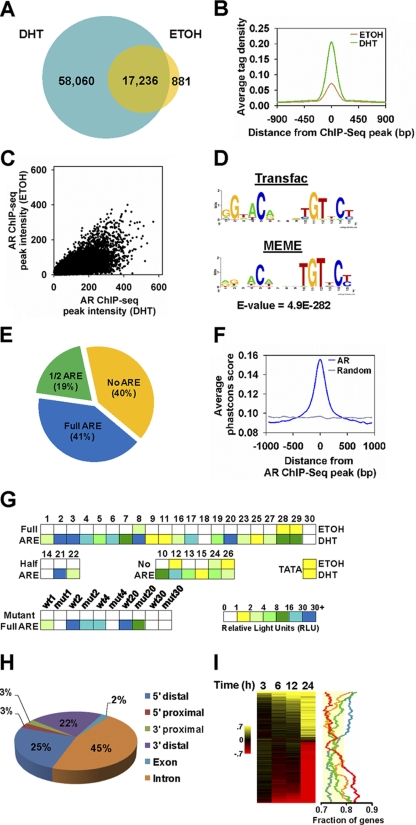
Genomic characterization of ARBS in prostate cancer cells. (A) Venn diagram depicting the overlap between ethanol (EtOH)- and DHT-treated AR ChIP-seq peaks. (B) Comparison between average tag densities of binding sites found within the EtOH- and DHT-treated AR binding maps after accounting for sequence depth difference. (C) Scatter plot of tag intensity of ARBS between the DHT- and EtOH-treated data sets after accounting for sequence depth difference. (D) Logos of canonical ARE from the Transfac database (top panel) and enriched within our top 500 ARBS (±50 bp from the DHT library peak) using MEME (bottom panel). (E) Pie chart illustrating the proportions of ARBS containing a full (a maximum of 3 mutations), half (0 mutation), or no ARE. (F) Graphical representation of the mean PhastCons sequence conservation score (alignment of 16 vertebrate genomes with human) for every position in a 2,000-bp window around the ChIP-seq peak of the DHT-treated AR library or around a randomly selected genomic region. (G) Putative ARBS were subcloned into the pGL4-TATA vector, transfected into androgen-deprived LNCaP cells, treated with EtOH or 100 nM DHT for 24 h, and then assayed for luciferase activity. Randomly selected ARBS binding sites were also mutated at the full ARE. The data are the average relative light units (RLU) for each AR binding site from at least 3 independent experiments. (H) Proportions of ARBS at different genomic locations relative to nearest transcription units from the Refseq database. 5′ proximal refers to 0 to 10 kb upstream of the TSS, 5′ distal refers to >10 kb upstream of the TSS; 3′ proximal refers to 0 to 10 kb downstream of the gene end, and 3′ distal refers to >10 kb downstream of the gene end. (I) Expression profile of all genes across time course DHT treatment (3, 6, 12, and 24 h) relative to their matched EtOH treatment. Induction and repression are represented by yellow and red, respectively. The graph represents 1,000-gene moving averages of the fraction of genes with at least one ARBS within 50 kb of the TSS. Genes at each time point were sorted according to the fold change upon DHT stimulation relative to the EtOH treatment. Blue, 3 h; orange, 6 h; green, 12 h; red, 24 h. ARBS association with nonregulated genes with a fold change of between 0.91 and 1.1 at the 12-h treatment time point was used to determine the 99% t-statistics confidence interval for moving average fraction of genes with AR binding (yellow band).
Using the de novo motif discovery algorithm MEME, we identified a canonical ARE motif that was significantly enriched (E value = 4.9E−282) in the ARBS from LNCaP cells treated with DHT (Fig. 1D). Recently, noncanonical AREs comprising different orientations of hexameric half sites with various spacer lengths were reported as in vivo AR recognition sites (67). We searched for these AREs but could not find any of them overrepresented in our LNCaP ARBS. Instead, we detected only the enrichment of the canonical ARE (up to 3-bp mismatch) (see Fig. S2 in the supplemental material). Overall, approximately 41% of the ARBS contain full AREs, whereas 19% harbor half AREs. Surprisingly, a large fraction of the ARBS (40%) lacked either motif (Fig. 1E), which suggests that AR may be recruited to these sites indirectly via other transcription factors. In the ARBS that contain AREs, the ARE was localized at the center of the ChIP-seq peak compared to randomly selected genomic sequences (see Fig. S3A in the supplemental material). Sequence conservation analysis of ARBS across different species showed that sequences near AR peaks tend to be more evolutionarily conserved than flanking sequences (Fig. 1F) and largely independent of the AR peak intensity (see Fig. S3B in the supplemental material). To assess whether the ARBS identified from our ChIP-seq experiment are functional, we subcloned 30 ARBS into a luciferase reporter containing a TATA box and transfected these constructs into LNCaP cells. We found that most constructs were activated by DHT and showed at least a 2-fold ligand response, including binding sites that lacked AREs (Fig. 1G). Mutating the AREs in the ARBS led to a significant decrease in androgen-dependent activity (Fig. 1G), indicating that these ARBS contain functional ARE motifs.
Next, we examined the location of AR binding relative to the TSS of Refseq genes. Unlike RNA Pol II, which bound distinctly at the TSS (see Fig. S3C in the supplemental material), we found that AR resided mainly at distal regions of known genes (Fig. 1H), suggesting that AR may regulate transcription by chromatin looping. To obtain a global view of gene transcription by AR, we associated ARBS with DHT-regulated genes from our microarray analysis of LNCaP cells treated with DHT at early (3 h), intermediate (6 and 12 h), and late (24 h) time points (Fig. 1I). In general, our results indicated that ARBS are associated with early and intermediate upregulated genes as well as intermediate and late downregulated genes. Interestingly, similar to estrogen receptor binding (11), we observed many more ARBS (>70,000) than the number of androgen-regulated genes (5,718). It is unclear why this is the case, but it is possible that many of these sites are nonfunctional under our experimental conditions or that multiple ARBS are involved in gene regulation through complex chromatin interactions. Taken together, our results suggest that DHT stimulates the binding of AR across the genome to directly regulate androgen target genes in prostate cancer cells.
NKX3-1 is colocalized with AR near androgen-regulated genes in prostate cancer cells.
Recent genomic studies showed that transcription factors such as FoxA1, Oct1, and Ets1 are important collaborative factors of AR transcriptional activity (15, 48, 67). These factors were identified based on the enrichment of their cognate DNA binding sequence within ARBS. To discover novel coregulators of AR, we analyzed our AR ChIP-seq peaks using an alternative approach called “center of distribution” (CentDist) (76). Briefly, CentDist is an algorithm that ranks motifs of collaborative factors according to their imbalanced distribution within the ChIP-seq peaks of a transcription factor. The concept of CentDist was developed based on our observations that collaborative factors and thus their cognate DNA binding sequences not only are enriched but also are frequently distributed near the center of the transcription factor binding site. Using CentDist on our AR ChIP-seq peaks, we observed high center-of-distribution scores for the DNA binding motifs of previously reported collaborative factors of AR, including Forkhead, ETS, Oct, and GATA (Fig. 2A). Interestingly, in addition to motifs of known AR collaborative factors, we also found highly ranked sequences belonging to the NKX family of transcription factors.
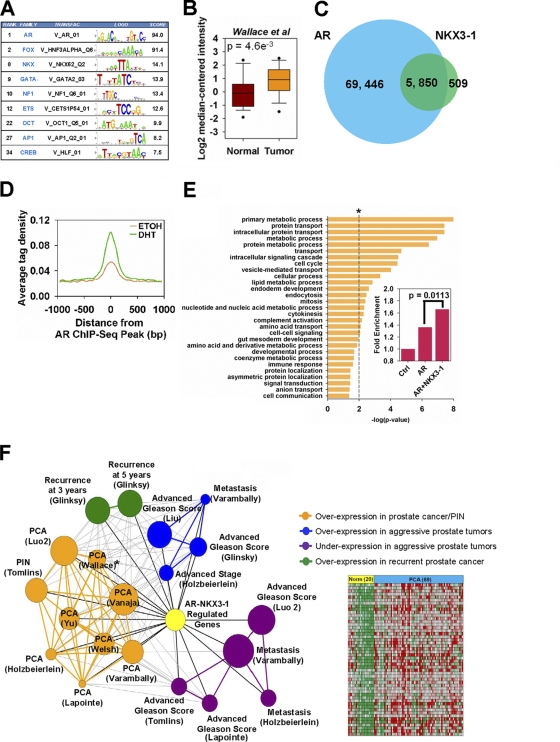
NKX3-1 is a novel collaborative factor of AR. (A) CentDist output of the top cooccurring transcription factor family motifs from DHT-stimulated AR ChIP-seq peaks. (B) Box plot comparing the transcript levels of NKX3-1 in normal prostate and prostate adenocarcinoma samples from the Oncomine microarray database (study by Wallace et al. in the database). The differential gene expression data are centered on the median of expression levels and plotted on a log2 scale. The P value was calculated using a Welch two-sample t test. (C) Venn diagram illustrating the overlap between DHT-stimulated AR and NKX3-1 ChIP-seq peaks. (D) Distribution of EtOH or DHT NKX3-1 ChIP-seq tags centered around AR (DHT) binding peaks. (E) GO analysis of androgen-dependent genes cooccupied by AR and NKX3-1 within 50 kb from the TSS using the Panther web tool. *, threshold was set at a P value of 0.01. Fold enrichment was calculated by dividing the observed number of genes by the expected number of genes within the same GO pathway. A two-tailed Fisher exact test was performed for the fold enrichment difference in genes from the “protein transport” pathway that are associated with AR only and those associated with both AR and NKX3-1. (F) Left panel, Oncomine concept map analysis illustrating the enrichment network between androgen-dependent genes cooccupied by AR and NKX3-1 and gene signatures in different prostate carcinoma subtypes. Each node represents one molecular concept, with the node size proportional to the number of genes within each gene set. Statistically significant overlap (P < 0.01) between genes in two linked nodes is represented by an edge. Concepts were categorized into 5 major clusters as indicated by the different colors. Right panel, expression level of the top 50 overexpressed genes in prostate carcinoma samples from the study by Wallace et al. in the Oncomine database (indicated by an asterisk in the concept map).
The NKX family belongs to the homeodomain class of transcription factors (58). There are 12 members in the human NKX family (58). Among them, NKX3-1, which is expressed in LNCaP cells, has been well documented to be important in prostate biology (2). NKX3-1 expression is largely prostate specific and is upregulated by androgen stimulation (8, 32, 53, 56). NKX3-1 is important in the development and differentiation of the prostate epithelium (7, 8, 32). It is also important in prostate cancer; however, its exact role is still unclear. For example, NKX3-1 has been implicated as a tumor suppressor, as its allelic locus maps to a hot spot on human chromosome 8p21 which undergoes loss of heterozygosity (1, 5, 32, 63, 64), but recent studies showed that it is highly expressed in advanced prostate cancer (16, 30). Furthermore, when we examined the Oncomine microarray database, we found that the level of NKX3-1 expression in prostate tumors is higher than that in normal prostate tissues (Fig. 2B; see Fig. S4 in the supplemental material). These observations suggest that perhaps NKX3-1 may actually function as an oncogene rather than a tumor suppressor in prostate cancer. With our finding that NKX motifs are enriched in ARBS, we hypothesized that NKX3-1 may collaborate with AR to regulate the expression of genes important in prostate cancer progression.
To begin examining this possibility, we first determined whether NKX3-1 functions together with AR to directly regulate the transcription of androgen-dependent genes in prostate cancer cells. For this, we mapped the binding sites of NKX3-1 in LNCaP cells using ChIP-seq under the same conditions as AR. We identified a total of 6,359 NKX3-1 binding sites in the presence of DHT (but none in vehicle) (see Table S1 in the supplemental material), of which 92% colocalized with AR (Fig. 2C). We validated the colocalized binding of NKX3-1 at ARBS by ChIP-qPCR before and after DHT stimulation. Our results showed that all the binding sites tested were enriched ≥5-fold compared to a control site (see Fig. S5 in the supplemental material). To examine the effect of DHT on global NKX3-1 binding at ARBS, we compared the average ChIP-seq tag density of NKX3-1 before and after DHT stimulation. As shown in Fig. 2D, DHT significantly enhanced the overall binding of NKX3-1 to chromatin.
Next, we examined the genes and biological processes that are associated with both NKX3-1 and AR in prostate cancer cells. In general, we noticed NKX3-1 binding sites are frequently colocalized together with AR near well-characterized model androgen-regulated genes, including TMPRSS2, FKBP5, and prostate-specific antigen (PSA) (Fig. 2D; see Fig. S6A and B in the supplemental material), suggesting that the expression of these known direct targets of AR may also require NKX3-1. We also performed GO analysis on androgen-regulated genes associated with binding sites containing both factors. Our results showed that NKX3-1- and AR-coassociated genes were overrepresented in categories similar to those for AR-associated genes, such as “metabolic processes” and “cell cycle” (Fig. 2E; see Fig. S7 in the supplemental material). However, NKX3-1- and AR-coassociated genes were preferentially enriched for categories related to “protein trafficking” (Fig. 2E, inset). In addition, we carried out molecular concept map (MCM) analysis to determine whether NKX3-1- and AR-coassociated genes have clinical significance in prostate tumors. In this analysis, associated genes were compared with predefined clinical prostate cancer gene sets in the Oncomine database. Overall, NKX3-1- and AR-coassociated genes shared concepts similar to but distinct from those for AR-associated genes (Fig. 2F; see Fig. S8 in the supplemental material). Notably, NKX3-1- and AR-coassociated genes were predominantly overexpressed in prostate carcinoma compared to normal counterparts (Fig. 2F). Moreover, these genes were, in particular, implicated in advanced and recurrent prostate cancer, thus further strengthening the biological significance of NKX3-1 in AR-dependent gene regulation and prostate cancer progression. Taken together, our results suggest that NKX3-1 is a potential novel collaborating transcription factor of AR that is important in mediating androgen-dependent gene transcription in prostate cancer cells.
A feed-forward regulatory loop between AR and NKX3-1 in prostate cancer cells.
NKX3-1 is a known androgen-regulated gene in prostate cancer cells and has been shown in clinical studies to be overexpressed in prostate cancer patients (32, 39, 53); however, the mechanism underlying this regulation or deregulation is currently unclear. With our genome-wide binding maps of AR, we asked whether NKX3-1 could be a direct target of AR. We scanned for ARBS near the NKX3-1 gene and observed the ligand-dependent recruitment of AR at +2 and +39 kb from the TSS of NKX3-1 (Fig. 3A and B). To determine if the expression of NKX3-1 is dependent on AR, we performed siRNA-mediated knockdown of AR in LNCaP cells and measured the expression level of NKX3-1. Our results showed that depleting AR levels reduced both the transcript (Fig. 3C; see Fig. S10A in the supplemental material) and protein (Fig. 3D; see Fig. S10B in the supplemental material) levels of NKX3-1. Taken together, our results suggest that NKX3-1 is likely a direct transcriptional target of AR in prostate cancer cells.

AR and NKX3-1 directly regulate each other in a feed-forward manner. AR, NKX3-1, and RNA Pol II colocalize at enhancer and promoter regions of the NKX3-1 gene. (A) Screenshots of AR, NKX3-1, and RNA Pol II ChIP-seq peaks surrounding the NKX3-1 gene. (B) ChIP-qPCR validation of AR and NKX3-1 at the enhancer and promoter of the NKX3-1 gene. DNA enrichment is represented as percentage of input chromatin immunoprecipitated. Data represent the means ± standard errors of the means (SEM) from at least three independent experiments. (C and D) Effect of AR silencing on NKX3-1 expression level. Hormone-depleted LNCaP cells were transfected with either control siRNA or siRNA targeting AR before treatment with EtOH or 10 nM DHT for 8 h. (C) Total RNA was isolated and amplified with real-time RT-qPCR primers for AR and NKX3-1. mRNA expression levels were normalized against GAPDH (glyceraldehyde-3-phosphate dehydrogenase). (D) Western blot assaying the protein levels of AR, NKX3-1, and α-tubulin after siRNA transfection. AR and NKX3-1 colocalize binding within the AR gene body. (E) Screenshots of AR and NKX3-1 ChIP-seq peaks around the AR gene. (F) ChIP-qPCR validation of AR and NKX3-1 ChIP-seq peaks within the intragenic region of AR. DNA enrichment is presented as the percentage of input chromatin immunoprecipitated. Data show the mean ± SEM from at least three independent experiments. (G and H) Effect of NKX3-1 depletion on AR expression level. Androgen-deprived LNCaP cells were transfected with either control siRNA or siRNA targeting NKX3-1 before treatment with EtOH or 10 nM DHT for 8 h. (G) Total RNA was isolated and amplified with real-time RT-qPCR primers for AR, NKX3-1, and GAPDH. mRNA expression levels were normalized against GAPDH. (H) Western analysis of AR, NKX3-1, and α-tubulin protein levels after siRNA knockdown.
Since our results indicate that AR maybe regulating the expression of NKX3-1 directly, we considered the possibility that these two factors could be involved in an autoregulatory network in which NKX3-1 may also directly regulate the transcription of AR. As shown in Fig. 3E and F, NKX3-1 is recruited in a ligand-dependent manner at an intragenic region 79 kb downstream from the TSS of the AR gene. Moreover, when we depleted the protein levels of NKX3-1 with siRNA, we observed a drastic decrease in both the AR transcript and protein levels (Fig. 3G and H; see Fig. S10C and D in the supplemental material), as well as a global change in the expression of androgen-regulated genes, suggesting that NKX3-1 regulates AR transcriptional signaling, in part by controlling the expression of AR (see Fig. S9 in the supplemental material). We also examined the binding and transcriptional regulation between AR and NKX3-1 in the AR-positive prostate cancer cell line VCaP and found that the autoregulatory network between AR and NKX3-1 also occurs in these cells (see Fig. S11 in the supplemental material). Interestingly, when we looked at the binding of AR and NKX3-1 in LNCaP and VCaP cells in closer detail, we found that these two factors are colocalized together at both the AR and NKX3-1 genes (Fig. 3A, B, E, and F; see Fig. S11A and C in the supplemental material). Taken together, our results reveal a highly integrated transcriptional network between AR and NKX3-1 in which these factors collaborate together to directly regulate not only downstream targets but also the expression of each other.
NKX3-1 converges at ARBS with FoxA1 to form functional androgen-regulated enhancers across the prostate cancer genome.
To understand how NKX3-1 collaborates with AR, we decided to search for additional players that might work in conjunction with NKX3-1 in mediating androgen-dependent transcription. To do this, we performed CentDist analysis on the NKX3-1 ChIP-seq peaks to screen for motifs with good center-of-distribution scores around NKX3-1. As expected, NKX motifs were highly ranked, and since the majority of NKX3-1 binding sites are colocalized with AR binding events, we also observed a good center-of-distribution score for ARE motifs (Fig. 4A). Surprisingly, the highest-ranked center-of-distribution score belonged to Forkhead motifs. Similar results were also obtained when we examined the NKX3-1 ChIP-seq peaks using de novo motif algorithms, including MEME and Amadeus (44) (see Fig. S12 in the supplemental material). Taken together, our results suggest that NKX3-1 may function together with members of the Forkhead family in regulating the transcriptional activity of AR.

Androgen stimulates the formation of a multitranscriptional complex consisting of NKX3-1, FoxA1, and AR. (A) CentDist result for the top cooccurring transcription factor family motifs within DHT-stimulated NKX3-1 ChIP-seq peaks. (B) Heat map of AR (red), NKX3-1 (blue), and FoxA1 (green) ChIP-seq tag intensity sorted according to NKX3-1 (DHT) tag intensity (top to bottom, highest to lowest) and centered on AR (DHT) peaks in a 2-kb window. Numbers 1 to 4 indicate groups of common or unique NKX3-1 binding sites identified: 1, AR/NKX3-1/FoxA1; 2, AR/NKX3-1; 3, NKX3-1/FoxA1; 4, NKX3-1 only. (C) Proportion of FoxA1 binding sites cobound by AR and/or NKX3-1 upon DHT stimulation. (D) Screenshots of AR, NKX3-1, FoxA1, and RNA Pol II ChIP-seq peaks around the PSA gene. (E) ChIP-qPCR of AR, NKX3-1, and FoxA1 occupancy at the PSA enhancer and promoter regions. The data represent the means ± SEM from at least 3 independent assays. (F) Reciprocal sequential ChIP re-ChIP was performed for AR, FoxA1, NKX3-1, or IgG. Fold enrichment represents the relative abundance of the PSA enhancer region compared to a randomly selected genomic control site. The data show the means ± SEM from at least 3 independent assays. (G) Androgen-deprived LNCaP cells were treated with EtOH or 100 nM DHT for 24 h prior to immunoprecipitation of whole-cell lysates with the indicated antibody or IgG, followed by Western blot (WB) analysis with the indicated antibody.
Recently, FoxA1, a member of the Forkhead family, was reported as an AR pioneer factor (26, 67). Moreover, FoxA1 was proposed to determine the lineage-specific recruitment of AR in prostate cancer cells (46). However, whether NKX3-1 functions together with FoxA1 in regulating AR-dependent transcription in prostate cancer cells has not been examined. Therefore, to address the potential interplay among these factors, we performed ChIP-seq analysis of FoxA1 in LNCaP cells before and after DHT stimulation and integrated these results with the ChIP-seq maps of AR and NKX3-1. We identified a total of 79,975 and 61,273 FoxA1 binding sites prior to and after DHT stimulation, respectively (see Table S1 and Fig. S13 in the supplemental material). When we examined FoxA1 localization with respect to AR and NKX3-1, we found that the majority of NKX3-1 and AR overlapping binding sites also have FoxA1 bound in close proximity (Fig. 4B; see Fig. S14 in the supplemental material). In agreement with previous studies, our ChIP-seq results also showed that FoxA1 binding to chromatin in general is independent of DHT stimulation (Fig. 4B; see Fig. S13 in the supplemental material). Moreover, we found that FoxA1 was more enriched at genomic regions cooccupied by AR and NKX3-1 than at unique AR or NKX3-1 binding sites (Fig. 4C). Taken together, our results suggest that NKX3-1 could potentially function together with FoxA1 in AR-dependent transcription.
Next, we examined whether NKX3-1 functions together with FoxA1 and AR on chromatin. For this, we carried out sequential ChIP re-ChIP assays by immunoprecipitating chromatin with antibodies recognizing AR, FoxA1, or NKX3-1, followed by immunoprecipitating the eluant with a second antibody against one of the remaining two proteins or IgG as a control. We then examined DNA enrichment at the enhancer ARBS of PSA, which has colocalization of all three factors (Fig. 4D and E). As shown in Fig. 4F, the PSA enhancer region was significantly enriched after DHT treatment in all the ChIP re-ChIP combinations tested, suggesting that NKX3-1 is bound at the enhancer of PSA at the same time with AR and FoxA1 after androgen stimulation. We also obtained similar results at three other genomic regions where ChIP-seq showed the colocalization of all three transcription factors (see Fig. S6C to E and S15 in the supplemental material). Taken together, our results suggest that NKX3-1 is recruited to ARBS with FoxA1 to regulate androgen-dependent transcription.
Previous studies showed that AR and FoxA1 are physically associated in LNCaP cells (26, 67). Our results described above suggest that NKX3-1 could also be another component of this protein complex. To test this, we performed coimmunoprecipitation assays in LNCaP cells with NKX3-1, AR, and FoxA1. As reported previously, we observed an androgen-independent association between AR and FoxA1 (Fig. 4G, top panel). More importantly, we found that NKX3-1 interacted with both AR (Fig. 4G, middle panel) and FoxA1 (Fig. 4G, bottom panel) in an androgen-dependent manner. Taken together, our results show that NKX3-1 is physically associated with AR and FoxA1 as a ternary complex and further suggest that NKX3-1 is an important collaborative factor in modulating the transcriptional activity of AR.
Our results thus far indicate an androgen signaling program that consists of an autoregulatory loop between AR and NKX3-1 in which each factor affects the other's transcript and protein levels. Because of this, we could not use a traditional siRNA knockdown strategy to examine the direct contributions of NKX3-1 in AR-mediated transcription in LNCaP cells. As an alternative approach, we instead introduced exogenous NKX3-1 in various combinations with AR and FoxA1 in the prostate cancer cell line PC3 (which expresses no or low endogenous levels of these factors) together with a luciferase reporter containing an upstream ARBS harboring motifs for all three factors. We decided to use the luciferase reporter ARBS8 (Fig. 1G), since FoxA1 and NKX3-1 bind strongly to this region (see Fig. S6F in the supplemental material) and it is highly androgen responsive. As shown in Fig. 5A and B, AR alone was sufficient to enhance the transcriptional activity of the ARBS8 luciferase reporter in a ligand-dependent manner. The addition of either NKX3-1 or FoxA1 only slightly increased the transcription activity of AR; however, when NKX3-1 was coexpressed with FoxA1, the transcriptional activity was increased significantly. Besides overexpression studies, we also explored the contribution of NKX3-1 and FoxA1 in AR-dependent transcription by mutating their cognate motifs in the ARBS8 luciferase construct and testing them in LNCaP cells and PC3 cells cotransfected with NKX3-1, AR, and FoxA1. As shown in Fig. 5C and D, mutating either the NKX3-1 or FoxA1 motif in the ARBS resulted in a drop in AR-dependent transcription similar to that after as mutating the ARE motif. Taken together, our results suggest that NKX3-1 facilitates AR transcriptional activity by partnering with FoxA1.

NKX3-1 is required for AR-dependent transcription and prostate cancer cell survival. (A) Schematic diagram of reporter constructs generated and used in transient-transfection assays. (B) Wild-type (WT) ARBS reporter plasmid was cotransfected with expression constructs for AR, FoxA1, and/or NKX3-1 into androgen-deprived PC3 cells. Cells were treated with EtOH or 100 nM DHT for 24 h and assessed for luciferase activity. (C) Mutations were generated at either the ARE, FKH, or NKX motifs in the ARBS construct as shown in panel A. The mutant reporter constructs were transfected and assayed for luciferase activity as described for panel B. (D) Wild-type and mutant ARBS constructs as shown in panel A were transfected into androgen-deprived LNCaP cells. The cells were treated with EtOH or 100 nM DHT for 24 h and assessed for luciferase activity. The data presented in panels B to D are mean RLU ± SEM from triplicates. (E) AR, FoxA1, and NKX3-1 were silenced with siRNA in various combinations in LNCaP cells under EtOH or 100 nM DHT treatment for 72 h. Cells were stained with propidium iodide and analyzed using flow cytometry. The analysis is presented as the percentage of cell apoptosis based on the percentage of the total number of gated cells in the sub-G1 phase. Data are presented as the means ± SEM from triplicate experiments. Statistical analysis was performed for differences between EtOH and DHT conditions using Student's t test (**, P < 0.05). (F) LNCaP cells were depleted of AR, NKX3-1, or FoxA1 in various combinations as described for panel E and assayed for caspase-3 activity. Harvested cell pellets were fixed and immunostained with FITC-conjugated antibody recognizing the active form of caspase-3. The flow cytometry analysis is represented as relative caspase-3 activity based on the percentage of the total number of gated cells with active caspase-3 upon transfection of target-specific siRNA compared to control siRNA. Data are presented as the means ± SEM from triplicate experiments. Statistical analysis was performed for the differences between EtOH and DHT conditions using Student's t test (**, P < 0.05).
NKX3-1 collaborates with AR to regulate apoptosis of prostate cancer cells.
Numerous studies have shown that AR is required for promoting prostate cancer cell survival (10, 42, 72) and proliferation (18, 22, 23, 70, 74). However, the role of NKX3-1 in prostate cancer biology is less certain. Previous reports showed that the NKX3-1 gene is a tumor suppressor gene in prostate cancer cells, but recent evidence suggests that it may also have oncogenic properties. The results from our genome-wide and molecular studies support the latter and suggest NKX3-1 functions together with AR to promote either cell proliferation or prostate cancer cell survival. To examine the role of NKX3-1 in prostate cancer, we depleted NKX3-1 in LNCaP cells with siRNA individually or in combination with either AR and/or FoxA1 and subsequently stained the cells with propidium iodide for fluorescence-activated cell sorting (FACS) analysis. We found that silencing of NKX3-1 and/or AR had minimal effect on S-phase entry (see Fig. S23 in the supplemental material). Interestingly, FoxA1 knockdown also did not significantly affect LNCaP cells entering into S phase. This result is in contrast to a recent report which showed that silencing of FoxA1 increased cell proliferation by promoting entry into S phase (65). The discrepancy between the studies could be due to numerous reasons, including differences in experimental conditions.
Although no effect on S-phase entry was found, depleting NKX3-1 alone was sufficient to cause an increase in the population of sub-G1 cells (Fig. 5E; see Fig. S16A in the supplemental material). Moreover, the depletion of all three factors resulted in the greatest cell death. To determine whether the accumulation of sub-G1 cells after siRNA knockdown of the three factors was due to apoptosis, we measured the level of caspase-3 activity via flow cytometry. Our results showed that knockdown of NKX3-1, AR, and FoxA1 resulted in caspase-3 activities that were very similar to the FACS result, suggesting that cell death caused by the loss of the three factors was mainly a result of apoptosis (Fig. 5F; see Fig. S16B in the supplemental material). Interestingly, we observed that DHT generally caused a decrease in cell death (Fig. 5E and F), which may indicate that androgen signaling has a potential role in promoting cell survival through transcriptional regulation. The effect of NKX3-1, AR, and FoxA1 on prostate cancer survival was also similar in VCaP cells (see Fig. S17 in the supplemental material). Taken together, our results suggest that NKX3-1 is a prosurvival factor that functions together with AR and FoxA1 to suppress the apoptotic pathway in prostate cancer cells.
NKX3-1 upregulates RAB3B to promote prostate cancer cell survival.
To define the NKX3-1 and AR regulatory network in further detail and identify the pathway in which NKX3-1 is linked to prostate cancer cell survival, we decided to examine the direct downstream target genes that are associated with both factors. As our GO analysis on NKX3-1- and AR-coassociated genes (Fig. 2E) already indicated, they are significantly more enriched in categories for “protein trafficking” (i.e., protein transport and intracellular protein transport), a process that is necessary in the integration of oncogenic signaling pathways. Interestingly, when we looked into this group of genes further, we found that a large number of RAB genes were associated with NKX3-1 and AR. RAB genes belong to the Ras oncogene superfamily and encode small monomeric GTPase molecules frequently reported to mediate intracellular vesicle trafficking and organelle-targeted membrane fusion (29, 45, 69). Furthermore, emerging studies also suggest that RAB genes are important players in cancer, including the proliferation, survival, and aggressiveness in breast cancer invasion as well as the metastasis of breast tumor cells (14, 35, 66). Thus, it is possible that the prosurvival properties of NKX3-1 that we observed in our studies could be explained by NKX3-1 driving the AR transcription network to directly control the expression of RAB genes.
To determine the regulation and function of RAB genes in prostate cancer, we began by examining the expression of RAB genes in LNCaP cells before and after androgen stimulation from our time course microarray analysis. Our results showed that DHT differentially regulated the expression of all the RAB genes in the “protein trafficking” category (Fig. 6A). Next, we checked the expression of these genes in clinical studies and found that they are also differentially expressed in prostate tumors compared to the normal counterparts (Fig. 6B and C; see Fig. S18 in the supplemental material). One gene that particularly drew our attention was the RAB3B gene, which we noticed was highly responsive and upregulated in LNCaP cells (Fig. 6A) and in comparison to other RAB genes was consistently overexpressed in multiple prostate cancer studies (Fig. 6B; see Fig. S18 in the supplemental material). In addition, overexpression of the RAB3B gene was restricted to prostate cancer, which is similar to what has been observed for NKX3-1 (Fig. 6C and data not shown). Finally, the expression profile of RAB3B also correlated with NKX3-1 and AR in at least two clinical studies, suggesting that the regulation of RAB3B transcript level in prostate tumors is likely dependent on AR and NKX3-1 (Fig. 6B).

NKX3-1 promotes prostate cancer cell survival by collaborating with AR and FoxA1 to regulate RAB3B gene expression. (A) Expression profile of NKX3-1- and AR-associated RAB GTPase genes that are androgen regulated, from our microarray analysis of LNCaP cells. Induction and repression are represented by yellow and red, respectively. All RAB genes are cobound by AR, NKX3-1, and FoxA1, unless otherwise stated. (B) Left panel, transcript levels of RAB GTPase genes across multiple Oncomine studies. Red and green represent over- and underexpression in prostate adenocarcinoma versus normal samples, respectively. NA denotes lack of expression data for the respective gene in the annotated study. * and **, P < 0.1 and P < 0.05, respectively. Right panel, box plot comparing the transcript levels of RAB3B in normal prostate and tumor samples from the study by Wallace et al. in the Oncomine database. The differential gene expression data are centered on the median of expression levels and expressed on a log2 scale. The P value was calculated using a Welch two-sample t test. (C) Box plot illustrating RAB3B transcript levels across different cancer types from the study by Bittner et al. in the Oncomine microarray database. The differential gene expression data are centered on the median of expression levels and expressed on a log2 scale. Parentheses depict the number of samples within each category. (D) Screenshots of AR, NKX3-1, FoxA1, and RNA Pol II ChIP-seq peaks around the RAB3B gene. (E) ChIP-qPCR of AR, NKX3-1, and FoxA1 occupancy at the RAB3B enhancer region. The data represent means ± SEM from at least 3 independent assays. (F) Reciprocal re-ChIP at the RAB3B enhancer was performed as described for Fig. 4F. (G) Transient-transfection analysis was performed as described for Fig. 5B with a reporter plasmid containing the wild-type RAB3B ARBS. (H) The half ARE, FKH, and NKX motifs in the ARBS construct from panel G were mutated and used in transient-cotransfection analysis on LNCaP cells as described for Fig. 5D. (I) Wild-type and mutant ARBS constructs from panel H were cotransfected with expression constructs for AR, FoxA1, and NKX3-1 into androgen-deprived PC3 cells as described for Fig. 5D. Data shown in panels G to I are mean RLU ± SEM for triplicates. (J) Two different siRNA duplexes were designed to specifically and independently target the RAB3B gene in LNCaP cells. Knockdown efficiency was assessed by Western blotting. (K) PI staining (left panel) and caspase-3 activity (right panel) as described for Fig. 5E and F on LNCaP cells after RAB3B knockdown. For both assays, data are represented as means ± SEM from triplicate experiments. Statistical analysis was performed for the differences between EtOH and DHT conditions using Student's t test (**, P < 0.05).
Next, we investigated whether NKX3-1 and AR are indeed directly linked to the Ras oncogenic pathway via RAB3B. For this we functionally characterized the NKX3-1 and AR binding site that is associated with the RAB3B gene. As shown in Fig. 6D, NKX3-1, AR, and FoxA1 are colocalized at an intronic region 42 kb downstream from the TSS of the gene. We validated the binding of the three factors at this site by ChIP-qPCR (Fig. 6E) and showed they all converge together as one complex after DHT stimulation (Fig. 6F). We also cloned the ARBS of RAB3B upstream of a luciferase reporter and tested it for enhancer activity in PC3 cells cotransfected with NKX3-1, AR, and FoxA1. As shown in Fig. 6G, NKX3-1 was able to stimulate AR-dependent transcription, and this was further enhanced in the presence of FoxA1. We also mutated the NKX3-1, AR, and FoxA1 motifs within the RAB3B ARBS and examined them in LNCaP (Fig. 6H) and PC3 (Fig. 6I) cells. Androgen robustly stimulated luciferase activity of the reporter construct harboring the wild-type RAB3B but not the mutant constructs. We also examined the expression of RAB3B after depleting NKX3-1 and AR. Although this result alone will not address whether these factors directly regulate RAB3B, we should still see an effect on this gene if it is regulated by these factors. Indeed, silencing of NKX3-1 and AR (and FoxA1) reduced the androgenic upregulation of both the transcript and protein levels of RAB3B (see Fig. S19 in the supplemental material). Similar to our observations in LNCaP cells, we also noticed the downregulation of the RAB3B transcript level upon AR or NKX3-1 silencing in VCaP cells (see Fig. S20 in the supplemental material). However, even though FoxA1 is colocalized with AR and NKX3-1 at the RAB3B enhancer region in VCaP cells, depletion of FoxA1 had only minimal effects on RAB3B expression, which may be due to transcriptional redundancy among the Forkhead family members. Taken together, our findings suggest that NKX3-1 together with AR and FoxA1 directly regulates RAB3B transcription in LNCaP cells.
Our findings thus far support that prostate cancer cell survival is linked to androgen signaling via the transcriptional regulation of RAB3B by NKX3-1, AR, and FoxA1. To test whether RAB3B is indeed involved in prostate cell survival, we performed FACS analysis and caspase-3 assays on LNCaP cells treated with and without RAB3B siRNA. Our results showed that RAB3B depletion led to an increase in the population of sub-G1 cells as well as caspase-3 activity under androgen-deprived and serum conditions, suggesting that RAB3B is required for maintaining cell survival by suppressing the apoptosis pathway (Fig. 6J and K; see Fig. S21 in the supplemental material). Similar to the case with NKX3-1, AR, and FoxA1 (Fig. 5E and F), cell death was significantly decreased after DHT stimulation. Furthermore, the effect of RAB3B on cell survival was also observed in VCaP cells (see Fig. S22 in the supplemental material). Taken together, our combined genomic and functional analyses on androgen signaling in prostate cancer cells has established the essential cooperation between NKX3-1, AR, and FoxA1 in directly regulating genes involved in promoting and maintaining prostate cancer cell survival.
DISCUSSION
In the present study, we have used ChIP-seq to interrogate the global residency of AR in prostate cancer cells. Overall, we identified a large number of AR binding events (75,296) across the genome of LNCaP cells upon 2 h of DHT stimulation (Fig. 1A to C). Recently, Yu et al. also generated ARBS maps in the same cell line, but in contrast to our study, they reported 37,193 ARBS after 16 h of treatment with R1881 (73). When we compared the two maps, we found that approximately 73% of their ARBS overlapped with our data set (data not shown). Large discrepancies in the number of binding sites between genome-wide studies are not uncommon and could be due to numerous parameters, including differences in the concentration and length of androgen stimulation, as well as sequencing depth and peak calling programs used to define binding sites. Despite the difference in the number of binding sites, both studies in general suggest that AR is recruited to a large number of regions in the prostate cancer genome upon androgen stimulation.
Besides using high-throughput technology to determine the number of ARBS in the prostate cancer genome, several groups have tried to refine the sequence of the ARE motif (9, 48, 67). Wang et al. described the presence of alternative AREs, including full AREs with variable spacer lengths (67). However, from our data set we found evidence of only the strong enrichment of a palindromic ARE even when a 3-bp deviation was allowed from the consensus (see Fig. S2A in the supplemental material). The differences between these studies could be due to the small number of ARBS that were identified in the study by Wang et al. Thus, our global analysis of ARBS suggests that the palindromic full ARE with up to 3-bp mismatches from the consensus sequence is the most frequently occurring AR motif in the ARBS of prostate cancer cells.
The intricate relationship between normal cell development and carcinogenesis has long been under scrutiny (51). As they are master regulators of embryogenesis, the deregulation of homeobox genes has profound effects on the cellular phenotype and may lead to tissue neoplasia (55, 57). This is consistent with the observation that a number of solid tumors have aberrant expression of homeobox genes, which could encode transcription factors that have oncogenic or tumor-suppressing properties (55, 57). NKX3-1, which is the first known marker for prostate epithelial differentiation (7, 40), is one of the few homeodomain proteins associated with PCa and that functions during prostate epithelial regeneration in stem cells (68). Although NKX3-1 has long been postulated to be a prostate-specific tumor suppressor, recent studies suggest that it may also support prostate cancer cell survival. Here, using genome-wide, molecular, and cell-based approaches, we identified NKX3-1 as a novel regulator of androgen signaling that is necessary in the formation of active enhancers and the regulation of AR target genes involved in prostate cancer survival. During the course of this study, He et al. reported the enrichment of NKX3-1 motifs at ARBS using dimethylated H3K4-marked nucleosomal ChIP-seq information (31) and showed NKX3-1 binding at several genomic regions. From the global analysis of AR and NKX3-1 binding, our work showed that NKX3-1 is found almost exclusively at ARBS upon androgen stimulation (Fig. 2C and and4B).4B). We also found that more than 90% of the NKX3-1 binding sites that contain AR are also colocalized with FoxA1 in close proximity, suggesting dynamic interplay between these three factors in prostate cancer (Fig. 4C; see also Fig. S14 in the supplemental material). We demonstrated that NKX3-1 is a positive regulator of AR transcriptional activity (Fig. 5B to D) and showed that NKX3-1 works synergistically with FoxA1 to modulate AR-mediated transcription of the RAB3B GTPase gene, which is essential in prostate cancer cell survival (Fig. 6; see Fig. S19 to S22 in the supplemental material). Overall, our study implicates NKX3-1 as novel collaborative factor of AR and a prosurvival factor in prostate cancer.
The NKX3-1 gene is a well-known androgen-regulated gene, and from our ChIP-seq profile of AR this is likely through the direct binding of AR to two regions located at +2 and +39 kb downstream of the TSS of the gene (Fig. 3A and B). However, the regulation of AR by NKX3-1 is less clear. For example, NKX3-1 has been reported not only to activate but also to repress the expression of AR (41, 52). From our ChIP-seq analysis of NKX3-1, we observed enhanced binding of NKX3-1 to an intronic region of AR (Fig. 3E and F). Moreover, in knockdown experiments using two independent siRNAs, we showed that NKX3-1 is required for upregulating the expression of AR in LNCaP cells (Fig. 3E to H; see Fig. S10C and D in the supplemental material). The inconsistent results for AR regulation by NKX3-1 could be due to differences in experimental conditions or to different cellular contexts. Despite these differences, our work has provided for the first time a direct physical link between NKX3-1 and AR, and it will be of interest in future studies to understand this feed-forward loop in greater detail.
NKX3-1 has long been proposed to be a tumor suppressor in PCa. Besides NKX3-1 residing on an allelic hot spot which frequently undergoes loss of heterozygosity in the majority of prostate tumors (1, 5, 32, 63, 64), stable knockdown of NKX3-1 has also been shown to promote LNCaP cell viability and proliferation, metastasis of PCa cells to the lymph node, and murine tumor growth (47, 75). Furthermore, Bhatia-Gaur et al. showed that NKX3-1 mutant mice were more prone to prostatic intraepithelial neoplasia (PIN) lesion formation, which is the precursor of PCa initiation (7). Nonetheless, recent emerging evidence, including our work, suggests that the role of NKX3-1 in PCa is still debatable. In contrast to the classical tumor suppressors, NKX3-1 is not regulated in a genetic manner whereby the remaining 8p monoallele is hardly lost and mutation of its coding region is not observed (6). Instead, downregulation of the NKX3-1 protein level may be attributed to selective CpG island methylation and protein degradation (3). NKX3-1 is haploinsufficient, and manifestation of prostate tumorigenesis requires the presence of other oncogenic alterations (27, 50). From this study, we showed that NKX3-1 directly regulates AR-dependent genes which are overexpressed in prostate carcinoma, deregulated in advanced PCa, and enriched in recurrent PCa, which indicates that NKX3-1 has a pivotal role in prostate tumorigenicity (Fig. 2F). We also found that NKX3-1 likely regulates genes that are active in promoting cell survival or preventing cell apoptosis (Fig. 2 and and66).
RAB GTPases have recently been implicated in signal transduction pathways and intrinsic cellular processes, including cell growth, proliferation, differentiation, survival, and the cell cycle (14). For instance, amplification of the Rab25 gene promotes proliferation, survival, and aggressiveness in breast and ovarian cells (14), while overexpression of Rab27 secretory proteins is associated with invasion and metastasis of breast cancer cells and poor clinical prognosis (35, 66). From our work, we found that the RAB3B gene, which is coregulated by NKX3-1 and AR, is consistently enriched in prostate carcinoma from multiple studies, suggesting that it may be functionally important in PCa (Fig. 6B; see Fig. S18 in the supplemental material). Indeed, depletion of RAB3B resulted in significant cell death (Fig. 6K; see Fig. S21 and S22 in the supplemental material). Hence, our work shows that RAB3B is a critical component of the PCa cell survival pathway. Interestingly, we also found that NKX3-1 and AR are associated with several other members of the RAB GTPase family (Fig. 6A and B). Whether these RAB GTPases are also important in PCa cell survival will be examined in future studies.
The complex dynamics of transcriptional interplay between two or more transcription factors are intriguing but not yet well understood. From our current work, we report an intimate hierarchical transcriptional interplay between AR, NKX3-1, and FoxA1 within “enhanceosomes.” We further showed that NKX3-1, AR, and FoxA1 promote PCa cell viability in a concerted manner, and we propose NKX3-1 to be an essential factor in prostate cancer carcinogenesis which prevents cell apoptosis either by direct modulation of gene targets or by indirect regulation of AR expression (Fig. 7). Interestingly, our work also revealed that FoxA1 may have dual, opposing actions in prostate cancer. While our work suggests that FoxA1 promotes PCa progression, a recent report by Wang et al. showed that loss of FoxA1 facilitates S-phase entry and that low FoxA1 expression is associated with a poor prognosis (65). How FoxA1 can have opposing roles in prostate cancer is unclear, but it is likely that the level of FoxA1 expression is critical in determining which pathway FoxA1 takes. Finally, our work has also delineated a novel functional role for RAB3B GTPase in PCa. Taking our results together, we propose that the convergence at RAB3B may mediate cross talk between AR signaling and other predominant survival signaling pathways and should therefore be considered for PCa therapeutics.

NKX3-1 regulates the AR transcriptional network by two distinct mechanisms. 1. In the presence of DHT, NKX3-1, which is androgen responsive, directly activates the expression of AR to constitute a feed-forward regulatory mechanism between the two factors. 2. NKX3-1 together with AR and FoxA1 form an “enhanceosome” that is recruited to specific genomic elements upon androgen signaling. This allows for the subsequent loading of histone modifiers and chromatin remodelers which facilitate downstream gene activities.
ACKNOWLEDGMENTS
We thank the Genome Technology and Biology group and the IT group at GIS for sequencing support.
This work was supported by the Biomedical Research Council/Science and Engineering Research Council of A*STAR (Agency for Science, Technology and Research), Singapore. P.Y.T., C.W.C., and K.R.C. are supported by A*STAR graduate scholarships.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
Articles from Molecular and Cellular Biology are provided here courtesy of Taylor & Francis
Full text links
Read article at publisher's site: https://doi.org/10.1128/mcb.05958-11
Read article for free, from open access legal sources, via Unpaywall:
https://mcb.asm.org/content/32/2/399.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/143200146
Article citations
The Use of Next-Generation Sequencing in Personalized Medicine.
Methods Mol Biol, 2866:287-315, 01 Jan 2025
Cited by: 0 articles | PMID: 39546209
Review
Rab3B Proteins: Cellular Functions, Regulatory Mechanisms, and Potential as a Cancer Therapy Target.
Cell Biochem Biophys, 25 Sep 2024
Cited by: 0 articles | PMID: 39320613
Review
R-loop resolution by ARIP4 helicase promotes androgen-mediated transcription induction.
Sci Adv, 10(29):eadm9577, 19 Jul 2024
Cited by: 0 articles | PMID: 39028815 | PMCID: PMC11259169
Rab3B enhances the stabilization of DDX6 to promote lung adenocarcinoma aggressiveness.
Mol Med, 30(1):75, 04 Jun 2024
Cited by: 1 article | PMID: 38834947 | PMCID: PMC11151598
Pan-cancer analysis reveals correlation between RAB3B expression and tumor heterogeneity, immune microenvironment, and prognosis in multiple cancers.
Sci Rep, 14(1):9881, 30 Apr 2024
Cited by: 2 articles | PMID: 38688977 | PMCID: PMC11061125
Go to all (113) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus (2)
- (1 citation) GEO - GSE28264
- (1 citation) GEO - GSE28596
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Androgen regulation of the prostatic tumour suppressor NKX3.1 is mediated by its 3' untranslated region.
Biochem J, 425(3):575-583, 15 Jan 2010
Cited by: 20 articles | PMID: 19886863
Transcriptional network involving ERG and AR orchestrates Distal-less homeobox-1 mediated prostate cancer progression.
Nat Commun, 12(1):5325, 07 Sep 2021
Cited by: 18 articles | PMID: 34493733 | PMCID: PMC8423767
Fibroblast-derived exosomal microRNA regulates NKX3-1 expression in androgen-sensitive, androgen receptor-dependent prostate cancer cells.
J Cell Biochem, 124(8):1135-1144, 19 Jun 2023
Cited by: 2 articles | PMID: 37334663
Shaping Chromatin States in Prostate Cancer by Pioneer Transcription Factors.
Cancer Res, 80(12):2427-2436, 24 Feb 2020
Cited by: 40 articles | PMID: 32094298 | PMCID: PMC7299826
Review Free full text in Europe PMC
 a,d,e
a,d,e



