Abstract
Free full text

Microtubule binding by KNL-1 contributes to spindle checkpoint silencing at the kinetochore
Abstract
Accurate chromosome segregation requires coordination between microtubule attachment and spindle checkpoint signaling at the kinetochore. The kinetochore-localized KMN (KNL-1/Mis12 complex/Ndc80 complex) network, which mediates microtubule attachment and scaffolds checkpoint signaling, harbors two distinct microtubule-binding activities: the load-bearing activity of the Ndc80 complex and a less well-understood activity in KNL-1. In this paper, we show that KNL-1 microtubule-binding and -bundling activity resides in its extreme N terminus. Selective perturbation of KNL-1 microtubule binding in Caenorhabditis elegans embryos revealed that this activity is dispensable for both load-bearing attachment formation and checkpoint activation but plays a role in checkpoint silencing at the kinetochore. Perturbation of both microtubule binding and protein phosphatase 1 docking at the KNL-1 N terminus additively affected checkpoint silencing, indicating that, despite their proximity in KNL-1, these two activities make independent contributions. We propose that microtubule binding by KNL-1 functions in checkpoint silencing by sensing microtubules attached to kinetochores and relaying their presence to eliminate generation of the checkpoint signal.
Introduction
During cell division, kinetochores assemble on the centromeric regions of chromosomes to form the primary attachment site for spindle microtubules (Cheeseman and Desai, 2008). The kinetochore also scaffolds the spindle checkpoint, the signaling pathway that ensures the fidelity of chromosome segregation by preventing anaphase onset until all chromosomes are properly connected to the spindle (Musacchio and Salmon, 2007). The KMN (Knl1/Mis12 complex/Ndc80 complex) network, comprised of three interacting conserved complexes, is the central hub of the outer kinetochore, where microtubule-binding and checkpoint signaling activities are coordinated (Burke and Stukenberg, 2008; Cheeseman and Desai, 2008; Santaguida and Musacchio, 2009). Components of the KMN network accumulate on kinetochores beginning in prophase and remain stably associated for the duration of mitosis. The three constituents of the KMN network—Knl1, the Mis12 complex, and the Ndc80 complex—have been studied using both in vivo and in vitro approaches in a variety of experimental systems (Cheeseman and Desai, 2008; Santaguida and Musacchio, 2009). Two important activities of this network are to form load-bearing microtubule attachments that segregate chromosomes and to recruit components essential for checkpoint signaling.
Two conserved microtubule-binding activities are present in the KMN network: the load-bearing activity in the Ndc80 complex, which has been analyzed in depth using cell biological, biochemical, biophysical, and structural approaches (Joglekar et al., 2010; Tooley and Stukenberg, 2011), and a second conserved activity in Knl1 family proteins (Cheeseman et al., 2006; Kerres et al., 2007; Pagliuca et al., 2009). The function of the microtubule-binding activity of Knl1 family proteins in chromosome segregation is not known. Knl1 recruits the checkpoint kinase Bub1 to kinetochores (Desai et al., 2003) potentially via a direct interaction with its N-terminal half (Kiyomitsu et al., 2007) and docks protein phosphatase 1 (PP1) through a conserved set of motifs in the extreme N terminus (Hendrickx et al., 2009; Liu et al., 2010); the C-terminal half of Knl1 participates in KMN network assembly (Kiyomitsu et al., 2007; Petrovic et al., 2010). PP1 docked by Knl1 is proposed to counteract Aurora B kinase enriched in the inner centromere and promote biorientation through dephosphorylation-mediated stabilization of attachments (Liu et al., 2010; Welburn et al., 2010).
PP1 has been implicated in spindle checkpoint silencing (Pinsky et al., 2009; Vanoosthuyse and Hardwick, 2009), and recent work in budding and fission yeast has shown that PP1 docked on Knl1 is critical for the silencing reaction (Meadows et al., 2011; Rosenberg et al., 2011). The checkpoint signal is generated by accumulation of a conserved set of proteins, most prominently Mad1 and Mad2, at unattached kinetochores and controls the activity of the anaphase-promoting complex/cyclosome in the cytoplasm. After microtubule attachment to kinetochores, generation of the checkpoint signal is silenced to promote anaphase onset.
To generate a switchlike transition into anaphase after attachment of the last kinetochore to spindle microtubules, the checkpoint signal must be continuously inactivated in the cytoplasm (Musacchio and Salmon, 2007). Thus, checkpoint silencing requires both microtubule attachment–dependent cessation of signal generation at kinetochores and inactivation of already generated signal in the cytoplasm. Dynein motor–dependent removal of checkpoint proteins from kinetochores after microtubule attachment (Howell et al., 2001; Wojcik et al., 2001) as well as a poorly understood dynein-independent mechanism (Chan et al., 2009; Gassmann et al., 2010) contribute to silencing checkpoint signal generation at the kinetochore.
Here, we use the early Caenorhabditis elegans embryo to investigate the function of the microtubule-binding activity of Knl1 family proteins. Specifically, we test whether this activity contributes to load-bearing attachment formation and/or to the regulation of checkpoint signaling at kinetochores. Using in vitro binding and two-hybrid approaches, we engineered mutants of C. elegans KNL-1 that selectively perturb its microtubule-binding activity. Analysis in the early embryo revealed that perturbing KNL-1 microtubule-binding activity did not affect formation of load-bearing microtubule attachments or spindle checkpoint activation but significantly delayed checkpoint silencing in cells with monopolar spindles. Comparison with PP1-docking motif mutants and analysis of double mutants indicate that microtubule binding and PP1 docking make independent contributions to checkpoint silencing. These results identify a microtubule-binding activity in the KMN network, whose inhibition significantly prolongs checkpoint-mediated cell cycle arrest without affecting other aspects of chromosome segregation. We propose that this activity functions as a sensor for the presence of microtubules at the kinetochore and relays their presence to turn off the checkpoint signal.
Results
KNL-1 microtubule-binding activity resides in its extreme N terminus
Purified C. elegans KNL-1 exhibits microtubule-binding and -bundling activity (Cheeseman et al., 2006); a similar activity has also been reported for Spc105, the budding yeast equivalent of KNL-1 (Pagliuca et al., 2009). However, the function of KNL-1’s microtubule-binding activity during chromosome segregation is not known. Microtubule-bundling experiments in vitro indicated that the N-terminal half of KNL-1 (KNL-11–505) exhibited similar activity to the full-length protein (unpublished data). This finding is consistent with the N-terminal but not the C-terminal half of Spc7 (the fission yeast ortholog of KNL-1) colocalizing with microtubules when overexpressed in vivo (Kerres et al., 2007). Sequence analysis revealed a short basic patch in the extreme KNL-1 N terminus (Fig. 1 A). To test whether this patch contributes to microtubule binding, we compared the activity of wild-type (WT) recombinant KNL-11–505 purified from bacteria with that of the same fragment with the four basic residues mutated to alanine (4A) or the first nine amino acids deleted (Δ9; Figs. 1 B and S1 A). The microtubule-bundling activity of the Δ9 and the 4A mutants was significantly reduced compared with WT KNL-11–505 (Fig. 1 C). All three proteins (WT, 4A, and Δ9) are primarily present in a large oligomeric state that elutes close to the void volume of a Superose 6 gel filtration column, indicating that reduced microtubule-bundling activity is not caused by a change in oligomeric state of the mutants (Fig. 1 D). This qualitative assay suggested that the 4A and Δ9 mutants are compromised in their microtubule-binding activity. Because of the oligomeric nature of KNL-1, we used a microscopic assay rather than sedimentation to compare microtubule binding of the WT and mutant proteins (Powers et al., 2009). Recombinant WT, 4A, and Δ9 KNL-11–505 proteins were immobilized on polystyrene beads using an anti-6xHis antibody and flowed into a chamber coated with microtubules, and the number of bound beads was quantified (Fig. 1 E). Beads coated with WT KNL-11–505 bound robustly to immobilized microtubules. In contrast, beads coated with the 4A and Δ9 mutant proteins exhibited no detectable binding, indicating that charge neutralization or removal of the N-terminal basic patch inhibits KNL-1 microtubule-binding activity.
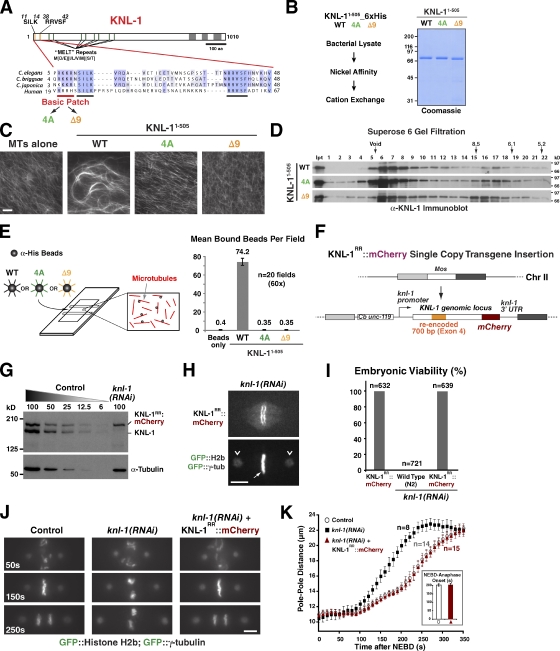
Generation of KNL-1 microtubule-binding mutants and development of an in vivo system to analyze KNL-1 functions. (A) Primary sequence features of KNL-1. A conserved basic patch is highlighted. Mutations engineered to neutralize the basic patch (4A) or delete it (Δ9) are indicated. (B) Purification of WT and mutant KNL-11–505 proteins from bacteria using the indicated steps. The final purified proteins were analyzed by SDS-PAGE and Coomassie staining. (C) Microtubule-bundling analysis of recombinant KNL-11–505 proteins. 1 µM taxol-stabilized rhodamine microtubules (MTs) was imaged either alone or in the presence of the indicated 2-µM KNL-11–505 variants. Bar, 10 µm. (D) Analysis of recombinant KNL-11–505 proteins by gel filtration chromatography. Fractions were analyzed by SDS-PAGE followed by immunoblotting with an anti–KNL-1 antibody. Ipt, input. (E) Bead assay to analyze microtubule-binding activity of KNL-1. 20 fields were photographed, and the number of bound beads was quantified. (F) A schematic of the KNL-1 transgene (KNL-1RR::mCherry) targeted to a single Mos transposon insertion on chromosome II (Chr II). The transgene has the endogenous knl-1 promoter and 3′ untranslated region (UTR), mCherry fused to the C terminus, and exon 4 modified to preserve coding information but alter nucleotide sequence, thereby enabling RNAi-mediated depletion of endogenous KNL-1. (G) dsRNA targeted to the recoded region selectively depletes >95% of endogenous KNL-1. α-Tubulin serves as a loading control. (H–K) Single-copy transgene insertion–encoded KNL-1RR::mCherry is fully functional. Kinetochore localization (H), embryonic viability (I; n = number of embryos scored), chromosome segregation phenotype (J), and kinetic analysis of spindle pole separation (K) are shown. (H and J) GFP::H2b and GFP::γ-tubulin were crossed into the KNL-1RR::mCherry transgenic strain to visualize chromosomes (arrow) and spindle poles (arrowheads), respectively. Bars, 5 µm. (J) Frames from time-lapse sequences aligned relative to NEBD (t = 0); the mCherry signal is not shown. (K) Pole–pole distance measured at 10-s intervals, aligned relative to NEBD, averaged for the indicated number (n) of embryos, and plotted versus time. Inset shows the time of anaphase onset in control (nontransgenic) and transgenic endogenous KNL-1–depleted one-cell embryos. Error bars represent the SEM with a 95% confidence interval.
These results indicate that KNL-1 binds to microtubules through an electrostatic interaction involving a basic patch at its extreme N terminus. Similar basic regions are present in Knl1 family proteins from other species (e.g., human Knl1; Fig. 1 A), suggesting that this mode of microtubule interaction is likely to be conserved.
A single-copy transgene insertion system to investigate KNL-1 microtubule-binding mutants
Prior work in the early C. elegans embryo established high-resolution imaging assays to monitor chromosome alignment and segregation, formation of load-bearing kinetochore–microtubule attachments, and spindle checkpoint signaling (Oegema et al., 2001; Desai et al., 2003; Cheeseman et al., 2004; Essex et al., 2009). Capitalizing on these assays, we analyzed KNL-1 mutants defective for microtubule binding. We used Mos single-copy insertion (Frøkjaer-Jensen et al., 2008) to integrate WT or mutant KNL-1RR::mCherry transgenes (RR denoting RNAi resistant) at a defined genomic location (Fig. 1 F). Transgenes were engineered to be RNAi resistant by altering the nucleotide sequence of exon 4 without affecting coding information; injection of a double-stranded RNA (dsRNA) homologous to exon 4 enabled penetrant and selective depletion of endogenous KNL-1 (Fig. 1 G). KNL-1 localizes to kinetochores, and its depletion leads to 100% embryonic lethality and a first-division kinetochore-null phenotype, characterized by clustering of chromosomes from the two pronuclei, failure of chromosome segregation, and premature spindle pole separation (Desai et al., 2003). The transgene-encoded WT KNL-1RR::mCherry localized similarly to endogenous KNL-1 and rescued the kinetochore-null phenotype and embryonic lethality caused by endogenous KNL-1 depletion (Fig. 1 [H–K] and Video 1). WT KNL-1RR::mCherry also rescued lethality of the knl-1(ok3457) deletion mutant (Fig. S1, B and C). Thus, single-copy KNL-1RR::mCherry transgene insertion provides a means to precisely compare engineered KNL-1 mutants after depletion of endogenous KNL-1.
KNL-1 microtubule-binding mutants support formation of normal load-bearing kinetochore–microtubule attachments
To determine the in vivo function of KNL-1 microtubule-binding activity, we generated strains with 4A or Δ9 mutant KNL-1RR::mCherry transgenes integrated at the same genomic location as the WT transgene. Both mutant KNL-1 fusions localized to kinetochores at levels similar to the WT fusion (Fig. 2 A). To compare the ability of the WT and mutant transgenes to support chromosome alignment and segregation, we first crossed them into a strain expressing GFP fusions with histone H2b and γ-tubulin and then depleted endogenous KNL-1 and monitored the dynamics of chromosomes and spindle poles throughout the first division. Quantitative analysis of spindle pole separation serves as a readout for the formation of load-bearing kinetochore–microtubule attachments, as interactions between astral microtubules and the cortex generate pulling forces that are resisted by load-bearing kinetochore–microtubule attachments within the spindle (Fig. 2 B; Oegema et al., 2001; Desai et al., 2003).
Microtubule-binding mutants of KNL-1 do not affect formation of load-bearing attachments or chromosome segregation. (A) An image of a metaphase plate in living one-cell embryos from strains harboring the indicated KNL-1RR::mCherry transgenes and depleted of endogenous KNL-1. Bar, 2 µm. (B) A schematic of analysis performed after crossing in GFP::histone H2b and GFP::γ-tubulin and depleting endogenous KNL-1. (C) Frames from time-lapse sequences of the first embryonic division for the indicated KNL-1 mutants. Bar, 3 µm. (D) Spindle pole separation kinetics for the indicated conditions. Error bars represent the SEM with a 95% confidence interval. The no transgene knl-1(RNAi) trace is reproduced from Fig. 1 K. (E) Timing of anaphase onset in the indicated conditions. The first visible sign of sister chromatid separation (based on the GFP::H2b signal) was scored as anaphase onset. The gray dashed line indicates the NEBD–anaphase interval for the WT transgene (reproduced from the inset in Fig. 1 K). Error bars represent the SEM with a 95% confidence interval. For comprehensive statistical analysis, see Table S1. (F) Embryonic viability analysis of KNL-1 microtubule-binding mutants. L4-stage worms were injected with dsRNA-targeting endogenous knl-1, and the embryos laid by the injected worms were collected 21–41 h after injection and scored for hatching to form larvae.
After endogenous KNL-1 depletion in the 4A and Δ9 microtubule-binding mutant strains, no significant defects in chromosome segregation were observed. In addition, the kinetics of spindle pole separation were normal until metaphase, but anaphase onset was delayed by 50 s (Fig. 2 [C–E] and Video 2). These observations suggested that the microtubule-binding activity of KNL-1 does not make a major contribution to the mechanics of chromosome segregation. Consistent with this, the 4A and Δ9 mutants rescued embryo viability after depletion of endogenous KNL-1 (Fig. 2 F), and both mutants rescued the lethality of the knl-1(ok3457) deletion allele (Fig. S1, B and C).
Mild defects in chromosome–microtubule attachment can be masked by the spindle checkpoint, which delays cell cycle progression until proper attachments are established. For both the 4A and Δ9 mutants, we observed a significant (P < 0.0001; Table S1) 50-s delay in anaphase onset (Fig. 2 E) that correlated with a plateau in the spindle pole separation profile at metaphase spindle length (Fig. 2 D, top graph). Therefore, we codepleted the checkpoint protein Mad2MDF-2 together with endogenous KNL-1 to determine whether a mild attachment defect resulting from the 4A and Δ9 mutations was being masked by checkpoint activity. Although Mad2MDF-2 depletion eliminated the 50-s delay, it did not lead to visible segregation defects or enhance embryonic lethality (note that in C. elegans, the one-cell embryo undergoes ~550 individual cell divisions before hatching, and chromosome segregation defects are associated with 100% penetrant embryonic lethality; Fig. 2, C–F). Cumulatively, the lack of a significant pole separation defect in the 4A and Δ9 mutants, the ability of these mutants to rescue a deletion allele of KNL-1, and their lack of phenotypic enhancement after Mad2MDF-2 codepletion suggest that the microtubule-binding activity of KNL-1 does not play a significant role in the formation of load-bearing attachments during chromosome segregation.
KNL-1 microtubule-binding mutants extend the spindle checkpoint–dependent cell cycle delay induced by monopolar spindles
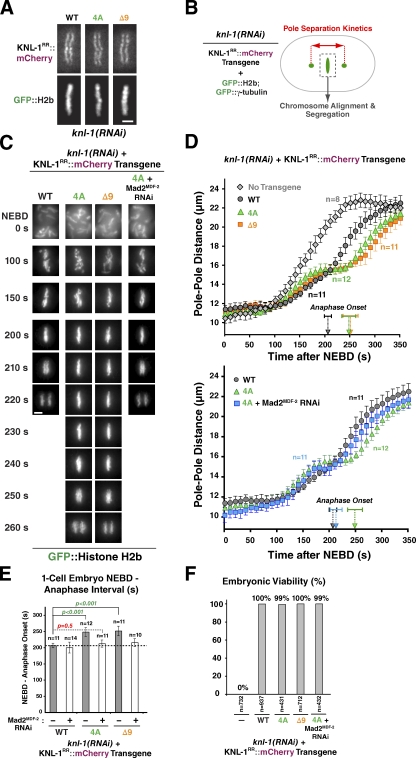
The spindle checkpoint does not act as a timer controlling mitotic duration in C. elegans embryos and is potentially only very mildly activated during the rapid embryonic divisions, as detectable kinetochore enrichment of checkpoint signaling proteins such as Mad1MDF-1 and Mad2MDF-2 is not observed during unperturbed mitosis (Essex et al., 2009). Therefore, we hypothesized that the anaphase onset delay observed with KNL-1 microtubule-binding mutants in the first cell division was a result of a compromised ability to silence a mild checkpoint signal. We previously developed controlled formation of monopolar spindles as a means to monitor checkpoint activation in the early embryo (Fig. 3 A; Essex et al., 2009); this assay circumvents the difficulty of treating normally impermeable embryos with microtubule-depolymerizing drugs. Second-division embryos with monopolar spindles exhibit a significant mitotic delay that requires the checkpoint pathway and the KMN network and correlates with visible transient enrichment of Mad2MDF-2 on unattached kinetochores (Essex et al., 2009).
Microtubule-binding mutants of KNL-1 significantly extend the spindle checkpoint–mediated cell cycle delay induced by monopolar spindles. (A) A schematic of the monopolar spindle–based checkpoint signaling assay in C. elegans embryos. Depletion of the centrosome duplication kinase ZYG-1 generates monopolar spindles in the second division, which trigger a spindle checkpoint–dependent cell cycle delay. (B) Mean time from NEBD to chromosome decondensation in the AB cell for the indicated conditions. Error bars represent the SEM with a 95% confidence interval. The gray dashed line marks the duration of AB cell mitosis for the WT transgene, and the red dashed line marks the duration of AB cell mitosis induced by monopolar spindles in the same strain. For comprehensive statistical analysis, see Table S2. (C) Stills from time-lapse sequences of the AB cell monopolar division in worm strains coexpressing GFP::Mad2MDF-2 and the indicated transgenes. Mad2MDF-2 accumulation on unattached kinetochores is marked with open arrowheads. Bar, 3 µm.
To determine whether the 4A and Δ9 mutations affected checkpoint signaling, especially whether they impaired checkpoint silencing, we generated monopolar spindles and measured the interval from nuclear envelope breakdown (NEBD) to chromosome decondensation. A GFP fusion of cyclin B has been generated in C. elegans, but the fluorescent signal is lost at anaphase of oocyte meiosis I and is not subsequently regained (Liu et al., 2004), likely because of insufficient time to mature GFP fluorescence. It is for this reason that we use decondensation or onset of cortical contractility, coupled with Mad2MDF-2 inhibition, to measure checkpoint-dependent cell cycle delays.
The transgene-encoded WT KNL-1::mCherry fusion supported normal checkpoint signaling (Fig. 3 B); the NEBD-to-decondensation interval was longer in the presence of monopolar spindles than in control cells with bipolar spindles, and this delay was abrogated by Mad2MDF-2 depletion. In cells expressing the microtubule-binding 4A and Δ9 mutants, the delay induced by monopolar spindles was significantly longer compared with the delay observed with WT KNL-1 (P < 0.0001; Fig. 3 B). This extended delay is a result of persistent checkpoint activation, as codepletion of Mad2MDF-2 caused the cell cycle timing to match that of control embryos with bipolar spindles (Fig. 3 B). Thus, perturbing KNL-1 microtubule binding extends the time that cells with monopolar spindles spend in a checkpoint-active state.
Confocal imaging of GFP::Mad2MDF-2 on chromosomes on the monopolar spindles revealed that the extended delay induced by the 4A mutation correlated with a significantly extended residence time for GFP::Mad2MDF-2 on kinetochores (WT residence time was 69 ± 13 s for 13 embryos, and 4A residence time was 118 ± 11 s for 11 embryos; P < 0.0001; Fig. 3 C). The extended persistence of Mad2MDF-2 on the kinetochores of monopolar spindles suggests that the checkpoint silencing defect observed in KNL-1 microtubule-binding mutants is a result of a kinetochore-localized reaction rather than an effect on cytoplasmic regulation of the checkpoint signal. This analysis of cells with monopolar spindles collectively with the results in one-cell embryos with bipolar spindles—where KNL-1 microtubule-binding mutants do not affect load-bearing attachment formation but nonetheless exhibit checkpoint-dependent extended mitotic duration—suggests that KNL-1 microtubule-binding activity contributes to checkpoint silencing at kinetochores.
KNL-1 microtubule-binding mutants do not extend the cell cycle in the absence of microtubules
In cells with bipolar spindles, checkpoint silencing occurs after formation of kinetochore–microtubule attachments on both sister chromatids. In cells with monopolar spindles, the mechanism that controls the exit from the checkpoint-activated state is not clear; the involvement of KNL-1 microtubule binding in this mechanism led us to hypothesize that noncentrosomal microtubules that assemble in the vicinity of chromosomes may interact with kinetochores facing away from the single pole, and this in turn promotes exit from the checkpoint-active state. This hypothesis makes two predictions. First, the checkpoint delay induced by microtubule depolymerization should be longer than that induced by monopolar spindles, as there would be no microtubules present to promote mitotic exit. Second, the checkpoint delay induced by microtubule depolymerization should be the same for WT and microtubule-binding mutants of KNL-1, as there would be no microtubules present to allow WT KNL-1 microtubule-binding activity to accelerate checkpoint silencing relative to the microtubule-binding mutants.
To examine checkpoint signaling in the absence of microtubules, we used a recently developed method to add drugs in a timed manner to C. elegans embryos (Carvalho et al., 2011). In this approach, inhibition of the perm-1 gene, which is involved in generating the eggshell permeability barrier, is used to permeabilize the eggshell to small molecules (Fig. 4 A). The fragile permeabilized embryos are placed in wells of a microchamber, enabling nocodazole addition while filming. To enable comparison with the monopolar spindle assay, nocodazole addition was performed 1–2 min before NEBD in the AB cell of the two-cell–stage embryo. As nocodazole treatment causes chromosomes to collapse together, decondensation cannot be used to monitor mitotic duration. Therefore, we quantified the interval between NEBD and the onset of cortical contractility, an alternative hallmark of Cdk1 inactivation (Essex et al., 2009).

KNL-1 microtubule-binding mutants do not extend the cell cycle in the absence of microtubules. (A) A schematic of the experimental approach used to analyze mitotic duration in the AB cell after microtubule (MT) depolymerization. (B) Mean time from NEBD to the onset of cortical contractility for the indicated conditions. Error bars represent the SEM with a 95% confidence interval.
Treatment of embryos with nocodazole resulted in a longer cell cycle delay compared with that induced by the formation of monopolar spindles, suggesting that microtubules do promote exit from the checkpoint-active state induced by monopolar spindles (Fig. 4 B). Next, we compared mitotic duration in WT and 4A mutant KNL-1 in the absence of microtubules and found that there was no significant difference between them (Fig. 4 B). We performed this experiment in a strain expressing GFP:Mad2MDF-2 to correlate the delay in mitotic exit with GFP:Mad2MDF-2 localization; however, chromosome compaction after nocodazole addition and diffuse Mad2MDF-2 signal in the nuclear environment prevented us from assessing Mad2MDF-2 kinetochore localization.
Thus, inhibiting KNL-1 microtubule-binding activity extends the checkpoint-active state in cells with bipolar and monopolar spindles but not in cells lacking microtubules. This result suggests that the effect of mutating KNL-1–dependent microtubule-binding activity on checkpoint silencing is a microtubule-dependent reaction in vivo.
KNL-1 microtubule-binding mutants do not affect PP1 docking and vice versa
The basic patch implicated in microtubule binding is adjacent to the conserved KNL-1 PP1-docking motifs (Fig. 5 A). As PP1 and the KNL-1–PP1 interaction have been implicated in checkpoint silencing (Pinsky et al., 2009; Vanoosthuyse and Hardwick, 2009; Meadows et al., 2011; Rosenberg et al., 2011) and the microtubule-binding mutants of KNL-1 that we engineered affected checkpoint silencing, we tested whether the mutations that disrupted microtubule binding also perturbed PP1 docking. For this purpose, we established biochemical and yeast two-hybrid–based assays for the KNL-1–PP1 interaction. C. elegans has four PP1 catalytic subunits: GSP-1, -2, -3, and -4 (Fig. S3 A). GSP-1 and -2 are partially redundant and essential for viability (Hsu et al., 2000), whereas GSP-3 and -4 are expressed during spermatogenesis (Chu et al., 2006). KNL-1 interacts with GSP-1 and -2 but not GSP-3 or -4 (Fig. 5 B); biochemical analysis confirmed that the interactions are direct (Fig. 5 E). Consistent with prior work, mutation of RRVSF to RRASA abolished binding in both assays (Fig. 5, C–E; Liu et al., 2010). Mutation of SILK to SAAA had no detectable effect in the biochemical assay, but a defect was observed at high stringency in the two-hybrid assay (Fig. S3 C). Notably, the 4A and Δ9 mutants that perturb microtubule binding/bundling did not affect PP1 docking (Fig. 5, D and E). Conversely, the RRASA mutant that abolished PP1 docking did not affect microtubule binding or bundling (Fig. 5 F; Liu et al., 2010). These results demonstrate that, despite their close proximity in the KNL-1 primary sequence, microtubule binding and PP1 docking are separable activities.
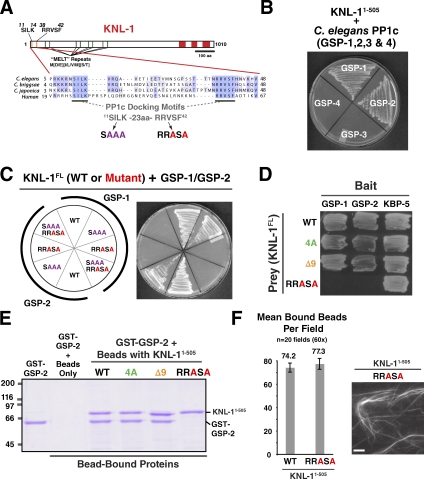
Microtubule-binding mutants of KNL-1 do not affect the interaction with PP1 and vice versa. (A) The conserved PP1 (PP1c)-docking motif SILK-RRVSF is highlighted, and mutations generated in the docking motif are indicated. (B) KNL-1 interacts with the C. elegans PP1 homologs GSP-1 and -2 but not GSP-3 and -4 in yeast two-hybrid assays (see also Fig. S3, A and B). (C) KNL-1 interaction with GSP-1/2 requires the RRVSF motif (see also Figs. 6 D and S3 C). (D) The 4A and Δ9 mutants do not perturb interaction of KNL-1 with GSP-1 and -2 in yeast two-hybrid analysis. KBP-5, a KNL-1–binding protein that interacts with the C-terminal half of KNL-1, serves as a positive control. (E) Biochemical analysis of KNL-1 interaction with GSP-2. Indicated variants of KNL-11–505–6xHis immobilized on nickel agarose beads were incubated with GST–GSP-2, and bead-bound proteins were analyzed by SDS-PAGE. Molecular mass is indicated in kilodaltons. (F) PP1-docking mutant of KNL-1 interacts normally with microtubules. Assays using purified KNL-11–505 variants were performed as described in Fig. 1 (C and E). The WT data are reproduced from Fig. 1 E. Error bars represent the SEM with a 95% confidence interval. Bar, 10 µm.
PP1-docking mutants of KNL-1 delay the formation of load-bearing attachments and extend mitotic duration in cells with monopolar spindles
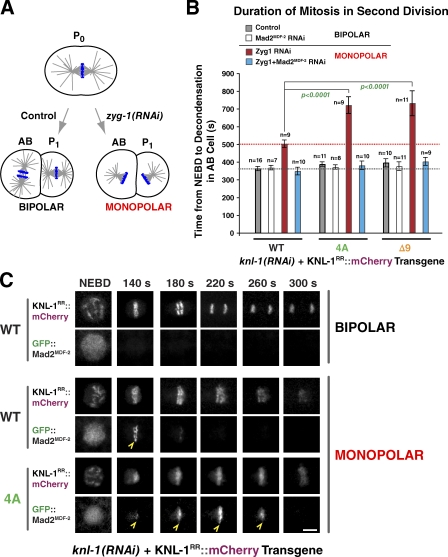
Next, we analyzed the in vivo phenotypes of a KNL-1 RRASA mutant, an SAAA mutant, and an SAAA;RRASA double mutant. All of these mutants localized normally to kinetochores in the absence of endogenous KNL-1 (Fig. 6 A). Three phenotypes were evident after endogenous KNL-1 depletion in the RRASA mutant: a kinetic delay in the formation of load-bearing kinetochore–microtubule attachments (Fig. 6, B and C), a delay in anaphase onset (Fig. 6 [B, C, and E] and Video 3), and partial embryonic lethality that decreased with time after dsRNA injection (Figs. 6 G and S2). The delay in load-bearing attachment formation is inferred from the pronounced bump in the pole-tracking analysis (Fig. 6 B); the front half of the bump represents the interval during which kinetochores provide no resistance to spindle pole separation induced by cortical pulling forces. Despite the delay in attachment formation, lagging anaphase chromosomes were not observed at appreciable frequency in the first division (unpublished data). The SAAA mutant, consistent with its mild effect on PP1 binding relative to the RRASA mutant, showed significantly milder phenotypes in vivo (Fig. 6, B, D, and E). Importantly, an SAAA;RRASA double mutant was quantitatively identical to the RRASA mutant in the pole-tracking assay (Fig. 6 B and Video 3) and in the extent to which it elongated the NEBD–anaphase onset interval in one-cell embryos (Fig. 6, B and E). Thus, the RRASA mutant alone behaves as a null for KNL-1–PP1 docking in vivo. In contrast to the RRASA and SAAA;RRASA mutants, depletion of the two catalytic PP1 subunits (GSP-1 and -2) resulted in highly pleiotropic defects in the early embryo (Fig. S4), likely reflecting multiple PP1 functions that are executed in association with different docking subunits.
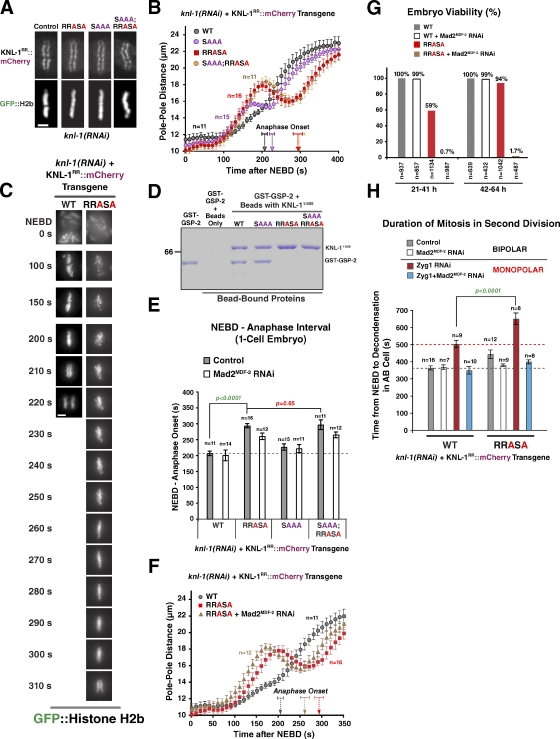
PP1-docking mutants of KNL-1 exhibit kinetic defects in load-bearing attachment formation and are synthetically lethal with checkpoint inhibition. (A) An image of a metaphase plate in living one-cell embryos from strains harboring the indicated KNL-1RR::mCherry transgenes and depleted of endogenous KNL-1; the control image is reproduced from Fig. 2 A. Bar, 2 µm. (B) Spindle pole separation kinetics for the indicated conditions. Error bars represent the SEM with a 95% confidence interval. The WT trace is reproduced from Fig. 2 D. (C) Frames from time-lapse sequences of the first embryonic division for the indicated KNL-1 mutants. Bar, 3 µm. (D) Biochemical analysis of KNL-1 interaction with GSP-2 performed as in Fig. 5 E. Molecular mass is indicated in kilodaltons. (E) NEBD–anaphase onset interval in one-cell stage embryos for the indicated conditions. See Table S1 for statistical analysis. (F) Spindle pole separation kinetics for the indicated conditions. Error bars represent the SEM with a 95% confidence interval. The WT trace is reproduced from Fig. 2 D. (G) Embryonic viability analysis for the indicated conditions. Lethality was measured during two intervals (21–41 and 42–64 h) after endogenous KNL-1 depletion. In the earlier time point, embryos are depleted of maternal load but potentially inherit some dsRNA that affects zygotic KNL-1 expression; at the later time point, the maternal load is depleted, but zygotic expression is likely unaffected (see the legend of Fig. S2). (H) NEBD–chromosome decondensation interval in AB cells with bipolar or monopolar spindles for the indicated conditions. The red dashed line marks the duration of AB cell mitosis induced by monopolar spindles in the same strain. See Table S2 for statistical analysis. (E and H) The gray dashed lines mark the duration of AB cell mitosis for the WT transgene. Error bars represent the SEM with a 95% confidence interval.
Codepletion of Mad2MDF-2 reduced the significant anaphase onset delay in the RRASA mutant (as well as the SAAA;RRASA mutant) but did not restore anaphase timing to that in controls (Fig. 6 [E and F] and Table S1). The reason why perturbing PP1 docking on the KNL-1 N terminus extends the interval between NEBD and anaphase onset independently of checkpoint signaling is unclear.
Importantly, Mad2MDF-2 codepletion severely enhanced the lethality of the RRASA mutant, indicating that checkpoint activation protects against the defect in load-bearing attachment formation created by perturbing PP1 docking on KNL-1 (Fig. 6 G). Thus, unlike the 4A and Δ9 microtubule-binding mutants, where attachment defects are not observed and checkpoint inhibition does not enhance lethality, the RRASA mutant exhibits enhanced lethality after checkpoint inactivation.
Next, we performed the monopolar spindle checkpoint assay for the RRASA mutant. The RRASA mutant exhibited a significantly extended cell cycle delay (Fig. 6 H) as well as increased residence of GFP::Mad2MDF-2 at unattached kinetochores (Fig. S5). The extended delay was largely abrogated by Mad2MDF-2 codepletion, indicating that it is a result of persistent checkpoint activation. As prior work in yeast has suggested a direct role for the KNL-1–PP1 interaction in checkpoint silencing (Meadows et al., 2011; Rosenberg et al., 2011), this observation is most consistent with the view that KNL-1–PP1 docking, in addition to affecting the kinetics of load-bearing attachment formation, participates in checkpoint silencing.
KNL-1 mutants affecting both microtubule binding and PP1 docking exhibit an additive phenotype
The aforementioned results indicate that KNL-1 microtubule-binding activity is dispensable for load-bearing attachment formation but participates in checkpoint silencing at the kinetochore and that PP1 docking on KNL-1 delays load-bearing attachment formation and potentially also contributes to checkpoint silencing. Given the close proximity of the microtubule-binding and PP1-docking site in the KNL-1 primary sequence and prior work demonstrating regulation of PP1 activity by regions adjacent to docking motifs in other PP1 regulators (Ragusa et al., 2010), an attractive model is that KNL-1 microtubule binding controls checkpoint silencing via modulation of KNL-1–docked PP1 activity. This model predicts that a double mutant affecting both microtubule binding and PP1 docking should not exhibit an additive phenotype. To test this prediction, we generated a strain expressing a KNL-1 transgene containing both the 4A and RRASA mutations. Then, we analyzed this double mutant, which localized normally to kinetochores (Fig. 7 A), in all of the functional assays. In contrast to the prediction, the 4A;RRASA mutant exhibited additive defects in the one-cell embryo (Fig. 7 [B and D] and Video 4) and in the second-division monopolar spindle assay (Fig. 7 E). To test whether the additive defect observed in the 4A;RRASA mutant was a result of the extended checkpoint signaling, we codepleted Mad2MDF-2 along with endogenous KNL-1. When Mad2MDF-2 was depleted, the 4A;RRASA mutant behaved similarly to the RRASA mutant; the timing of the NEBD–anaphase onset interval in the one-cell embryo was not significantly different, and the pole-tracking profiles were similar (Fig. 7, C and D). Thus, in the absence of checkpoint signaling, the attachment defect of the RRASA mutant is not significantly enhanced by addition of the 4A mutation; a corollary to this conclusion is that the delay and higher peak amplitude observed in the 4A;RRASA mutant relative to the RRASA mutant without Mad2MDF-2 depletion (Fig. 7 B) reflect a consequence of extended checkpoint signaling. Collectively, these results suggest that the additive phenotype is a result of independent contributions to checkpoint silencing of the microtubule-binding and PP1-docking activities located within close proximity in the N terminus of KNL-1.
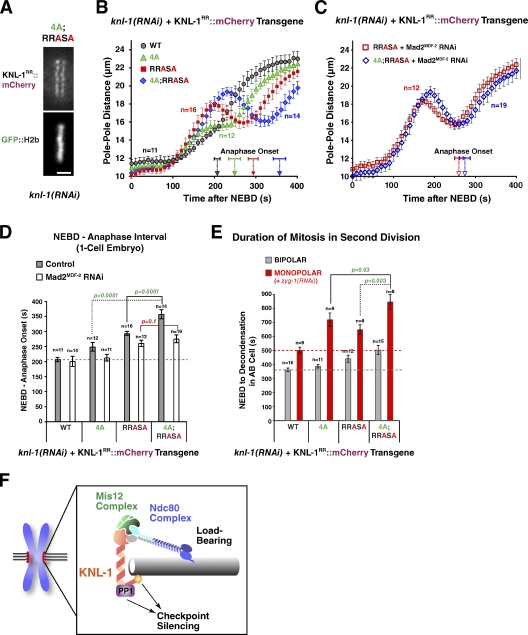
Disrupting both microtubule-binding and PP1-docking activities of KNL-1 has an additive phenotype. (A) A still image of a living one-cell metaphase embryo from a strain expressing a 4A;RRASA KNL-1 mutant; endogenous KNL-1 was depleted. Bar, 2 µm. (B and C) Spindle pole separation kinetics for the indicated conditions. WT and single mutant traces (4A and RRASA) are reproduced from Figs. 2 D and 6 B. Error bars represent the SEM with a 95% confidence interval. Anaphase onset times are marked on the x axis. (D) Timing of anaphase onset in the first embryonic division for the indicated conditions. The gray dashed line is drawn as in Fig. 2 E. For statistical analysis, see Table S1. WT and single-mutant timing data are reproduced from Figs. 2 E and 6 E. Error bars represent the SEM with a 95% confidence interval. (E) Mean duration of AB cell mitosis for the indicated conditions. Dashed lines are drawn as in Fig. 3 B; WT and single-mutant data are from Figs. 3 B and 6 H. For statistical analysis, see Table S2. Error bars represent the SEM with a 95% confidence interval. (F) A schematic summarizing the conclusion that the microtubule-binding activity located in the N terminus of KNL-1 participates in checkpoint silencing.
Discussion
In vitro reconstitution of the conserved KMN network, which provides the core microtubule-binding activity at kinetochores and scaffolds spindle checkpoint signaling, identified two microtubule-binding activities, the first in the Ndc80 complex and the second in KNL-1 (Cheeseman et al., 2006). Although microtubule binding by the Ndc80 complex is critical to form load-bearing attachments, the functional significance of KNL-1 microtubule-binding activity is unknown. Here, we define the function of this activity by coupled in vitro and in vivo approaches. Our results indicate that KNL-1 microtubule-binding activity is dispensable for the formation of load-bearing attachments and instead participates in silencing the spindle checkpoint at the kinetochore.
The second conserved microtubule-binding activity of the KMN network is dispensable for load-bearing microtubule attachment formation
KNL-1 depletion in C. elegans or mutation of the KNL-1 ortholog Spc105 in budding yeast severely inhibits the formation of load-bearing attachments (Desai et al., 2003; Pagliuca et al., 2009; Akiyoshi et al., 2010). In C. elegans, this is a result of an essential role for KNL-1 in outer kinetochore assembly (Desai et al., 2003). The initial hypothesis for the function of KNL-1 microtubule binding was that it synergized with the Ndc80 complex to generate a dynamic yet stable kinetochore–microtubule interface (Cheeseman et al., 2006). The data presented here indicate that this is not the case, as mutants that abrogate KNL-1 microtubule binding in vitro exhibit no significant defects in chromosome segregation or load-bearing attachment formation and are not enhanced by inhibiting checkpoint signaling. These results were in contrast to perturbing PP1 docking on KNL-1, which caused kinetic defects in load-bearing attachment formation and strong synergistic lethality with checkpoint inhibition. Thus, we conclude that microtubule binding by C. elegans KNL-1 does not make an essential contribution to the ability of kinetochores to form load-bearing attachments.
KNL-1 microtubule binding functions in checkpoint silencing at the kinetochore in the presence of microtubules
Although perturbing KNL-1 microtubule binding did not affect chromosome segregation, it led to a significantly increased monopolar spindle–induced cell cycle delay and an extended NEBD–anaphase duration in the presence of bipolar spindles. The full magnitude of both delays was a result of checkpoint signaling, and, on monopolar spindles, the delay was correlated with increased persistence of Mad2MDF-2 at kinetochores. In light of the lack of detectable attachment or segregation defects, these observations suggested that KNL-1 microtubule binding participates in silencing the spindle checkpoint signal. The extended checkpoint signaling observed in KNL-1 microtubule-binding mutants is not a general feature of perturbing kinetochore–microtubule interactions, as mutations affecting Ndc80 complex microtubule binding do not extend the duration of the monopolar spindle–induced cell cycle delay (unpublished data). Notably, KNL-1 microtubule-binding mutants only exhibited an increased checkpoint-dependent cell cycle delay relative to WT KNL-1 in the presence of microtubules, suggesting that KNL-1 microtubule-binding activity participates in a microtubule-dependent manner in checkpoint silencing at the kinetochore. We propose that KNL-1 microtubule binding senses the presence of microtubules attached to the kinetochore, potentially via the closely associated Ndc80 complex, and relays their presence to shut off generation of the checkpoint signal (Fig. 7 F).
KNL-1 microtubule binding: A dynein-independent means for sensing presence of microtubules at kinetochores?
Work in Drosophila melanogaster and vertebrate cells has indicated that kinetochore-localized dynein motor contributes to checkpoint silencing by removing checkpoint proteins from kinetochores after microtubule attachment (Howell et al., 2001; Wojcik et al., 2001). However, many species, including fungi and higher plants, silence the checkpoint after microtubule attachment without the involvement of dynein. Recent work in human cells suggests that even in systems with dynein involvement, the checkpoint can be silenced via a dynein-independent pathway (Chan et al., 2009; Barisic et al., 2010; Gassmann et al., 2010). Microtubule-binding KNL-1 mutants do not affect dynein recruitment to unattached kinetochores, suggesting that the effect we observe is not a result of a loss of kinetochore dynein (unpublished data). As the microtubule-binding and PP1-docking mutants of KNL-1 have an additive effect on checkpoint silencing and prior work has implicated PP1 in dynein regulation at the kinetochore (Whyte et al., 2008), we speculate that KNL-1 microtubule binding may affect checkpoint silencing through a dynein-independent mechanism. As KNL-1 family proteins are present throughout eukaryotic evolution, including in species that do not have kinetochore dynein, KNL-1 microtubule-binding–based silencing may represent an evolutionarily ancient mechanism that has been enhanced/superseded by the dynein-based mechanism in animal cells. Testing this idea will require elucidating whether KNL-1 microtubule binding functions in parallel to or in the same pathway as dynein in checkpoint silencing, mapping and mutating KNL-1 microtubule-binding activity in yeast/higher plants that lack kinetochore dynein or dynein altogether, and analyzing mammalian Knl1 microtubule-binding mutants under conditions in which kinetochore dynein has been largely removed (Gassmann et al., 2010).
Mechanism of checkpoint silencing at the kinetochore
A major challenge with respect to elucidating the mechanism by which KNL-1 microtubule binding and PP1 docking participate in checkpoint silencing is our poor understanding of how the checkpoint signal is generated at the kinetochore. Recent work has suggested that the kinetochore neighborhood, as opposed to a specific protein binding–based allosteric mechanism, activates the Mad1–Mad2 complex for checkpoint signaling (Maldonado and Kapoor, 2011). However, the mechanism by which the kinetochore neighborhood activates signaling is not known and is a necessary prerequisite to understanding the silencing reaction. In addition to sites for microtubule binding and PP1 docking, the KNL-1 N terminus also provides an interaction surface for BUB-1, a conserved kinase that is critical for checkpoint signal generation and participates in chromosome segregation. The relationship between microtubule binding, PP1 docking, and scaffolding of BUB-1 on the KNL-1 N terminus is currently unclear; neither the microtubule-binding nor the PP1-docking mutants of KNL-1 affect BUB-1 targeting (unpublished data). We have not observed direct interactions between KNL-1 and BUB-1 in C. elegans using two-hybrid analysis; the sequence divergence of KNL-1 family proteins has precluded modeling BUB-1 interaction–defective alleles based on homology. KNL-1 is also directly involved in recruitment of the Ndc80 complex and the Rod–Zwilch–Zw10 complex, both of which participate in load-bearing attachment formation and are required for checkpoint activation. As the 4A and Δ9 mutants do not significantly affect load-bearing attachment formation, these mutants are unlikely to broadly affect Bub1, Ndc80 complex, and Rod–Zwilch–Zw10 complex function. However, it remains possible that KNL-1 microtubule binding may specifically affect the checkpoint activation functions of these components, which are currently poorly understood. Thus, mechanistic insight into the reactions that activate checkpoint signaling at the kinetochore and testing the effect of KNL-1 microtubule binding on these reactions as well as elucidating whether KNL-1 mutants participate in parallel to or in the same pathway as dynein in checkpoint silencing are important future goals.
In summary, engineering of precise mutations in KNL-1 guided by in vitro biochemistry combined with single-copy–targeted transgene insertions and high-resolution phenotypic analysis in the early C. elegans embryo has revealed an unexpected function for the second conserved microtubule-binding activity of the KMN network in checkpoint silencing. This finding provides insight into the network of relationships between the mechanical and checkpoint signaling activities of the kinetochore that underlies accurate segregation of chromosomes during cell division.
Materials and methods
Worm strains, RNAi, time-lapse microscopy, and yeast two hybrid
C. elegans strains used in this study are listed in Table S3. All strains were maintained at 20°C. For KNL-1RR::mCherry transgenes, exon 4 in the genomic locus was recoded, and the mCherry sequence was introduced just before the stop codon. The engineered locus and mutant variants were cloned into pCFJ151 (Frøkjaer-Jensen et al., 2008) and injected into strain EG4322 to obtain stable single-copy integrants. Integration of transgenes was confirmed by PCR and homogeneous KNL-1::mCherry fluorescence in all progeny. Transgenes were crossed into various marker strains before analysis. Immunoblots were performed on whole-worm extracts with affinity-purified antibodies to KNL-1 (Desai et al., 2003) using anti–α-tubulin as a loading control (DM1-α; Sigma-Aldrich). For RNAi, L4 worms were injected with dsRNAs (Table S4) and incubated for 38–43 h at 20°C. For double depletions, dsRNAs were mixed to obtain equal concentrations of ≥0.75 mg/ml for each dsRNA.
Chromosome segregation was followed in embryos expressing GFP::H2b/GFP:γ-tubulin using a deconvolution microscope (DeltaVision; Applied Precision) equipped with a charge-coupled device camera (CoolSnap; Roper Scientific) at 20°C. At 10-s intervals, five z sections were acquired at 2-µm steps using a 100× 1.3 NA U-Plan Apochromat objective (Olympus) with 2 × 2 binning and a 480 × 480–pixel area. Illumination was attenuated using a 10% neutral density filter, and each exposure was 100 ms. Quantitative analysis of spindle pole elongation was performed on videos in which the two poles remained in the same or neighboring focal planes. Z stacks were projected and imported into MetaMorph software (Molecular Devices), and spindle poles were manually tracked. Mean plots were generated by aligning sequences using NEBD as a time reference. NEBD was defined as the time when GFP fluorescence was equalized between both pronuclei and the cytoplasm. Tracking data were analyzed and plotted using a custom macro in Excel (Microsoft). Error bars represent the SEM with a confidence interval of 0.95. Measurements of the NEBD–decondensation intervals were performed in the same conditions with the same microscope, but the z sections were acquired at 20-s intervals.
For GFP::Mad2MDF-2 localization, embryos were filmed using a spinning disk confocal mounted on an inverted microscope (TE2000-E; Nikon) equipped with a 60× 1.4 NA Plan Apochromat lens (Nikon), a solid-state laser combiner (Andor Technology) with 491- and 561-nm lines, a CSU10 head (Yokogawa), and an electron multiplication back-thinned charge-coupled device camera (iXon; Andor Technology). Acquisition parameters, shutters, and focus were controlled by iQ 1.10.0 software (Andor Technology). 5 × 2–µm mCherry/GFP z series with no binning were collected every 20 s at 20°C. Exposures were 300 ms for GFP and 400 ms for mCherry. Analysis of acquired images was performed with MetaMorph software.
Two-hybrid analysis was performed according to the manufacturer’s instructions (Matchmaker; Takara Bio Inc.). KNL-1 and GSP-1/2/3/4 ORFs were amplified from N2 cDNA and cloned into pGBKT7 and pGADT7 plasmids. For nocodazole treatment of embryos, L4-stage larvae were injected with dsRNAs and incubated at 20°C for 21–24 h. One to three worms were placed on a dissection board of a microdevice specifically designed for drug treatment of C. elegans embryos (Carvalho et al., 2011) after filling it with 60 µl of 0.7× Egg Salts (1× Egg Salts: 118 mM NaCl, 40 mM KCl, 3.4 mM MgCl2, 3.4 mM CaCl2, and 5 mM Hepes, pH 7.4). Worms were dissected with a scalpel, and the released early embryos were swept into microwells with an eyelash tool. The microdevice was transferred to the microscope, and the embryos were imaged. 1–2 min before AB cell NEBD, the medium in the microdevice was replaced with fresh medium containing 33 µM nocodazole (M1404; Sigma-Aldrich).
Protein purification and pull-down assays
KNL-11–505 and GSP-2 ORFs were amplified from N2 cDNA and cloned into pET21A and pGEX6P-1, respectively. Expression in Rosetta (DE3) pLysS Escherichia coli was induced with 0.3 mM IPTG for 4 h at 20°C, and 6xHis and GST purifications were performed using standard procedures. For KNL-11–505, imidazole-eluted protein was exchanged into SP buffer (30 mM MOPS, pH 7.0, 50 mM NaCl, 0.5 mM EDTA, and 2 mM β-mercaptoethanol), bound to HiTrap SP Sepharose (GE Healthcare), eluted with a gradient from 50 to 500 mM NaCl, and dialyzed to reduce the salt concentration to 150 mM NaCl or exchanged into BRB80 (80 mM Pipes, pH 6.8, 1 mM EGTA, and 1 mM MgCl2) + 80 mM KCl. For GSP-2, glutathione-eluted protein was exchanged into 30 mM Hepes, pH 7.0, 50 mM NaCl, 0.1 mM EDTA, and 2 mM β-mercaptoethanol, bound to HiTrap SP Sepharose, and eluted with a gradient from 50 to 500 mM NaCl followed by dialysis to 100 mM NaCl.
For pull-down assays, 3 µg KNL-11–505 was immobilized on 20 µl Ni–nitrilotriacetic acid agarose resin (QIAGEN) for 1 h at 4°C. The resin was then washed three times (wash buffer: 50 mM Hepes, pH 7.5, 100 mM NaCl, 2 mM MnCl2, 0.1 mM EDTA, 0.1% Tween 20, 30 mM imidazole, 10% glycerol, and 5 mM β-mercaptoethanol) and incubated for 1 h at room temperature with 12 µg of purified GST–GSP-2 protein in a 55-µl volume. The resin was then washed three times, eluted with sample buffer, and analyzed by SDS-PAGE.
Microtubule-bundling and bead assays
For microtubule-bundling assays, 1 µM taxol-stabilized rhodamine microtubules was mixed with 0.5 µM KNL-11–505 variants or control buffer. After 5 min, the mixture was imaged using a 60× 1.4 NA Plan Apochromat objective.
For the bead microtubule–binding assay, 3 µg of KNL-11–505 variants was incubated with 10 µl of 12-pM polystyrene beads (SVP-05-10; Spherotech, Inc.) coated with anti-6xHis antibody (no. 34440; QIAGEN) for 45 min at 4°C. After sonication in ice water for 2 min, the beads were flowed through a microscope flow chamber coated with taxol-stabilized rhodamine microtubules, 20 fields of view were imaged at 60× magnification, and images were analyzed to measure the mean number of beads bound per field.
Online supplemental material
Fig. S1 shows purification of KNL-1 variants and rescue of a knl-1 deletion allele. Fig. S2 shows an analysis of embryonic viability. Fig. S3 shows PP1 catalytic subunit sequence alignments and controls for two-hybrid assays. Fig. S4 shows the phenotype of PP1 catalytic subunit depletions. Fig. S5 shows Mad2MDF-2 enrichment on monopolar spindles in the PP1-docking motif mutants. Tables S1–S4 show statistical analysis of NEBD–anaphase onset interval in one-cell embryos (Table S1) and of the monopolar spindle–induced cell cycle delay in two-cell embryos (Table S2) and the C. elegans strains (Table S3) and dsRNAs (Table S4) used in this study. Videos 1–4 show the NEBD–anaphase interval in one-cell C. elegans embryos expressing GFP::histone H2b and GFP::γ-tubulin to mark chromosomes and spindle poles, respectively. Video 1 shows rescue of the kinetochore-null phenotype by WT KNL-1::mCherry. Video 2 shows the 4A and Δ9 KNL-1 mutants. Video 3 shows the PP1-docking motif mutants of KNL-1. Video 4 shows the 4A, RRASA, and 4A;RRASA double mutant. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201111107/DC1.
Acknowledgments
We are indebted to Christian Frøkjær-Jensen and Erik Jorgensen (University of Utah, Salt Lake City, UT) for their development of Mos single-copy insertion and their generosity with reagents and expertise. We thank the C. elegans Gene Knockout Consortium for the knl-1(ok3457) mutant, Andy Powers and Chip Asbury for help with the bead assay, Sue Biggins and members of the Desai and Oegema laboratories for useful discussions, and Becky Green for comments on the manuscript.
This work was supported by a grant from the National Institutes of Health to A. Desai (GM074215). K. Oegema and A. Desai receive salary support from the Ludwig Institute for Cancer Research.
Footnotes
Abbreviations used in this paper:
- dsRNA
- double-stranded RNA
- NEBD
- nuclear envelope breakdown
- WT
- wild type
References
- Akiyoshi B., Sarangapani K.K., Powers A.F., Nelson C.R., Reichow S.L., Arellano-Santoyo H., Gonen T., Ranish J.A., Asbury C.L., Biggins S. 2010. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature. 468:576–579 10.1038/nature09594 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Barisic M., Sohm B., Mikolcevic P., Wandke C., Rauch V., Ringer T., Hess M., Bonn G., Geley S. 2010. Spindly/CCDC99 is required for efficient chromosome congression and mitotic checkpoint regulation. Mol. Biol. Cell. 21:1968–1981 10.1091/mbc.E09-04-0356 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Burke D.J., Stukenberg P.T. 2008. Linking kinetochore-microtubule binding to the spindle checkpoint. Dev. Cell. 14:474–479 10.1016/j.devcel.2008.03.015 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Carvalho A., Olson S.K., Gutierrez E., Zhang K., Noble L.B., Zanin E., Desai A., Groisman A., Oegema K. 2011. Acute drug treatment in the early C. elegans embryo. PLoS ONE. 6:e24656 10.1371/journal.pone.0024656 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Chan Y.W., Fava L.L., Uldschmid A., Schmitz M.H., Gerlich D.W., Nigg E.A., Santamaria A. 2009. Mitotic control of kinetochore-associated dynein and spindle orientation by human Spindly. J. Cell Biol. 185:859–874 10.1083/jcb.200812167 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Cheeseman I.M., Desai A. 2008. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9:33–46 10.1038/nrm2310 [Abstract] [CrossRef] [Google Scholar]
- Cheeseman I.M., Niessen S., Anderson S., Hyndman F., Yates J.R., III, Oegema K., Desai A. 2004. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 18:2255–2268 10.1101/gad.1234104 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., Desai A. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997 10.1016/j.cell.2006.09.039 [Abstract] [CrossRef] [Google Scholar]
- Chu D.S., Liu H., Nix P., Wu T.F., Ralston E.J., Yates J.R., III, Meyer B.J. 2006. Sperm chromatin proteomics identifies evolutionarily conserved fertility factors. Nature. 443:101–105 10.1038/nature05050 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Desai A., Rybina S., Müller-Reichert T., Shevchenko A., Shevchenko A., Hyman A., Oegema K. 2003. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 17:2421–2435 10.1101/gad.1126303 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Essex A., Dammermann A., Lewellyn L., Oegema K., Desai A. 2009. Systematic analysis in Caenorhabditis elegans reveals that the spindle checkpoint is composed of two largely independent branches. Mol. Biol. Cell. 20:1252–1267 10.1091/mbc.E08-10-1047 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Frøkjaer-Jensen C., Davis M.W., Hopkins C.E., Newman B.J., Thummel J.M., Olesen S.P., Grunnet M., Jorgensen E.M. 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40:1375–1383 10.1038/ng.248 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Gassmann R., Holland A.J., Varma D., Wan X., Civril F., Cleveland D.W., Oegema K., Salmon E.D., Desai A. 2010. Removal of Spindly from microtubule-attached kinetochores controls spindle checkpoint silencing in human cells. Genes Dev. 24:957–971 10.1101/gad.1886810 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Hendrickx A., Beullens M., Ceulemans H., Den Abt T., Van Eynde A., Nicolaescu E., Lesage B., Bollen M. 2009. Docking motif-guided mapping of the interactome of protein phosphatase-1. Chem. Biol. 16:365–371 10.1016/j.chembiol.2009.02.012 [Abstract] [CrossRef] [Google Scholar]
- Howell B.J., McEwen B.F., Canman J.C., Hoffman D.B., Farrar E.M., Rieder C.L., Salmon E.D. 2001. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 155:1159–1172 10.1083/jcb.200105093 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Hsu J.Y., Sun Z.W., Li X., Reuben M., Tatchell K., Bishop D.K., Grushcow J.M., Brame C.J., Caldwell J.A., Hunt D.F., et al. 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 102:279–291 10.1016/S0092-8674(00)00034-9 [Abstract] [CrossRef] [Google Scholar]
- Joglekar A.P., Bloom K.S., Salmon E.D. 2010. Mechanisms of force generation by end-on kinetochore-microtubule attachments. Curr. Opin. Cell Biol. 22:57–67 10.1016/j.ceb.2009.12.010 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Kerres A., Jakopec V., Fleig U. 2007. The conserved Spc7 protein is required for spindle integrity and links kinetochore complexes in fission yeast. Mol. Biol. Cell. 18:2441–2454 10.1091/mbc.E06-08-0738 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Kiyomitsu T., Obuse C., Yanagida M. 2007. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev. Cell. 13:663–676 10.1016/j.devcel.2007.09.005 [Abstract] [CrossRef] [Google Scholar]
- Liu J., Vasudevan S., Kipreos E.T. 2004. CUL-2 and ZYG-11 promote meiotic anaphase II and the proper placement of the anterior-posterior axis in C. elegans. Development. 131:3513–3525 10.1242/dev.01245 [Abstract] [CrossRef] [Google Scholar]
- Liu D., Vleugel M., Backer C.B., Hori T., Fukagawa T., Cheeseman I.M., Lampson M.A. 2010. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J. Cell Biol. 188:809–820 10.1083/jcb.201001006 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Maldonado M., Kapoor T.M. 2011. Constitutive Mad1 targeting to kinetochores uncouples checkpoint signalling from chromosome biorientation. Nat. Cell Biol. 13:475–482 10.1038/ncb2223 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Meadows J.C., Shepperd L.A., Vanoosthuyse V., Lancaster T.C., Sochaj A.M., Buttrick G.J., Hardwick K.G., Millar J.B. 2011. Spindle checkpoint silencing requires association of PP1 to both Spc7 and kinesin-8 motors. Dev. Cell. 20:739–750 10.1016/j.devcel.2011.05.008 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Musacchio A., Salmon E.D. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393 10.1038/nrm2163 [Abstract] [CrossRef] [Google Scholar]
- Oegema K., Desai A., Rybina S., Kirkham M., Hyman A.A. 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153:1209–1226 10.1083/jcb.153.6.1209 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Pagliuca C., Draviam V.M., Marco E., Sorger P.K., De Wulf P. 2009. Roles for the conserved spc105p/kre28p complex in kinetochore-microtubule binding and the spindle assembly checkpoint. PLoS ONE. 4:e7640 10.1371/journal.pone.0007640 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Petrovic A., Pasqualato S., Dube P., Krenn V., Santaguida S., Cittaro D., Monzani S., Massimiliano L., Keller J., Tarricone A., et al. 2010. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J. Cell Biol. 190:835–852 10.1083/jcb.201002070 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Pinsky B.A., Nelson C.R., Biggins S. 2009. Protein phosphatase 1 regulates exit from the spindle checkpoint in budding yeast. Curr. Biol. 19:1182–1187 10.1016/j.cub.2009.06.043 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Powers A.F., Franck A.D., Gestaut D.R., Cooper J., Gracyzk B., Wei R.R., Wordeman L., Davis T.N., Asbury C.L. 2009. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell. 136:865–875 10.1016/j.cell.2008.12.045 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Ragusa M.J., Dancheck B., Critton D.A., Nairn A.C., Page R., Peti W. 2010. Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites. Nat. Struct. Mol. Biol. 17:459–464 10.1038/nsmb.1786 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Rosenberg J.S., Cross F.R., Funabiki H. 2011. KNL1/Spc105 recruits PP1 to silence the spindle assembly checkpoint. Curr. Biol. 21:942–947 10.1016/j.cub.2011.04.011 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Santaguida S., Musacchio A. 2009. The life and miracles of kinetochores. EMBO J. 28:2511–2531 10.1038/emboj.2009.173 [Abstract] [CrossRef] [Google Scholar]
- Tooley J., Stukenberg P.T. 2011. The Ndc80 complex: Integrating the kinetochore’s many movements. Chromosome Res. 19:377–391 10.1007/s10577-010-9180-5 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Vanoosthuyse V., Hardwick K.G. 2009. A novel protein phosphatase 1-dependent spindle checkpoint silencing mechanism. Curr. Biol. 19:1176–1181 10.1016/j.cub.2009.05.060 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Welburn J.P., Vleugel M., Liu D., Yates J.R., III, Lampson M.A., Fukagawa T., Cheeseman I.M. 2010. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell. 38:383–392 10.1016/j.molcel.2010.02.034 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Whyte J., Bader J.R., Tauhata S.B., Raycroft M., Hornick J., Pfister K.K., Lane W.S., Chan G.K., Hinchcliffe E.H., Vaughan P.S., Vaughan K.T. 2008. Phosphorylation regulates targeting of cytoplasmic dynein to kinetochores during mitosis. J. Cell Biol. 183:819–834 10.1083/jcb.200804114 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Wojcik E., Basto R., Serr M., Scaërou F., Karess R., Hays T. 2001. Kinetochore dynein: Its dynamics and role in the transport of the Rough deal checkpoint protein. Nat. Cell Biol. 3:1001–1007 10.1038/ncb1101-1001 [Abstract] [CrossRef] [Google Scholar]
Articles from The Journal of Cell Biology are provided here courtesy of The Rockefeller University Press
Full text links
Read article at publisher's site: https://doi.org/10.1083/jcb.201111107
Read article for free, from open access legal sources, via Unpaywall:
http://jcb.rupress.org/content/jcb/196/4/469.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1083/jcb.201111107
Article citations
Plant kinetochore complex: composition, function, and regulation.
Front Plant Sci, 15:1467236, 10 Oct 2024
Cited by: 0 articles | PMID: 39464281 | PMCID: PMC11503545
Review Free full text in Europe PMC
Kinetochore dynein is sufficient to biorient chromosomes and remodel the outer kinetochore.
Nat Commun, 15(1):9085, 21 Oct 2024
Cited by: 0 articles | PMID: 39433738 | PMCID: PMC11494143
Argonaute protein CSR-1 restricts localization of holocentromere protein HCP-3, the C. elegans CENP-A homolog.
J Cell Sci, 137(18):jcs261895, 18 Sep 2024
Cited by: 0 articles | PMID: 39037215 | PMCID: PMC11423810
Meiosis-specific functions of kinetochore protein SPC105R required for chromosome segregation in Drosophila oocytes.
Mol Biol Cell, 35(8):ar105, 12 Jun 2024
Cited by: 1 article | PMID: 38865189 | PMCID: PMC11321039
The outer kinetochore components KNL-1 and Ndc80 complex regulate axon and neuronal cell body positioning in the C. elegans nervous system.
Mol Biol Cell, 35(6):ar83, 24 Apr 2024
Cited by: 0 articles | PMID: 38656792 | PMCID: PMC11238089
Go to all (92) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The outer kinetochore protein KNL-1 contains a defined oligomerization domain in nematodes.
Mol Biol Cell, 26(2):229-237, 19 Nov 2014
Cited by: 8 articles | PMID: 25411336 | PMCID: PMC4294671
KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans.
Genes Dev, 17(19):2421-2435, 01 Oct 2003
Cited by: 172 articles | PMID: 14522947 | PMCID: PMC218079
The outer kinetochore components KNL-1 and Ndc80 complex regulate axon and neuronal cell body positioning in the C. elegans nervous system.
Mol Biol Cell, 35(6):ar83, 24 Apr 2024
Cited by: 0 articles | PMID: 38656792 | PMCID: PMC11238089
Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore.
Nat Rev Mol Cell Biol, 14(1):25-37, 01 Jan 2013
Cited by: 409 articles | PMID: 23258294 | PMCID: PMC3762224
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NIGMS NIH HHS (2)
Grant ID: R01 GM074215
Grant ID: GM074215






