Abstract
Free full text

Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs
Associated Data
SUMMARY
Small RNAs target invaders for silencing in the CRISPR-Cas pathways that protect bacteria and archaea from viruses and plasmids. The CRISPR RNAs (crRNAs) contain sequence elements acquired from invaders that guide CRISPR-associated (Cas) proteins back to the complementary invading DNA or RNA. Here, we have analyzed essential features of the crRNAs associated with the Cas RAMP module (Cmr) effector complex, which cleaves targeted RNAs. We show that Cmr crRNAs contain an 8-nucleotide 5’ sequence tag (also found on crRNAs associated with other CRISPR-Cas pathways) that is critical for crRNA function and can be used to engineer crRNAs that direct cleavage of novel targets. We also present data that indicates that the Cmr complex cleaves an endogenous complementary RNA in Pyrococcus furiosus, providing direct in vivo evidence of RNA targeting by the CRISPR-Cas system. Our findings indicate that the CRISPR RNA-Cmr protein pathway may be exploited to cleave RNAs of interest.
INTRODUCTION
The CRISPR-Cas systems are RNA-based immune systems that protect prokaryotes from viruses, plasmids, and other invaders (see recent general reviews: (Deveau et al., 2010; Horvath and Barrangou, 2010; Jore et al., 2011a; Karginov and Hannon, 2010; Makarova et al., 2011; Marraffini and Sontheimer, 2010; Terns and Terns, 2011; van der Oost et al., 2009)). The CRISPR (Clustered Regularly Interspaced Short Palindromic Repeat) loci found in prokaryotic genomes acquire short fragments of invader sequence (~30-40 base pairs in length) that are inserted between short CRISPR repeat sequences. The CRISPR loci give rise to CRISPR RNAs (crRNAs) that each contain an invader-derived (guide) sequence and interact with CRISPR-associated or Cas proteins to form effector complexes that recognize and silence the corresponding invader (Brouns et al., 2008; Hale et al., 2008; Hale et al., 2009; Jore et al., 2011b; Lintner et al., 2011; Wiedenheft et al., 2011). There are multiple modules of Cas proteins (termed for example Cse and Csy proteins) that can function independently with crRNAs to effect CRISPR-Cas defense (Haft et al., 2005; Makarova et al., 2006; Makarova et al., 2011; Terns and Terns, 2011); thus distinct CRISPR-Cas pathways function in various prokaryotes.
CRISPR-Cas-mediated silencing appears to occur through sequence-specific targeting of RNA or DNA. In Streptococcus thermophilus (Barrangou et al., 2007; Garneau et al., 2010), Escherichia coli (Brouns et al., 2008; Semenova et al., 2011), Staphylococcus epidermidis (Marraffini and Sontheimer, 2008), and Sulfolobus solfataricus (Gudbergsdottir et al., 2011; Manica et al., 2011), there is evidence that CRISPR-Cas systems target foreign DNA. Indeed, sequence-specific cleavage of both plasmid and bacteriophage DNA was recently documented in S. thermophilus (Garneau et al., 2010). At the same time, the Cmr-type crRNA-Cas protein complex from Pyrococcus furiosus specifically cleaves complementary RNAs (Hale et al., 2009). Thus, the various CRISPR-Cas systems (formed by distinct modules of Cas proteins) may function by different mechanisms, including DNA and RNA cleavage. Interestingly, many organisms possess more than one module of Cas proteins and may resist invaders by multiple mechanisms to provide robust protection from diverse invaders.
CRISPR RNAs (previously also referred to as prokaryotic silencing (psi)RNAs) are utilized by all versions of the CRISPR-Cas system characterized to date (Brouns et al., 2008; Deltcheva et al., 2011; Gudbergsdottir et al., 2011; Hale et al., 2009; Lintner et al., 2011; Manica et al., 2011; Marraffini and Sontheimer, 2008; Wiedenheft et al., 2011). The CRISPR loci are transcribed from a leader sequence region of 100-500 basepairs found at one end of each locus (Jansen et al., 2002), where promoter sequences have been identified in a few organisms (Lillestol et al., 2009; Pul et al., 2010). Insertion of new invader-derived sequences into CRISPR loci most frequently occurs immediately downstream of the leader (Andersson and Banfield, 2008; Barrangou et al., 2007; Pourcel et al., 2005; Shah et al., 2009). The CRISPR locus transcripts are cleaved within the repeat sequence to release the individual embedded crRNAs. A series of structurally related proteins from various CRISPR-Cas systems process CRISPR RNA transcripts: Cas6 (Carte et al., 2010; Carte et al., 2008; Wang et al., 2011), Cse3 (Brouns et al., 2008; Gesner et al., 2011; Sashital et al., 2011), and Csy4 (Haurwitz et al., 2010; Przybilski et al., 2011). Cleavage within the repeats by these Cas endonucleases produces 1X intermediate RNAs generally comprised of 8 nucleotides of repeat sequence (called the 5’ tag), a guide sequence, and the remaining ~20-25 nucleotides (nt) of repeat sequence at the 3’ end (the 3’ tag) (Brouns et al., 2008; Gesner et al., 2011; Hale et al., 2008; Haurwitz et al., 2010). These 1X intermediates undergo various amounts of 3’ end trimming (Hale et al., 2008; Hale et al., 2009; Jore et al., 2011b; Lintner et al., 2011; Marraffini and Sontheimer, 2008; Wiedenheft et al., 2011). However, an equivalent 8-nt 5’ tag is present on mature crRNAs from a variety of bacterial and archaeal species including E. coli (Brouns et al., 2008), S. epidermidis (Marraffini and Sontheimer, 2008), Pseudomonas aeruginosa (Haurwitz et al., 2010), P. furiosus (Hale et al., 2008; Hale et al., 2009), S. solfataricus ((Lintner et al., 2011), our unpublished data), Sulfolobus tokadai (our unpublished data), and Thermococcus kodakarensis (our unpublished data). In S. epidermis, the 5’ tag of the crRNAs provides for discrimination between self (i.e., the complementary DNA strand in the CRISPR locus, which can basepair with the tag) and non-self (invader DNA, which does not basepair with the tag) (Marraffini and Sontheimer, 2010), but other functions of this crRNA signature sequence are not known.
There are 3 modules of Cas proteins (Cmr, Csa, and Cst) and 7 CRISPR loci encoded in the genome of the hyperthermophilic archaeon P. furiosus. Previously, we identified and characterized a complex comprised of the Cas module RAMP or Cmr proteins (Cmr1-6) and 2 crRNA species in P. furiosus (Hale et al., 2009). The Cmr-crRNA protein complex is unique among CRISPR-Cas systems characterized to date in that it targets RNA; the complex specifically cleaves RNAs that are complementary to the crRNAs, 14 nt from the 3’ end of the crRNAs (Hale et al., 2009). Activity was observed in vitro using both native and reconstituted complexes, but has not been described in vivo. Here we have investigated the functionality of the crRNAs that direct cleavage by the Cmr complex. Deep sequencing of the RNAs associated with the complex as well as total crRNAs unexpectedly revealed the cleavage of a fortuitous endogenous target RNA by the Cmr complex in vivo. While multiple species of crRNAs are present in P. furiosus, the Cmr complex selectively includes 45- and 39-nt crRNAs regardless of the size of the invader-derived guide sequence encoded in the genome. We show that the 8-nt 5’ tag sequence is not only necessary for Cmr-directed cleavage, but can be used to target the complex to cleave novel RNAs, providing the potential for manipulation of the CRISPR-Cas system for experimental or pharmaceutical gene silencing.
RESULTS
Key features of the Cmr complex RNAs
To delineate functional features of the crRNAs that guide cleavage by the Cmr complex, we isolated active Cmr complexes using antibodies against the Cmr2 protein. Polyclonal antibodies raised against recombinant P. furiosus Cmr2 specifically immunoprecipitated Cmr2 (assessed by Western blotting, Figure 1A), as well as the five other Cmr complex proteins (Cmr1 and Cmr3-6; assessed by mass spectrometry, unpublished data), and 39- and 45-nt species of a crRNA (assessed by Northern blotting, Figure 1B). Moreover, the immunoprecipitated RNA-protein complex replicated the cleavage activity observed with both the chromatographically-purified and reconstituted Cmr complexes (Hale et al., 2009), cleaving a target RNA (complementary to one of the endogenous crRNAs) at the two sites located 14 nt upstream of the 3’ ends of the complementary 39- and 45-nt crRNA species (Figure 1C).
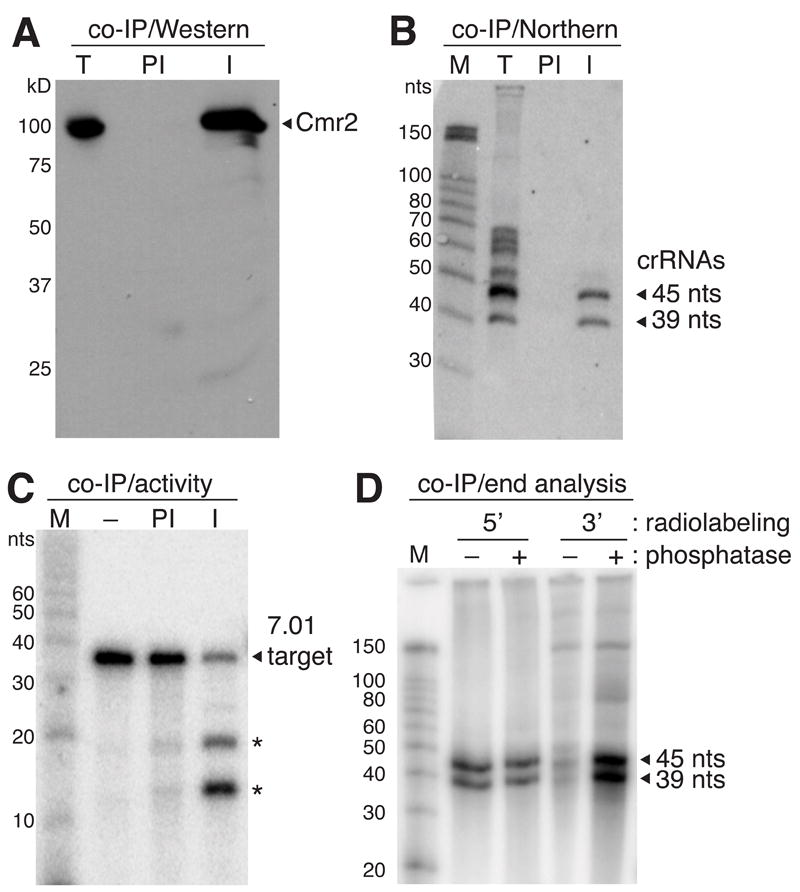
A) Cmr2 antibodies recognize and specifically immunoprecipitate ~100 kD protein. P. furiosus S100 extract (T) and PfCmr2 antibody immune (I) and pre-immune (PI) immunoprecipitation (IP) samples were analyzed by Western blotting with Cmr2 antibodies. Positions of size markers are indicated. ~100 kD protein specifically recognized by immunoblotting and immunoprecipitated by Cmr2 antibodies is indicated by arrow. B) 45- and 39-nt crRNAs are immunopurified with Cmr2 antibodies. RNAs extracted from P. furiosus S100 extract (T) and immune (I) and pre-immune (PI) immunoprecipitation (IP) samples were analyzed by Northern blotting with a probe against P. furiosus crRNA 7.01. The 45- and 39-nt species of 7.01 that are co-IPed with the Cmr2 antibodies are indicated by arrows. Sizes of RNA markers (M) are shown. C) Immunoprecipitated Cmr complexes cleave a target RNA. 5’ end-labeled 7.01 target RNA (complementary to crRNA 7.01, indicated with arrow) was incubated in the absence (−) of proteins or in the presence of immune (I) and pre-immune (PI) immunoprecipitation samples. Cleavage products are marked with asterisks. D) Co-immunopurified crRNAs possess 5’ OH and 3’ phosphate ends. Immunoprecipitated crRNAs were isolated and 5’ or 3’ labeled with (+) and without (−) prior phosphatase treatment. The 45- and 39-nt crRNA species are indicated by arrows and RNA size markers (M) are shown.
We probed the nature of the end groups of the crRNAs immunopurified with Cmr complexes by end radiolabeling. 5’ end labeling of the 39- and 45-nt RNAs was not affected by prior enzymatic treatment with phosphatase, whereas phosphatase pre-treatment significantly increased 3’ end labeling of the RNAs (Figure 1D), indicating that the majority of both species of crRNAs in the Cmr complex contain 5’ hydroxyl and 3’ phosphate end groups.
In order to further characterize the Cmr-associated crRNAs, we deep-sequenced the RNAs immunopurified with the complex, as well as total P. furiosus small RNAs. We isolated RNAs smaller than ~70 nt by gel purification, and based on the results of end group analysis (Figure 1D), treated the RNAs with phosphatase to optimize cloning. The cDNA libraries were subject to Illumina sequencing. Figure 2A shows the numbers of small RNA sequence reads that map to each of the seven P. furiosus CRISPR loci from the total RNA and Cmr complex samples. A total of 2,632,773 uniquely mapped small RNA reads were obtained from the total RNA sample, of which 54.5% mapped to the CRISPR loci. In contrast, 91.2% of the 990,060 uniquely mapped reads from the Cmr complex sample mapped to the CRISPR loci.
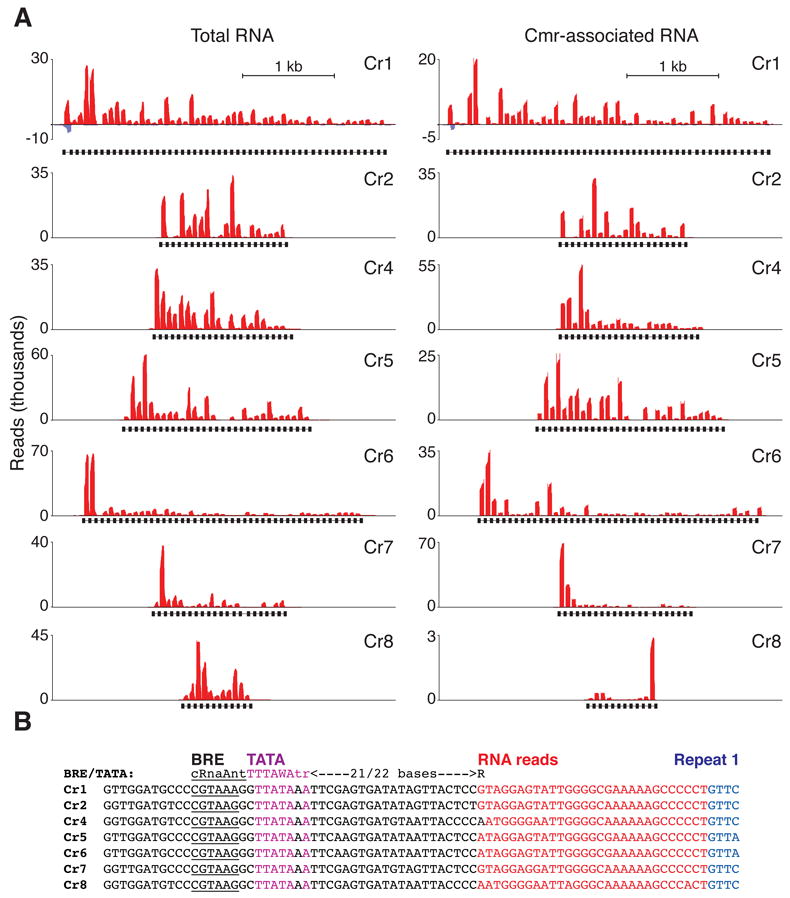
A) Total and Cmr-associated small RNA sequencing reads that map to the seven P. furiosus CRISPR loci. Y-axis scale bars indicate the number of reads (thousands) that contain nucleotides that map to the indicated position within the CRISPR locus. Red indicates sequence reads that map to the sense strand relative to the CRISPR leader region, and blue corresponds to reads that map to the antisense strand. The positions of CRISPR repeats are shown as black bars below the graph. 1 kb x-axis scale is indicated. Images were generated using the UCSC archaeal genome browser (Karolchik et al., 2003). B) Putative promoter sequences found in the P. furiosus CRISPR leader regions. Alignment of the regions upstream of the first repeat of each P. furiosus CRISPR locus are shown. The repeats are shown in blue, the initial transcribed regions (detected by sequencing) are shown in red, the potential TATA elements are shown in pink, the potential BRE elements are underlined, and other leader sequences are shown in black. Consensus BRE/TATA elements are shown above the alignment.
The total RNA profiles indicate that RNAs are expressed from all seven P. furiosus CRISPR loci (Figure 2A, total RNA) as limited sequencing data had previously suggested (Hale et al., 2009). We analyzed the leader sequences of the seven CRISPR loci and found canonical BRE/TATA promoter sequences at a conserved position within the leaders (Figure 2B). A population of RNAs was detected that begins ~21 nt downstream of these promoter elements (and includes ~29 nt of the leader sequence at the 5’ end, indicated in red in Figure 2B). (These RNAs do not represent the major crRNA species, which are described below.) These findings suggest that the CRISPR loci are transcribed by archaeal RNA polymerase via recognition of the BRE/TATA promoters within the leader sequences. The deep sequencing data also indicate that, in general, crRNAs from the leader-proximal regions of the CRISPR loci are more abundant than crRNAs encoded downstream, as had been hypothesized based on previous data (Hale et al., 2008). The molecular basis for the gradient in steady state levels of the crRNAs from each locus is currently unknown, but could reflect differences in transcription, processing, or stability. The expression gradient is noteworthy because the crRNAs that arise from the leader-proximal end of a CRISPR locus generally target the most recent invaders confronted by the system (Andersson and Banfield, 2008; Barrangou et al., 2007; Pourcel et al., 2005; Shah et al., 2009)
Deep sequencing and Northern analysis indicate that a series of species of crRNAs of different lengths are present in P. furiosus, and that the Cmr complex specifically loads two of these crRNA species. The 5’ ends of the vast majority of crRNAs in P. furiosus, in both the total and Cmr complex samples, are comprised of an identical 5’ end tag sequence (AUUGAAAG) derived from the upstream repeat element (Figure 3A). This end is generated by Cas6 cleavage of CRISPR transcripts at this site (Carte et al., 2010; Carte et al., 2008; Wang et al., 2011). In total RNA, a mixture of 3’ ends corresponding to crRNAs 30 to 70 nt in length is observed (Figure 3A), consistent with a series of RNA species detected in total RNA by Northern analysis of one of the crRNAs (Figure 1B, T lane). Strikingly, the 39- and 45-nt crRNA species selectively co-purify with the Cmr complex (Figures 1B and and3A).3A). The 45-nt crRNAs constitute 46% of the crRNAs associated with the Cmr complex, and the 39-nucleotide species make up 38% (Figure 3A).
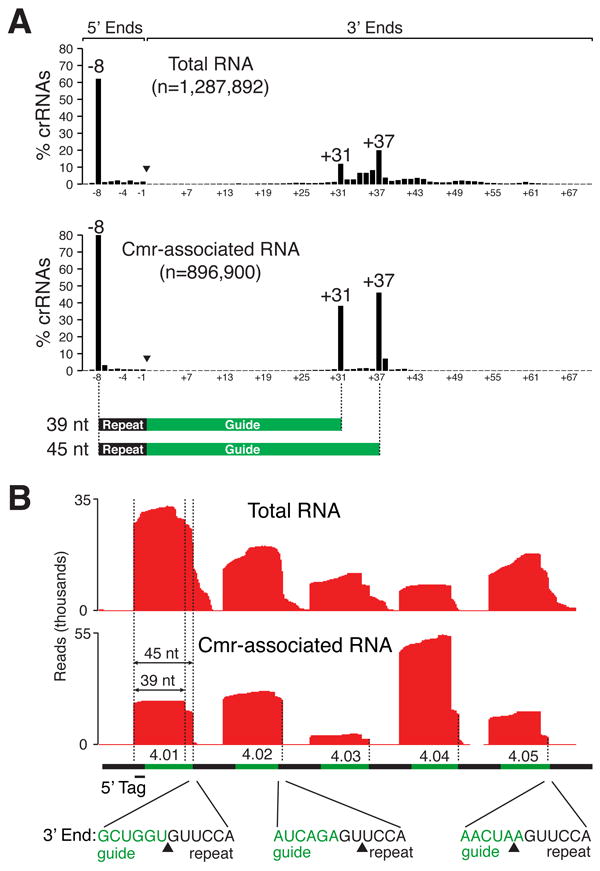
A) The 5’ and 3’ ends of Cmr-associated crRNAs are defined relative to the upstream repeat element. Graphs show the % of crRNAs sequenced in total P. furiosus RNA (upper panel) and immunopurified Cmr complexes (lower) with ends that map at indicated positions relative to the 5’ repeat-guide junction (arrowhead). The Cmr-associated crRNA species are illustrated below with 5’ repeat tag (black) and guide sequence (green) indicated. RNAs from CRISPR8 were excluded from this analysis due to a repeat sequence polymorphism (see text). Read numbers analyzed (n) are indicated. B) The length of the Cmr complex crRNAs is independent of the length of the genome-encoded guide sequence. Deep sequence profiles of crRNAs 4.01 – 4.05 in total P. furiosus RNA (upper panel) and immunopurified Cmr complexes (lower) are shown relative to repeat (black) and guide (green) sequence elements (see also Figure 2A). Dashed lines mark the 3’ ends of the 45-nucleotide crRNA species. (The 5’ end and 3’ end of the 39-nucleotide species are also indicated for crRNA 4.01.) The sequence at the 3’ guide-repeat junction is shown below for crRNAs 4.01, 4.02 and 4.05. Guide sequences are shown in green, and repeat sequences are shown in black. Arrowheads indicate the locations of the 3’ ends of the 45-nucleotide crRNA species.
Interestingly, the deep sequence analysis indicates that the 3’ ends of the Cmr complex-associated crRNAs are defined by distance from the 5’ tag or absolute length of the crRNA (rather than, for example, by the length of the guide sequence encoded in the CRISPR locus or by position relative to the 3’ repeat element). Guide sequences in P. furiosus CRISPRs are most commonly 37 nt long, in which case the 45-nt crRNA species is comprised of an 8-nt repeat tag sequence and a full 37-nt guide sequence. (The 39-nt species simply lacks 6 nt of the guide sequence at the 3’ end, see Figure 3A.) However some guide sequences are as short as 34 nt and as long as 59 nt in P. furiosus, and Cmr complex crRNAs are nonetheless consistently 39 and 45 nt in length. For example, while three of the first 5 guide sequences in CRISPR locus 4 are 37 nt long, the second guide sequence in the locus (termed 4.02) is 35 nt; yet the 4.02 crRNAs found in the Cmr complex are 45 and 39 nt in length (Figure 3B). The 45-nt 4.02 crRNA includes 2 nt of the downstream repeat sequence. Conversely, the fifth CRISPR 4 guide sequence is one nt longer than standard (38 nt), and the 45-nt 4.05 crRNA lacks the last encoded nt of the guide sequence (Figure 3B). Similarly, the first guide sequence in CRISPR 1 is 48 nt long (11 nt longer than is typical), and the 45-nt 1.01 crRNA found in the Cmr complex includes only 37 nt of the 48-nt guide sequence (see Figure 4B). Thus, regardless of the size of the guide sequence encoded in the genome, the crRNAs of the Cmr complex are comprised of an 8-nt 5’ repeat tag and a 37- or 31-nt guide sequence.
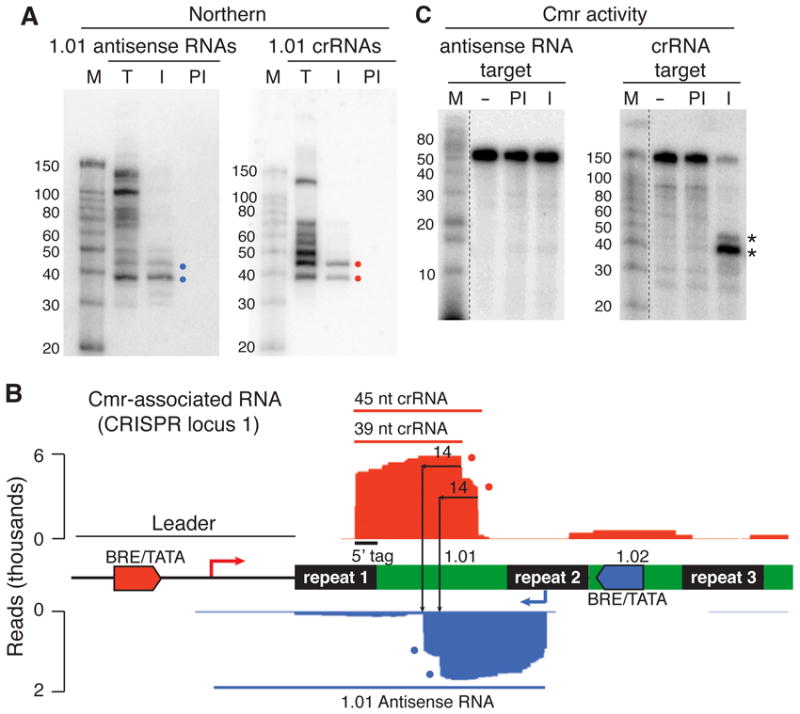
A) Antisense RNA from the region of crRNA 1.01 is detected by Northern analysis, and two species are associated with the Cmr complex. Northern analysis of total P. furiosus RNA (T) and RNAs isolated from immune (I) or pre-immune (PI) Cmr2 immunopurifications was probed for antisense (left panel) or sense 1.01 crRNAs (right panel). Colored dots indicate the primary species of antisense RNAs (blue dots) and crRNAs (red dots) specifically associated with the Cmr complex (and correspond with dots in panel B). Sizes of radiolabeled RNA marker (M) are noted. B) The two antisense RNAs associated with the Cmr complex map downstream of a potential fortuitous promoter and correspond to the expected products of crRNA 1.01-guided cleavage. Deep sequencing profiles of Cmr-associated crRNAs in the region encoding crRNA 1.01 are shown. Red indicates RNAs transcribed from the leader region, and blue corresponds to reads from the opposite strand. The positions of CRISPR repeats (black), guide sequences (green) and BRE/TATA promoters (red and blue) are indicated. A blue line represents the apparent full-length antisense RNA observed by deep sequencing total RNA (see Figure S1). Cleavage of the antisense RNA by the Cmr complex 14 nucleotides from the 3’ ends of the 45- and 39-nt species of crRNA 1.01 (red dots) would produce antisense RNA products of the sizes observed by deep sequencing and Northern analysis (blue dots). C) Immunopurified Cmr complexes cleave the antisense RNA. Radiolabeled RNA targets for the antisense RNAs (left panel) and crRNAs (right panel) were incubated in the absence (−) of proteins or in the presence of immune (I) or pre-immune (PI) immunourified preparations, and the products were analyzed following denaturing gel electrophoresis. (The target for the crRNAs is 140 nucleotides in length and corresponds to the full-length antisense RNA (see panel C).) The cleavage products obtained by incubation of the crRNA target with the immunopurified Cmr complex (asterisks) are approximately the same sizes as the endogenous antisense RNA species that are immunopurified with the Cmr complexes (A, left panel). Non-contiguous lanes from the same gel are indicated by dashed lines.
Cleavage of a complementary RNA in vivo
Very few RNAs were detected that were transcribed from the opposite (non-leader) ends of the CRISPR loci in P. furiosus; however, we did detect significant antisense transcription from an apparently fortuitous promoter within CRISPR locus 1. A BRE/TATA promoter sequence is present in the antisense orientation within the second guide sequence of CRISPR locus 1, and a significant number of antisense RNAs that begin distinctly 21 nt downstream of the promoter are present in the sequence from the total RNA sample (Figure S1, blue reads). The number of antisense RNA reads is approximately 1/3 the number of reads observed for the sense strand crRNA reads (crRNA 1.01; Figure S1, red reads). Northern analysis reveals a series of antisense RNAs up to ~140 nt in length in total P. furiosus RNA (Figure 4A, 1.01 antisense, total (T) lane).
The antisense CRISPR transcripts found in P. furiosus would be predicted to be recognized and targeted by Cmr complexes containing the crRNA encoded on the opposite strand of CRISPR locus 1 (crRNA 1.01). Previous in vitro studies established that the Cmr complex cleaves complementary RNAs 14 nt upstream of the 3’ ends of the 39- and 45-nt crRNAs (Hale et al., 2008). Accordingly, in this case, the 1.01 crRNAs would be predicted to generate 5’ products of 45 and 39 nt (as well as 3’ products of various lengths) upon cleavage of the antisense transcripts. And indeed, distinct antisense RNAs of 45 and 39 nt were observed in Northern analysis of the total RNA (with a probe to the 5’ region of the antisense transcript; Figure 4A, 1.01 antisense, total (T) lane), suggesting that the Cmr complex targets and cleaves the endogenous CRISPR1 antisense transcript in P. furiosus.
We also found in both RNA sequencing and Northern analysis that the 45- and 39-nt putative 5’ cleavage products are specifically co-immunopurified with the Cmr complex (Figure 4A, 1.01 antisense, immune (I) vs. preimmune (PI) lanes; and Figure 4B, blue reads vs. Figure S1, blue reads), suggesting that the products remain associated with the complex to some extent following cleavage. The 45- and 39-nt antisense RNAs do not possess the 5’ repeat tag sequence, which we hypothesize is important for recognition and function of crRNAs with the Cmr complex. However, because of the coincidence in the size of these RNAs with Cmr complex crRNA species, we tested the possibility that the antisense RNAs are functional guide RNAs. We found that the immunopurified Cmr complexes do not cleave a target RNA complementary to the antisense RNAs, indicating that the antisense RNAs do not guide cleavage by the Cmr complex (Figure 4C, antisense RNA target). At the same time, the immunopurified complexes do cleave an RNA corresponding to the ~140-nt antisense RNA transcript, generating 5’ products of the same sizes observed in vivo (45 and 39 nt; Figure 4C, crRNA target). Together, the findings indicate that the Cmr complex recognizes and cleaves endogenous complementary RNAs in vivo.
The CRISPR repeat tag sequence is required for functional Cmr complex crRNAs
Its conservation suggests that the 5’ CRISPR repeat tag is an important element of functional Cmr complex crRNAs. We investigated the importance of the tag by testing several tag sequence mutants for the ability to guide cleavage by reconstituted Cmr complexes. Removal of the repeat tag sequence from crRNA 7.01 eliminates cleavage of a complementary target RNA (Figure 5A). Substitution of the entire tag sequence to its complement (AUUGAAAG → UAACUUUC) or of the first two nucleotides of the tag sequence (AUUGAAAG → GAUGAAAG) also prevents function of the crRNA, as does the addition of a G at the 5’ end of the tag (Figure 5B).
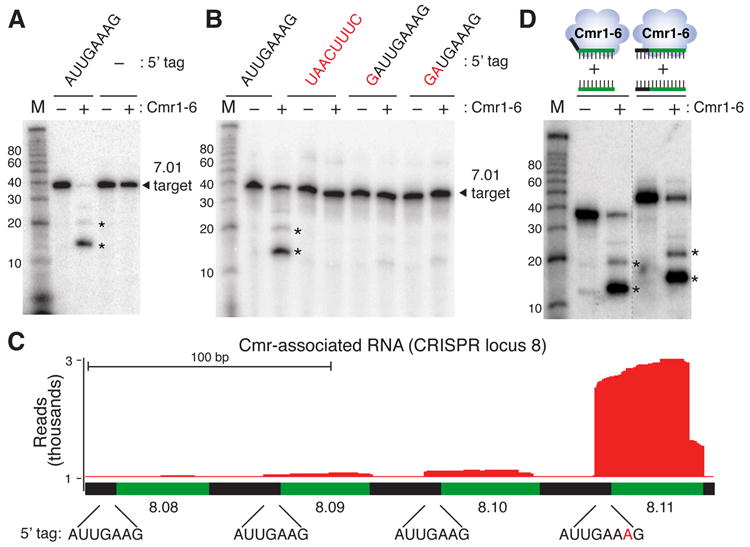
A) The 5’ repeat tag is required for Cmr complex function. Reconstitutions were performed with recombinant Cmr1-6 proteins and the 45-nt. wildtype 7.01 crRNA (AUUGAAAG tag) or crRNA 7.01 lacking the tag (−), and activity was tested against the radiolabeled 7.01 target RNA. RNA marker sizes are indicated. The cleavage product observed with the wildtype crRNA is indicated with an asterisk. B) Mutations to the tag sequence prevent function. Reconstitutions were performed with the 45-nucleotide wildtype 7.01 crRNA (AUUGAAAG tag) or crRNA 7.01 with the indicated mutations in the tag sequence (mutated or inserted nucleotides are highlighted in red) and tested in cleavage assays as described in A. The cleavage product observed with the wildtype crRNA is indicated with an asterisk. C) crRNAs with variant tag sequences are underrepresented in the Cmr complex. Deep sequencing profiles of Cmr-associated crRNAs in the region encoding crRNAs 8.08 – 8.11 are shown (see also Figure 2A). The positions of CRISPR repeats (black) and guide sequences (green) are indicated. The 5’ tag sequence is shown below for crRNAs 8.08 - 8.11. An additional nucleotide present in the wildtype tag of crRNA 8.11 is highlighted in red D) Complementarity to the tag does not prevent function. Cmr complexes reconstituted with wildtype crRNA 7.01 were tested for the ability to cleave the 7.01 target RNA or a target with complementarity to the tag sequence (as illustrated). Cleavage products are indicated with asterisks. Non-contiguous lanes from the same gel are indicated with a dashed line.
Analysis of CRISPR locus 8 provided further insight into the basis for the importance of the 5’ repeat tag in vivo. crRNAs from locus 8 were significantly underrepresented in the Cmr complex (Figures 2A and S2). The reduced presence of CRISPR locus 8 RNAs in the Cmr complex correlates with a difference in the 5’ repeat tag found on these RNAs. Most of the crRNAs from locus 8 possess a 7-nt AUUGAAG (rather than 8-nt AUUGAAAG) repeat tag due to a variation in the repeat sequences within this CRISPR locus (Figure 5C). The exception is crRNA 8.11 (the last crRNA encoded by locus 8), which retains the canonical 8-nt repeat tag sequence, and this crRNA is selectively enriched in the Cmr complex relative to the other crRNAs from CRISPR locus 8 (Figures 2A and and5C).5C). Together, these results indicate that the CRISPR repeat tag is critical for the formation of functional Cmr complexes by crRNAs.
For CRISPR-Cas systems that target DNA, the 5’ repeat tag on the crRNAs provides a key function in distinguishing self from non-self. In organisms with these systems, it is important for crRNAs to avoid targeting the complementary DNA sequence found in the host CRISPR locus itself, and evidence indicates that, in these systems, potential targets with complementarity to the tag sequence (found in the CRISPR locus but not in invader targets) are not silenced (Marraffini and Sontheimer, 2010). We tested the effect of target complementarity to the tag sequence on RNA cleavage by the Cmr complex, and observed no significant reduction in activity (Figure 5D). The antisense transcript that is cleaved in vivo by the Cmr complex also is complementary to the tag of crRNA 1.01 (Figure 4). Thus, RNA targeting by the Cmr complex does not require a lack of complementarity between the target and the crRNA repeat tag.
crRNAs can be engineered to direct the Cmr complex to cleave novel target RNAs
Our findings indicate that a 5’ repeat tag is necessary for a crRNA to function to guide cleavage by the Cmr complex. To determine whether the Cmr complex can be directed to cleave novel target RNAs by engineered crRNAs, we designed crRNAs with the 5’ AUUGAAAG tag sequence and guide sequences complementary to a random target sequence (X’) or to a sequence near the 5’ end of the β-lactamase (bla) mRNA. Cmr complexes reconstituted with the crRNA designed to cleave the random target sequence specifically cleaved the X’ target RNA but not a bla target sequence (Figure 6A). Similarly, the crRNA designed against the 5’ region of the bla mRNA directed cleavage of the bla target sequence but not the X’ target RNA (Figure 6A). The cleavage product sizes are consistent with target cleavage 14 nt from the 5’ ends of the crRNAs. The 5’ bla crRNA and a crRNA designed against an internal region of the bla mRNA were also tested for the ability to direct cleavage in the context of the full-length bla mRNA. The 5’ radiolabeled bla mRNA (~860 nt) was cleaved by Cmr complexes reconstituted with both bla crRNAs, yielding products of the expected sizes (~17 nt for the 5’ bla crRNA and ~236 nt for the internal bla crRNA) (Figure 6B). These results demonstrate that the Cmr complex can be directed to cleave novel target RNAs at a desired site by crRNAs designed with an appropriate repeat tag sequence and a guide region complementary to the target cleavage site.
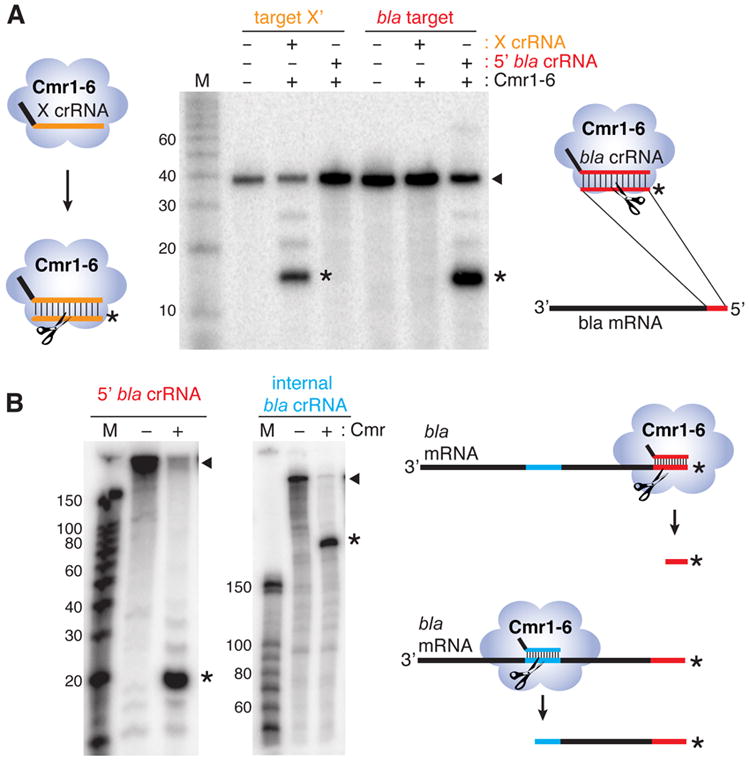
A) Specific cleavage of novel target RNAs. Cmr complexes were reconstituted with crRNAs designed to target a random sequence (X crRNA, X’ target, orange) or a fragment from the 5’ end of the β-lactamase (bla) mRNA (5’ bla crRNA, bla target, red), and tested for the ability to cleave both target RNAs. Cleavage products are indicated with asterisks. Diagrams illustrate engineered crRNAs with 5’ repeat tag (black) and guide sequence elements (orange or red) interacting with and cleaving the intended 5’ radiolabeled target. B) Directed cleavage of predicted sites in β-lactamase mRNA. Engineered crRNAs target two locations on the full-length bla mRNA. The full length bla mRNA was subject to cleavage by Cmr complexes reconstituted with crRNAs targeting a region near the 5’ end of the mRNA (5’ bla crRNA, red) or a sequence approximately 200 nucleotides from the 5’ end of the mRNA (internal bla crRNA, blue). Cleavage products are indicated by asterisks. Diagrams illustrate engineered crRNAs interacting with and cleaving the bla mRNA.
DISCUSSION
The RNA targeting branch of the CRISPR-Cas immune system
There is tremendous diversity in the CRISPR-Cas immune pathways that protect prokaryotes from invaders (recently reviewed in: (Deveau et al., 2010; Garrett et al., 2011; Horvath and Barrangou, 2010; Jore et al., 2011a; Karginov and Hannon, 2010; Makarova et al., 2011; Marraffini and Sontheimer, 2010; Terns and Terns, 2011; van der Oost et al., 2009)). The different CRISPR-Cas systems are populated by distinct modules of proteins, and the mechanisms of key steps in the pathways, including crRNA biogenesis and invader silencing, can be fundamentally different. In this work, we investigated the Cmr effector complex, which is unique among systems characterized to date in that it targets RNAs rather than DNAs (Hale et al., 2009). This RNA-targeting branch of the CRISPR-Cas immune system appears to play an important role in the war against viruses and other mobile genetic elements given that approximately 70% of the archaea and 30% of the bacteria with CRISPR-Cas systems include the Cmr module (Shah and Garrett, 2011). Our results provide direct evidence that the CRISPR-Cmr system cleaves complementary RNAs in vivo (Figure 4). Moreover, we have identified the key structural features of the crRNAs associated with active Cmr complexes (Figures 1--3,3, ,5)5) and engineered crRNAs capable of directing Cmr complexes to efficiently cleave selected RNA targets at predictable sites (Figure 6).
Anatomy of the Cmr complex guide RNAs
The crRNAs associated with the Cmr complex are of two sizes, 39 and 45 nt in length, both with an 8-nt CRISPR repeat sequence at the 5’ end (5’ tag) followed by either 31 or 39 nt of invader guide sequence (Figures 1 and and3,3, and (Hale et al., 2009)). Unlike other crRNAs found in P. furiosus, the Cmr crRNAs contain little or no CRISPR repeat sequence at the 3’ end (Figure 3). Our studies revealed that the crRNAs present in Cmr complexes are essentially restricted to an absolute length of 39 or 45 nt, which may be defined by a mechanism that measures from the 5’ end of the crRNA or from the 5’ tag sequence, but does not depend on the position of the guide sequence-3’ repeat sequence boundary (Figure 3). Both the 39- and 45-nt crRNAs have 5’ hydroxyl and 3’ phosphate end groups (Figure 1D). Overall, the 39- and 45-nt forms are found in roughly equal proportion in the purified Cmr complexes (Figure 3A); however, the proportion of the two forms for an individual crRNA can vary significantly (see crRNAs 4.01 and 4.05, Figure 3B), and the basis for this variation is unclear. It is not yet known whether individual Cmr complexes contain one or more crRNAs, however, functional Cmr complexes can be reconstituted with either the 39- or 45-nt crRNA species, indicating that both forms are not simultaneously required for cleavage activity and hinting that each Cmr complex may harbor a single crRNA.
While the 5’ ends of the Cmr complex crRNAs are presumably generated by cleavage of CRISPR transcripts by the site-specific riboendonuclease Cas6 (Carte et al., 2010; Carte et al., 2008; Wang et al., 2011), we do not yet know how the 3’ ends of mature crRNAs are generated or how the 39- and 45-nt crRNAs are selectively incorporated into Cmr complexes. This exquisite selectivity may simply be because the Cmr complex proteins preferentially bind crRNAs of these lengths. Alternatively, the proteins may associate with longer crRNA species (e.g., the 1X intermediate generated by Cas6 cleavage that includes an additional 22 nt of the repeat at the 3’ end) and the 3’ ends may be generated following formation of the complexes. The nuclease(s) responsible for crRNA 3’ end trimming could be an intrinsic subunit of the Cmr complex or an extrinsic enzyme that may or may not be specific for crRNA processing. The precisely measured 3’ ends of the Cmr complex crRNAs could result, for example, from a combination of non-specific nuclease activity and specific RNA protection by Cmr proteins. The 45-nt crRNA and the other crRNA size forms found in P. furiosus (which contain longer 3’ ends; see Figures 1 and and3A)3A) are components of other RNP complexes currently under investigation ((Hale et al., 2008), and our unpublished data). The P. furiosus genome encodes 21 Cas proteins in addition to the 6 Cmr proteins, including Csa and Cst subtype proteins that are also expected to associate with crRNAs to form additional effector complexes. Future work will elucidate the mechanisms by which distinct crRNA species are sorted among CRISPR-Cas complexes within a cell.
The 5’ tag is critical for Cmr complex crRNA function
The results presented here indicate that the 5’ tag sequence is necessary for crRNA function in Cmr complexes (Figure 5). We found that the sequence, and perhaps size, of the 5’ tag is important for Cmr complex crRNA function; even subtle mutations are detrimental to Cmr activity. We hypothesize that the 5’ tag is important as a binding site for one or more Cmr proteins. Consistent with this idea, we found that crRNAs from CRISPR locus 8 with variant 5’ repeat tags are preferentially excluded from Cmr complexes in vivo (Figure 5C). As discussed above, the basis for exclusion of longer crRNA species, which possess the 5’ tag, from Cmr complexes in P. furiosus is not yet clear. The presence of a 5’ tag sequence is a conserved feature of crRNAs in many CRISPR-Cas systems (Brouns et al., 2008; Hale et al., 2008; Hale et al., 2009; Haurwitz et al., 2010; Lintner et al., 2011; Marraffini and Sontheimer, 2008). A correlation exists between specific CRISPR-Cas modules and the CRISPR repeat sequences found together in various organisms, suggesting that the different systems may recognize characteristic 5’ tags. However, in P. furiosus the crRNAs encoded by the 7 CRISPR loci generally share the same 5’ tag sequence, which evidently functions with the Csa and Cst as well as Cmr modules. We hypothesize that the 5’ tag is also important in effector complex function in the other systems where it is found. Our results reveal the functional importance of the 5’ tag in Cmr effector complex function.
Directing Cmr complexes to cleave RNAs of choice
In this work, we have demonstrated that crRNAs can be designed to target the Cmr complex to cleave novel RNA substrates (Figure 6). crRNAs comprised of a 37-nt complementary guide sequence and 5’-tag were used to direct specific cleavage of RNAs including the β-lactamase mRNA in vitro. Cleavage product sizes indicate that the target was predictably cut 14 nt downstream of the 3’ end of the engineered crRNA. These findings will enable efforts to exploit the site-specific cleavage function of Cmr-crRNA complexes for in vivo gene knockdown experiments. Our studies have defined the minimal components for Cmr complex function (Cmr1-6 and a crRNA; Figure 6 and (Hale et al., 2009)), making it conceivable to express functional Cmr complexes in eukaryotes as well as additional prokaryotes to provide a novel method of RNA silencing. Our results indicate that Cmr-crRNA complexes have the potential to target destruction of mRNAs encoding any gene of interest and to become important research tools with industrial and biomedical applications.
RNA targeting vs. DNA targeting CRISPR-based gene silencing systems
Our findings demonstrate that there are significant differences in the modes of target nucleic acid recognition for CRISPR-based gene silencing systems that target RNA and DNA. For example, while DNA targeting by at least some CRISPR-Cas systems requires a short PAM (protospacer adjacent motif) in the invading DNA target (Deveau et al., 2008; Garneau et al., 2010; Gudbergsdottir et al., 2011; Mojica et al., 2009; Semenova et al., 2011), our work suggests that Cmr complexes function independently of PAMs. Cmr-crRNA complexes can cleave target RNAs that completely lack flanking sequences or contain a variety of flanking sequences (that do not match predicted PAMs; Figures 1, ,4,4, and and66 and (Hale et al., 2009)). For DNA-targeting systems, the requirement for a PAM sequence in the invader avoids damage to the host genome at the site of the corresponding guide sequence in the CRISPR locus (where PAMs are not found in the flanking repeat sequences). The DNA-targeting system of Staphylococcus epidermidis further demonstrates an explicit requirement for mismatches between the 5’ tag of the crRNA and the target DNA strand, which also prevents targeting the host CRISPR locus (Marraffini and Sontheimer, 2010). In contrast, for the RNA-targeting Cmr complex (where host genome damage is not a risk) target RNAs containing sequences with perfect homology to the 5’ tag of the crRNA are efficiently cleaved (Figures 4 and and5D).5D). Evidence indicates that there are also differences in the interactions with the guide region of the crRNA. A specific 7-8 nt region of the crRNA located immediately downstream of the 5’ tag (dubbed the “seed sequence”) was found to be important for initial high affinity recognition of targets for certain DNA-targeting CRISPR-Cas systems (Semenova et al., 2011; Wiedenheft et al., 2011). Base-pairing between the 3’ end of the crRNA and target DNA was dramatically less important. The opposite trend is observed for RNA targeting, where interaction with the 3’ end of the crRNA is critical for function and target RNAs that lack the “seed sequence” region are efficiently cleaved (Hale et al., 2009). The molecular bases of these differences in CRISPR-Cas RNA and DNA targeting pathways remain to be explored. The lack of restrictions in the flanking sequence content in RNA targeting provides for more flexibility in substrate interaction.
EXPERIMENTAL PROCEDURES
Antibody preparation and co-immunoprecipitation reactions
IgY antibodies were raised against recombinant P. furiosus Cmr2 as described (Carte et al., 2010). Pre-immune and immune Cmr2 antibodies were either coupled to anti-IgY conjugated agarose beads (Gallus Immunotech) for Northern analysis, end analysis or small RNA sequencing, or to CarboLink beads (Pierce) for Western analysis.
Co-IP/Western analysis
Western blotting of co-IP material was performed by standard procedures.
Co-IP/Northern analysis
Trizol LS reagent (Invitrogen) was used to isolate RNAs from the co-IP samples and P. furiosus extract (total RNA). Northern analysis was performed as described previously (Hale et al., 2008) with modifications (see Supplemental Experimental Procedures). Probe sequences can be found in Supplemental Table 1.
Co-IP/Activity assays
Co-IP beads were washed into 40 mM Hepes pH 7.0, 500 mM KCl and added to a 20 μl cleavage reaction containing radiolabeled target RNA (5,000 cpm). Reactions were incubated for 3 hours at 70°C with mixing. RNAs were isolated by proteinase K treatment, phenol extraction and ethanol precipitation and separated on 7M urea 15% polyacrylamide gels and visualized by phosphor imaging.
Co-IP/End analysis
Trizol LS was used to isolate small RNAs from co-IP samples. The RNAs were treated with thermostable shrimp alkaline phosphatase (Promega). Treated and non-treated RNAs was subject to either: 1) 5’ radiolabeling with γ32P-ATP (MP Biomedicals) via polynucleotide kinase (Ambion), or 2) 3’ radiolabeling via T4 ssRNA Ligase 1 (NEB) with cytidine-3’,5’-bisphosphate [5’-32P] (3,000 ci/mmol, MP Biomedicals). Both labeling reactions were carried out for 1 hour at 37°C and the RNAs were isolated, separated by denaturing gel electrophoresis, and visualized by phosphor imaging.
Small RNA library preparation
For preparation of total and Cmr-associated small RNA libraries, Trizol was used to extract RNAs from P. furiosus cells or from a co-IP reaction, respectively. 50 μg of total RNA was used for total small RNA library preparation. Briefly, the RNAs were phosphatase treated to remove 3’ phosphates, followed by 3’ ligation to an adaptor. These products were treated with T4 polynucleotide kinase before 5’ ligation to an additional adaptor. The 5’ and 3’ ligated products were subject to reverse transcription, RNase H digestion and PCR.
Illumina sequencing and analysis
cDNA libraries were visualized by electrophoresis, quantitated by Agilent Bioanalyzer and Nano-drop, diluted to 2 pM, and subject to 76 cycles of sequencing on the Illumina Genome Analyzer IIx. Sequence reads were trimmed of the 3’ linker and reads 18-76 nt in length were aligned to the P. furiosus genome using Bowtie (Langmead et al., 2009). The 5’ and 3’ ends of reads mapping to the CRISPR loci were calculated using a custom PERL script available upon request.
Recombinant Cmr protein production and activity assays
Expression and purification of recombinant Cmr proteins was performed as described (Hale et al., 2009) with modifications (See Supplemental Experimental Procedures). RNA cleavage assays were performed as described (Hale et al., 2009). Briefly, all six recombinant Cmr proteins (500 nM each) were pre-incubated with unlabeled crRNA (0.05 pmol) prior to addition of 5’ radiolabeled target RNAs (5,000 cpm). Reactions were incubated for 1-3 hours at 70°C and isolated RNAs analyzed by denaturing gel electrophoresis and phosphor imaging.
RNA preparation
Sequences of RNA/DNA oligos used for RNA preparation are found in Supplemental Table 1, and detailed information about RNA preparation is found in the Supplemental Experimental Procedures.
Acknowledgments
We thank the UCHC Translational Genomics Core Facility for use of the Illumina Genome Analyzer IIx. This work was supported by NIH grants RO1GM54682 (including ARRA funds) to M.P.T. and R.M.T. and R01GM062516 to B.R.G.
Footnotes
SUPPLEMENTAL INFORMATION Supplemental information includes two figures, one table and Supplemental Experimental Procedures.
ACCESSION NUMBERS RNA sequences and alignments are available at GEO (GSE33256).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersson AF, Banfield JF. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320:1047–1050. [Abstract] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. [Abstract] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. [Europe PMC free article] [Abstract] [Google Scholar]
- Carte J, Pfister NT, Compton MM, Terns RM, Terns MP. Binding and cleavage of CRISPR RNA by Cas6. RNA. 2010;16:2181–2188. [Europe PMC free article] [Abstract] [Google Scholar]
- Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. [Europe PMC free article] [Abstract] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. [Europe PMC free article] [Abstract] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–1400. [Europe PMC free article] [Abstract] [Google Scholar]
- Deveau H, Garneau JE, Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol. 2010;64:475–493. [Abstract] [Google Scholar]
- Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. [Abstract] [Google Scholar]
- Garrett RA, Shah SA, Vestergaard G, Deng L, Gudbergsdottir S, Kenchappa CS, Erdmann S, She Q. CRISPR-based immune systems of the Sulfolobales: complexity and diversity. Biochem Soc Trans. 2011;39:51–57. [Abstract] [Google Scholar]
- Gesner EM, Schellenberg MJ, Garside EL, George MM, Macmillan AM. Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nat Struct Mol Biol 2011 [Abstract] [Google Scholar]
- Gudbergsdottir S, Deng L, Chen Z, Jensen JV, Jensen LR, She Q, Garrett RA. Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol Microbiol. 2011;79:35–49. [Europe PMC free article] [Abstract] [Google Scholar]
- Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. [Europe PMC free article] [Abstract] [Google Scholar]
- Hale C, Kleppe K, Terns RM, Terns MP. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA. 2008;14:2572–2579. [Europe PMC free article] [Abstract] [Google Scholar]
- Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. [Europe PMC free article] [Abstract] [Google Scholar]
- Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. [Europe PMC free article] [Abstract] [Google Scholar]
- Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. [Abstract] [Google Scholar]
- Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–1575. [Abstract] [Google Scholar]
- Jore MM, Brouns SJ, van der Oost J. RNA in Defense: CRISPRs Protect Prokaryotes against Mobile Genetic Elements. Cold Spring Harb Perspect Biol 2011a [Europe PMC free article] [Abstract] [Google Scholar]
- Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul U, Wurm R, Wagner R, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol 2011b [Abstract] [Google Scholar]
- Karginov FV, Hannon GJ. The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol Cell. 2010;37:7–19. [Europe PMC free article] [Abstract] [Google Scholar]
- Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ, et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. [Europe PMC free article] [Abstract] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. [Europe PMC free article] [Abstract] [Google Scholar]
- Lillestol RK, Shah SA, Brugger K, Redder P, Phan H, Christiansen J, Garrett RA. CRISPR families of the crenarchaeal genus Sulfolobus: bidirectional transcription and dynamic properties. Mol Microbiol. 2009;72:259–272. [Abstract] [Google Scholar]
- Lintner NG, Kerou M, Brumfield SK, Graham S, Liu H, Naismith JH, Sdano M, Peng N, She Q, Copie V, et al. Structural and functional characterization of an archaeal CASCADE complex for CRISPR-mediated viral defense. J Biol Chem 2011 [Europe PMC free article] [Abstract] [Google Scholar]
- Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. [Europe PMC free article] [Abstract] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–477. [Europe PMC free article] [Abstract] [Google Scholar]
- Manica A, Zebec Z, Teichmann D, Schleper C. In vivo activity of CRISPR-mediated virus defence in a hyperthermophilic archaeon. Mol Microbiol 2011 [Abstract] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. [Europe PMC free article] [Abstract] [Google Scholar]
- Marraffini LA, Sontheimer EJ. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature. 2010;463:568–571. [Europe PMC free article] [Abstract] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. [Abstract] [Google Scholar]
- Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. [Abstract] [Google Scholar]
- Przybilski R, Richter C, Gristwood T, Clulow JS, Vercoe RB, Fineran PC. Csy4 is responsible for CRISPR RNA processing in Pectobacterium atrosepticum. RNA Biol. 2011;8 [Abstract] [Google Scholar]
- Pul U, Wurm R, Arslan Z, Geissen R, Hofmann N, Wagner R. Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS. Mol Microbiol. 2010;75:1495–1512. [Abstract] [Google Scholar]
- Sashital DG, Jinek M, Doudna JA. An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat Struct Mol Biol 2011 [Abstract] [Google Scholar]
- Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, van der Oost J, Brouns SJ, Severinov K. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci U S A. 2011;108:10098–10103. [Europe PMC free article] [Abstract] [Google Scholar]
- Shah SA, Garrett RA. CRISPR/Cas and Cmr modules, mobility and evolution of adaptive immune systems. Res Microbiol. 2011;162:27–38. [Abstract] [Google Scholar]
- Shah SA, Hansen NR, Garrett RA. Distribution of CRISPR spacer matches in viruses and plasmids of crenarchaeal acidothermophiles and implications for their inhibitory mechanism. Biochem Soc Trans. 2009;37:23–28. [Abstract] [Google Scholar]
- Terns MP, Terns RM. CRISPR-based adaptive immune systems. Curr Opin Microbiol 2011 [Europe PMC free article] [Abstract] [Google Scholar]
- van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci. 2009;34:401–407. [Abstract] [Google Scholar]
- Wang R, Preamplume G, Terns MP, Terns RM, Li H. Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage. Structure. 2011;19:257–264. [Europe PMC free article] [Abstract] [Google Scholar]
- Wiedenheft B, van Duijn E, Bultema J, Waghmare S, Zhou K, Barendregt A, Westphal W, Heck A, Boekema E, Dickman M, et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci U S A 2011 [Europe PMC free article] [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.molcel.2011.10.023
Read article for free, from open access legal sources, via Unpaywall:
http://www.cell.com/article/S1097276511009555/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1016/j.molcel.2011.10.023
Article citations
A positive contribution to nitrogen removal by a novel NOB in a full-scale duck wastewater treatment system.
Water Res X, 24:100237, 10 Jul 2024
Cited by: 1 article | PMID: 39155949 | PMCID: PMC11327836
From Gene Editing to Biofilm Busting: CRISPR-CAS9 Against Antibiotic Resistance-A Review.
Cell Biochem Biophys, 82(2):549-560, 03 May 2024
Cited by: 0 articles | PMID: 38702575
Review
Unity among the diverse RNA-guided CRISPR-Cas interference mechanisms.
J Biol Chem, 300(6):107295, 18 Apr 2024
Cited by: 1 article | PMID: 38641067 | PMCID: PMC11127173
Review Free full text in Europe PMC
Comprehensive review of CRISPR-based gene editing: mechanisms, challenges, and applications in cancer therapy.
Mol Cancer, 23(1):9, 09 Jan 2024
Cited by: 12 articles | PMID: 38195537 | PMCID: PMC10775503
Review Free full text in Europe PMC
Histones direct site-specific CRISPR spacer acquisition in model archaeon.
Nat Microbiol, 8(9):1682-1694, 07 Aug 2023
Cited by: 0 articles | PMID: 37550505 | PMCID: PMC10823912
Go to all (195) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (1 citation) GEO - GSE33256
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Three CRISPR-Cas immune effector complexes coexist in Pyrococcus furiosus.
RNA, 21(6):1147-1158, 22 Apr 2015
Cited by: 36 articles | PMID: 25904135 | PMCID: PMC4436667
The RNA- and DNA-targeting CRISPR-Cas immune systems of Pyrococcus furiosus.
Biochem Soc Trans, 41(6):1416-1421, 01 Dec 2013
Cited by: 23 articles | PMID: 24256230 | PMCID: PMC3996508
Review Free full text in Europe PMC
RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex.
Cell, 139(5):945-956, 01 Nov 2009
Cited by: 647 articles | PMID: 19945378 | PMCID: PMC2951265
DNA targeting by the type I-G and type I-A CRISPR-Cas systems of Pyrococcus furiosus.
Nucleic Acids Res, 43(21):10353-10363, 30 Oct 2015
Cited by: 32 articles | PMID: 26519471 | PMCID: PMC4666368
Funding
Funders who supported this work.
NIGMS NIH HHS (6)
Grant ID: R01 GM062516-09
Grant ID: R01GM062516
Grant ID: R01 GM062516
Grant ID: R01 GM054682-10A1
Grant ID: R01GM54682
Grant ID: R01 GM054682





