Abstract
Free full text

Characterization of a selective inhibitor of the Parkinson’s disease kinase LRRK2
Abstract
Mutations in leucine-rich repeat kinase 2 (LRRK2) are strongly associated with late-onset autosomal dominant Parkinson’s disease. We employed a novel, parallel, compound-centric approach to identify a potent and selective LRRK2 inhibitor LRRK2-IN-1, and demonstrated that inhibition of LRRK2 induces dephosphorylation of Ser910/Ser935 and accumulation of LRRK2 within aggregate structures. LRRK2-IN-1 will serve as a versatile tool to pharmacologically interrogate LRRK2 biology and study its role in Parkinson’s disease.
Parkinson’s disease (PD) is the second most common neurodegenerative disease in the world, with a prevalence of more than 1% after the age of 65 years1. Mutations in the Leucine-rich repeat kinase-2 (LRRK2) gene were first linked to PD in 20042,3 and extensive genetic, cellular and biochemical studies suggest that it is a key driver of the disease. Among the genes associated with PD, such as parkin and PINK1, LRRK2 is unique because a missense mutation, G2019S, is frequently found in not only familial but also sporadic PD cases4,5.
The most common and extensively characterized mutation, G2019S, enhances kinase activity, suggesting that small molecule LRRK2 kinase inhibitors may serve as potential therapeutic agents for the treatment of PD6. To date, only relatively nonspecific LRRK2 inhibitors have been reported such as staurosporine7,8, K252A7,8, sunitinib7–9, H-11529, and GW507410. Here we report the identification of a selective inhibitor of LRRK2 and demonstrate its ability both in vitro and in vivo to inhibit LRRK2.
In an effort to discover small molecules that can selectively inhibit LRRK2 kinase activity, we applied a novel, highly parallel, compound-centric strategy11. We screened a 300-membered compound library designed to target the ATP-binding site against a panel of 442 diverse kinases using an in vitro ATP-site competition binding assay12,13. In contrast to the traditional, linear process of inhibitor discovery, this high-throughput kinase profiling enables rapid identification of compounds capable of selectively inhibiting LRRK2 and other kinases of interest. A series of 2-amino-5,11-dimethyl-5H-benzo[e]pyrimido-[5,4-b][1,4]diazepin-6(11H)-one analogs emerged as potential LRRK2 inhibitors with strong binding affinities against both wild-type LRRK2 and LRRK2 mutant G2019S. LRRK2-IN-1 was identified as a promising inhibitor following the synthesis of approximately 50 analogs guided by iterative rounds of synthesis and testing in biochemical and cellular assays (to be described elsewhere) (Fig. 1a and Supplementary Methods). LRRK2-IN-1 inhibits both wild-type and G2019S mutant LRRK2 kinase activity with IC50 values of 13 nM and 6 nM (Fig. 1b), respectively with 0.1 mM ATP in the assay. Consistent with LRRK2-IN-1 being an ATP competitive inhibitor, the IC50 for G2019S mutant was increased when ATP was increased (Supplementary Fig. 1). Previous work identified a LRRK2[A2016T] mutant that is normally active, but 32-fold less sensitive to H-1152 and 12-fold less sensitive to sunitinib9. Remarkably, LRRK2[A2016T] and LRRK2[A2016T+G2019S] mutant were ~400-fold more resistant to LRRK2-IN-1 (Fig. 1b). A molecular model of LRRK2-IN-1 binding to the ATP-site of LRRK2 demonstrates that the A2016T mutation likely introduces a disfavorable steric interaction with the anthranilic acid phenyl group of LRRK2-IN-1 (Supplementary Fig. 2).
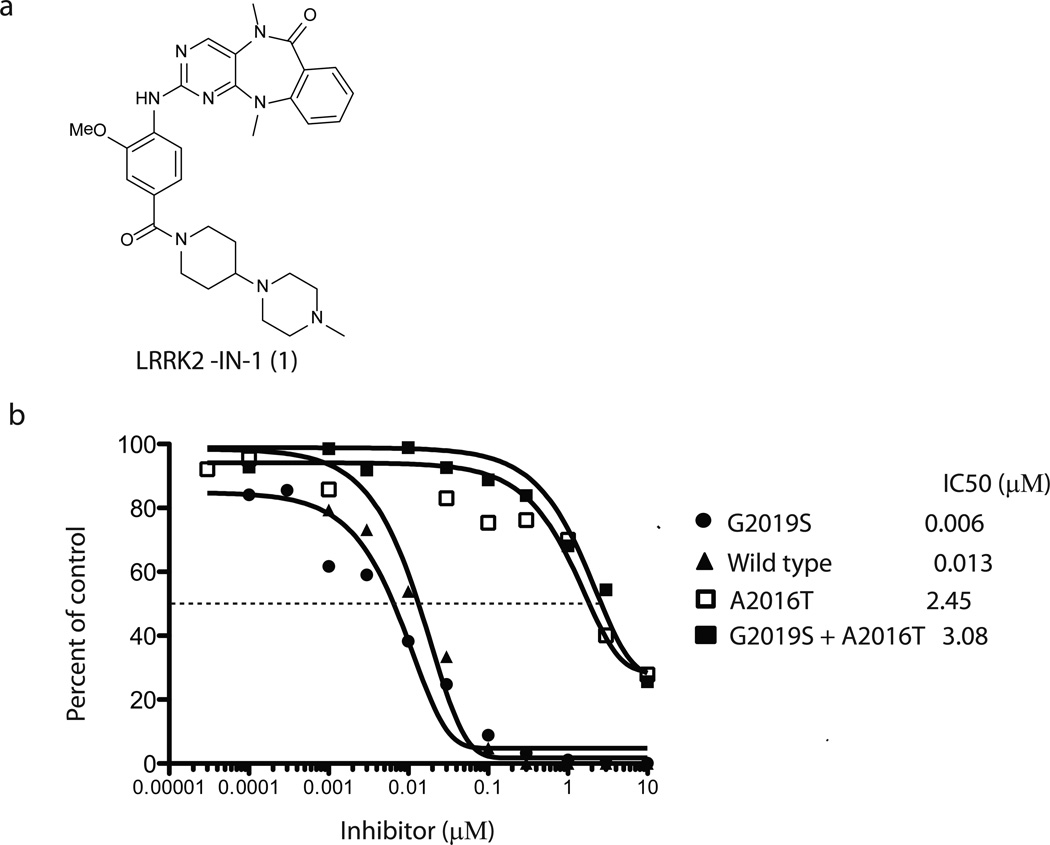
(a) Chemical structure of the LRRK2 inhibitor LRRK2-IN-1. (b) GST-LRRK2 (1326-2517), GST-LRRK2 [G2019S] (1326-2527), GST-LRRK2 [A2016T] (1326-2517) and GST-LRRK2 [A2016T+G2019S] (1326-2517) were assayed using 20µM Nictide in the presence of 100 µM ATP with the indicated concentrations of LRRK2-IN-1. The results are presented as percentage of kinase activity relative to the DMSO treated control. Results are averages of duplicate reactions with similar results obtained in at least one other experiment. The IC50 values, in µM, were derived from the graphs. (c) The combined kinase profiling hits with biochemical activities. Kinase hits from Ambit profiling with a score of less than 10% of the DMSO control at a concentration of 10 µM, ActivX profiling with higher than 50% inhibition at 1 µM, and Dundee profiling with greater than 50% inhibition at 1 µM are listed. The biochemical IC50 values of hits were measured by Invitrogen Adapta®, Z´-Lyte®, Lantha-Screen® assays (http://www.invitrogen.com/site/us/en/home/Products-and-Services/Services/Screening-and-Profiling-Services/SelectScreen-Profiling-ervice/SelectScreen-Kinase-Profiling-Service.html) and Dundee radioactive-base enzyme assay14. *Assessment of PLK1 cellular activity of LRRK2-IN-1 (see Supplementary Results).
The kinase selectivity of LRRK2-IN-1 was comprehensively evaluated using three independent and complementary experimental approaches that in aggregate assessed the selectivity against >470 kinases. These approaches included kinase-binding assays against a panel of 442 distinct kinases using the KINOMEscan™ methodology (Ambit Biosciences, San Diego, CA)12,13, standard radioactive-based enzymatic assays against a panel of 105 kinases (Dundee profiling)14, and activity-based proteomics against 260 kinases using the KiNativ™ technology (ActivX Biosciences, San Diego, CA)15. This analysis revealed that LRRK2-IN-1 possessed a selective profile (see Supplementary Dataset), inhibiting only 12 kinases among the ambit 442 diverse kinases panel with a score of less than 10% of the DMSO control at a concentration of 10 µM. In the biochemical assays, LRRK2-IN-1 possessed an IC50 of 45 nM for inhibition of DCLK2 and exhibited an IC50 of greater than 1 µM when evaluated in biochemical assays for AURKB, CHEK2, MKNK2, MYLK, NUAK1, and PLK1 (Supplementary Table 1). Biochemical assays are not available for MAPK7, DCLK1, PLK4, RPS6KA2 (Kin.Dom.2-C-terminal), and RPS6KA6 (Kin.Dom.2-C-terminal), therefore the dissociation constants (Kds) were measured by KINOMEscan binding assay (Supplementary Table 1). The selectivity score (S (3 µM)) for LRRK2-IN-1 calculated by dividing the number of kinases found to bind with a dissociation constant (Kd < 3 µM) by the total number of kinases tested is 0.029 (13/442), which represents high selectivity (see Supplementary Dataset)13. Further exploration of the selectivity of LRRK2-IN-1 was performed by profiling directly against all kinases detectable in human peripheral blood mononuclear cells (hPBMC, 180 kinases) and mouse brain tissue (195 kinases) using KiNativ™. This approach confirmed inhibition of LRRK2, DCLK1 and MAPK7, and supported >1 µM IC50s for CHEK2, and MYLK (smMLCK) and RPS6KA2. LRRK2-IN-1 was confirmed to inhibit MAPK7 with an EC50 of 160 nM (Supplementary Fig. 3). We are unable to evaluate whether LRRK2-IN-1 inhibits LRRK1 as LRRK1 kinase activity cannot be measured under the same conditions employed for LRRK2 (Supplementary Fig. 4). Overall the selectivity of LRRK2-IN-1 is clearly sufficient for use as a tool compound for assessing the effects of LRRK2 inhibition in cellular systems.
LRRK2 binds to isoforms of 14-3-3 proteins and this interaction depends on the phosphorylation of LRRK2 at Ser910 and Ser93516. Recent work suggested that inhibition of LRRK2 kinase activity stimulates an as yet uncharacterized regulatory feedback loop promoting the release of 14-3-3 and dephosphorylation of LRRK2 at Ser910 and Ser93517. Indeed, LRRK2-IN-1 induced a dose dependent inhibition of Ser910 and Ser935 phosphorylation accompanied by loss of 14-3-3 binding to both wild type LRRK2 (Fig. 2a) and LRRK2[G2019S] stably transfected into HEK293 cells(Fig. 2b). Significant dephosphorylation of Ser910/Ser935 was observed at 1-3 µM of LRRK2-IN-1 for wild type LRRK2 and at slightly lower doses for LRRK2[G2019S] (Fig. 2a & 2b). Crucially, LRRK2-IN-1 failed to induce dephosphorylation of Ser910/Ser935 in the drug resistant LRRK2[A2016T] (Fig. 2c) and LRRK2[A2016T+G2019S] (Fig. 2d) mutants, establishing that the effects of LRRK2-IN-1 on LRRK2 phosphorylation are indeed mediated via inhibition of LRRK2 kinase activity and not due to off target effects.
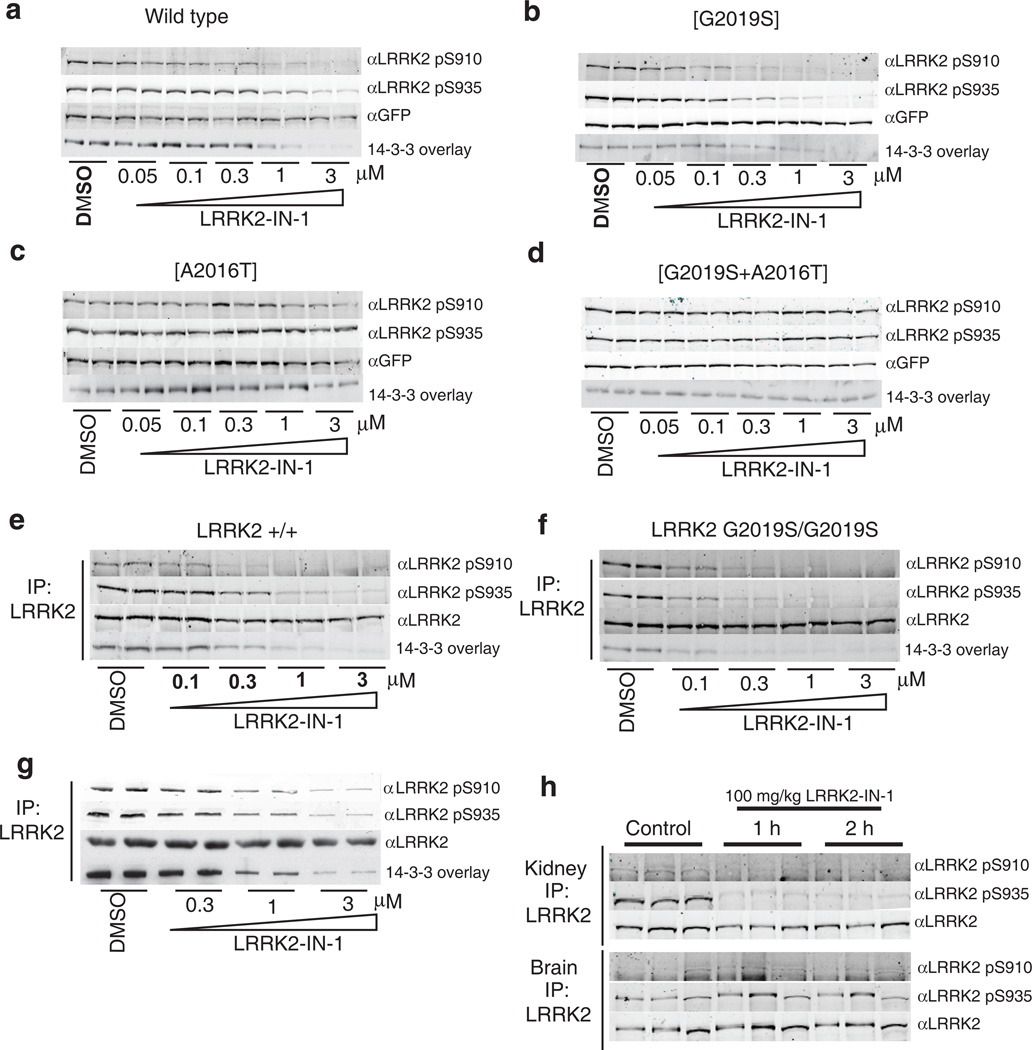
(a) HEK 293 cells stably expressing wild type GFP-LRRK2 were treated with DMSO or increasing concentrations of LRRK2-IN-1 for 90 min. Cell lysates were subjected to immunoblotting for detection of LRRK2 phosphorylated at S910 and S935 and for total GFP-LRRK2. 14-3-3 binding was measured by overlay assay following immunoprecipitation of LRRK2 with GFP-binder Sepharose. (b) As in (a) except HEK 293 cells stably expressing GFP-LRRK2 [G2019S] were utilized. (c) As in (a) except HEK 293 cells stably expressing drug resistant GFP-LRRK2 [A2016T] were utilized. (d) As in (a) except HEK 293 cells stably expressing drug resistant GFPLRRK2[ A2016T+G2019S] cells were utilized. (e) Endogenous LRRK2 was immunoprecipitated with anti-LRRK2 100-500 from EBV immortalized human lymphoblastoid cells from a control subject and a Parkinson’s disease patient homozygous for the LRRK2 [G2019S] mutation. (f) Following treatment of the cells with DMSO or the indicated concentrations of LRRK2-IN-1 for 90 min. Immunoprecipitates were subjected to immunoblot analysis with the indicated antibody as well as 14-3-3 overlay for western analysis. Immunoprecipitations were performed in duplicate and results are representative of at least two independent experiments. (g) As in (e) except that human SHSY5Y cells were utilized. (h) 100 mg/kg LRRK2-IN-1 was administered by intraperitoneal injection into wild type mice (n=3 per group). Tissues were collected and endogenous LRRK2 was immunoprecipitated and analysed for S910 and S935 phosphorylation as above. Quantitation of immunoblots along with original images are available in Supplementary Fig. 11
Near maximal effects of 1 µM LRRK2-IN-1 on dephosphorylation of Ser910/Ser935 and 14-3-3 binding to wild type LRRK2 and LRRK2[G2019S] were observed within 15 min and sustained for at least 2 h (Supplementary Fig. 5). Phosphorylation of Ser910/Ser935 and 14-3-3 binding was restored to near control levels within 2 hours of removal of LRRK2-IN-1 from the tissue culture medium (Supplementary Fig. 5). We observed that kinase inactive LRRK2 was phosphorylated at Ser910/Ser935 to a slightly lower level than wild type LRRK2 and treatment with LRRK2-IN-1 did not induce significant dephosphorylation of kinase-inactive LRRK2 (Supplementary Fig. 6).
We next examined the effect of LRRK2-IN-1 on endogenous LRRK2 phosphorylation and 14-3-3 binding in human lymphoblastoid cells derived from a control individual (Fig. 2e) and a Parkinson’s disease patient homozygous for the LRRK2 G2019S mutation (Fig. 2f). We found that increasing doses of LRRK2-IN-1 led to similar dephosphorylation of endogenous LRRK2 at Ser910 and Ser935, accompanied by loss of 14-3-3 binding, as was observed in HEK 293 cells stably expressing wild-type LRRK2 (compare Fig. 2a and 2e) or LRRK2[G2019S] (compare Fig. 2b and 2f). Moreover, endogenous LRRK2[G2019S] was also more sensitive to LRRK2-IN-1 than wild type LRRK2 (compare Fig. 2e and 2f). We also found that LRRK2-IN-1 induced a similar dose dependent Ser910/Ser935 dephosphorylation and loss of 14-3-3 binding to endogenous LRRK2 in human-derived neuroblastoma SHSY5Y cells (Fig. 2g) as well as mouse Swiss 3T3 cells (Supplementary Fig. 7). LRRK2-IN-1 exhibits favourable pharmacokinetics in mice with a T1/2 of 4.5 hr, AUC of 14758 hr*ng/mL and %F of 49.3% (Supplementary Table 2). Following intraperitoneal injection of LRRK2-IN-1 into mice (100 mg/kg) we observed complete Ser910/Ser935 dephosphorylation of LRRK2 in the kidney at the 1 h and 2 h time points (Fig. 2h). In contrast, no dephosphorylation of LRRK2 Ser910 or Ser935 was observed in brain, indicating that LRRK2-IN-1 is likely unable to efficiently cross the blood brain barrier (Fig. 2h).
Previous work suggested that dephosphorylation of Ser910/Ser935 and loss of 14-3-3-binding, promoted relocalization of LRRK2 to more aggregate and fibrillar-like structures17 that might resemble inclusion bodies that certain mutant forms of LRRK2 have been reported to accumulate within16,18,19. To verify how LRRK2-IN-1 affected LRRK2 localization, we treated HEK293 cell lines stably expressing forms of full length GFP-LRRK2 with 1 µM LRRK2-IN-1 over a 2 h time course. Prior to addition of LRRK2-IN-1, mutant LRRK2[G2019S] (Fig. 3a) and wild type LRRK2 (Supplementary Fig. 8a) were diffusely localized in the cytosol consistent with previous work17. Following addition of LRRK2-IN-1, we observed a significant accumulation of LRRK2[G2019S] (Fig. 3a, Movie 1 and Supplementary Fig. 9) and wild type LRRK2 (Supplementary Fig. 8a and Movie 2) into aggregate structures. Importantly, LRRK2-IN-1 did not affect the localization of drug resistant LRRK2[A2016T] (Supplementary Fig. 8b and Movie 3) or LRRK2[A2016T+G2019S] (Supplementary Fig. 8c and Movie 4) mutants, that remained diffusely localized in the cytoplasm. Consistent with LRRK2 becoming re-phosphorylated on Ser910 and Ser935 following inhibitor removal (Supplementary Fig. 5), LRRK2[G2019S] mutant (Fig. 3a & Movie 2) and wild type LRRK2 (Supplementary Fig. 8a and Movie 1) returned to a largely diffuse cytoplasmic re-localization within 2 hours of LRRK2-IN-1 removal. LRRK1 does not bind 14-3-3 and the localization of LRRK1 does not change with LRRK2-IN-1 treatment (Supplementary Fig. 10).
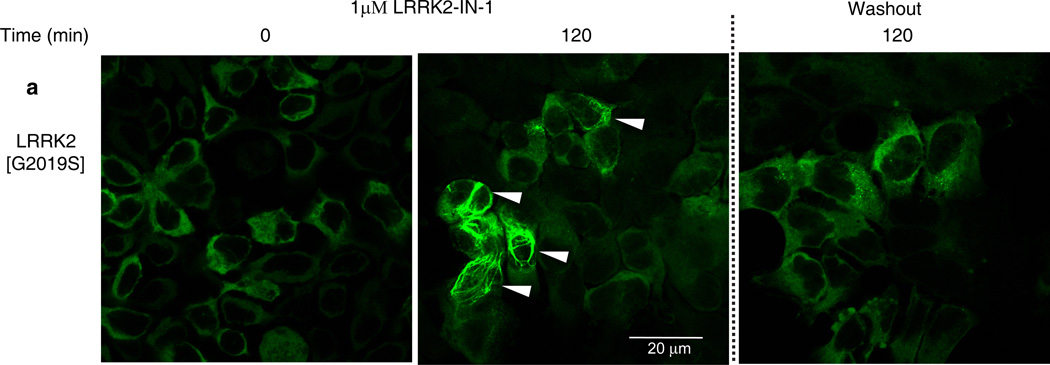
HEK 293 cells stably expressing GFP-LRRK2 [G2019S] were treated with 1 µM LRRK2-IN-1 for 2 h. Cells were then washed and fixed with 4% paraformaldehyde for fluorescence imaging. To visualize LRRK2 localization following removal of LRRK2-IN-1, inhibitor-containing media was removed 2 h after treatment. Cells were then washed twice and incubated with fresh media without inhibitor for 2 h before again being washed and then fixed for fluorescence imaging. Representative micrographs of three independent experiments are presented.
In conclusion, we report upon the discovery of LRRK2-IN-1, the first selective and potent inhibitor of LRRK2. We establish that LRRK2-IN-1 rapidly suppresses LRRK2 kinase activity in vivo leading to dephosphorylation of Ser910/Ser935, loss of 14-3-3 binding and accumulation of LRRK2 within aggregate fibrillar structures. LRRK2-IN-1 has the potential to transform our understanding of LRRK2 function, in the same way that other signal transduction inhibitors including PD9805920, SB20358021 and LY29400222 have aided with elucidation of the roles of Mek, p38 and PI 3-kinase respectively. We recommend use of drug resistant LRRK2[A2016T] mutant for validating that observed effects of LRRK2-IN-1 are indeed mediated via inhibition of LRRK2 kinase activity. LRRK2-IN-1 provides a useful first-generation ‘tool’ compound and provides a starting point for developing a compound that could ultimately be used to address the potential therapeutic benefit of inhibiting LRRK2 in Parkinson’s patients harboring the LRRK2 G2019S mutation.
Supplementary Material
1
2
3
4
5
Acknowledgements
We wish to thank staff at the National Centre for Protein Kinase Profiling (www.kinase-screen.mrc.ac.uk) for undertaking Dundee kinase specificity screening, Life Technologies Corporation, SelectScreen® Kinase Profiling Service for performing enzymatic biochemical kinase profiling, Ambit Bioscience for performing KinomeScan™ profiling, ActivX for performing KiNativ™ profiling, SAI Advantium for performing pharmacokinetic studies, Faycal Hentati (Institut National de Neurologie, Tunis, Tunisia) and Alastair Reith (GlaxoSmithKline Pharmaceuticals R&D) for providing EBV immortalised human lymphoblastoid cells, Paul Bamborough (GlaxoSmithKline Pharmaceuticals R&D) for providing LRRK2 homology model and the antibody purification teams [Division of Signal Transduction Therapy (DSTT), University of Dundee] coordinated by Hilary McLauchlan and James Hastie for generation of antibodies. This work was supported by NIH grant P41 GM079575-03 (N. Gray), the MRCT Industry Collaboration Award and a NHMRC postdoctoral fellowship (N. Dzamko), the Medical Research Council (D. Alessi), the Michael J Fox foundation for Parkinson’s disease research (D. Alessi), the pharmaceutical companies supporting the DSTT (AstraZeneca, Boehringer-Ingelheim, GlaxoSmithKline, Merck KgaA and Pfizer) (D. Alessi), the NIH grant CA079871 and CA114059 (J.-D. Lee) and funds from the Tobacco-Related Disease, Research Program of the University of California, 19XT-0084, (J.-D. Lee).
Footnotes
Author contributions
N.S.G. conceived and directed the chemistry effort. X.D. performed the chemical synthesis and SAR analysis. D.R.A. conceived and directed the biology effort. N.D. performed the biology experimental research with assistance from A.P. (confocal microscopy and live cell imaging). P.D performed the SHSY5Y experiment. Q.L. performed the modeling study. Q.Y. and J.D.L. performed the MAPK7 cellular assay. M.P.P. and T.K.N. performed the KiNativ selectivity profiling and data analysis. X.D, N.D. D.R.A and N.S.G. co-wrote the paper. All authors read and edited the manuscript.Competing financial interests
M.P.P. and T.K.N are employees of AcitivX Biosciences.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nchembio.538
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3287420
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
LRRK2 G2019S Promotes Colon Cancer Potentially via LRRK2-GSDMD Axis-Mediated Gut Inflammation.
Cells, 13(7):565, 23 Mar 2024
Cited by: 1 article | PMID: 38607004 | PMCID: PMC11011703
Sperm Toolbox-A selection of small molecules to study human spermatozoa.
PLoS One, 19(2):e0297666, 20 Feb 2024
Cited by: 1 article | PMID: 38377053 | PMCID: PMC10878532
Efficacy of a benzothiazole-based LRRK2 inhibitor in oligodendrocyte precursor cells and in a murine model of multiple sclerosis.
CNS Neurosci Ther, 30(1):e14552, 01 Jan 2024
Cited by: 1 article | PMID: 38287523 | PMCID: PMC10808848
Pharmacology of LRRK2 with type I and II kinase inhibitors revealed by cryo-EM.
Cell Discov, 10(1):10, 23 Jan 2024
Cited by: 3 articles | PMID: 38263358 | PMCID: PMC10805800
Kinome-wide siRNA screen identifies a DCLK2-TBK1 oncogenic signaling axis in clear cell renal cell carcinoma.
Mol Cell, 84(4):776-790.e5, 10 Jan 2024
Cited by: 3 articles | PMID: 38211588
Go to all (256) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The IkappaB kinase family phosphorylates the Parkinson's disease kinase LRRK2 at Ser935 and Ser910 during Toll-like receptor signaling.
PLoS One, 7(6):e39132, 18 Jun 2012
Cited by: 137 articles | PMID: 22723946 | PMCID: PMC3377608
Inhibition of LRRK2 kinase activity leads to dephosphorylation of Ser(910)/Ser(935), disruption of 14-3-3 binding and altered cytoplasmic localization.
Biochem J, 430(3):405-413, 01 Sep 2010
Cited by: 262 articles | PMID: 20659021 | PMCID: PMC3631100
Measurement of LRRK2 and Ser910/935 phosphorylated LRRK2 in peripheral blood mononuclear cells from idiopathic Parkinson's disease patients.
J Parkinsons Dis, 3(2):145-152, 01 Jan 2013
Cited by: 29 articles | PMID: 23938344
Current understanding of LRRK2 in Parkinson's disease: biochemical and structural features and inhibitor design.
Future Med Chem, 4(13):1701-1713, 01 Sep 2012
Cited by: 18 articles | PMID: 22924508 | PMCID: PMC3569718
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Medical Research Council (2)
Understanding the functions of LKB1 and other protein kinases mutated in inherited diseases
Professor Dario Alessi, MRC Protein Phosphorylation Unit
Grant ID: MC_U127070193
Characterisation of the LRRK2 protein kinase, mutated in inherited Parkinson s disease
Professor Dario Alessi, University of Dundee
Grant ID: G0700656
NCI NIH HHS (4)
Grant ID: CA079871
Grant ID: CA114059
Grant ID: R01 CA079871
Grant ID: R01 CA114059
NIGMS NIH HHS (5)
Grant ID: P41 GM079575-03S1
Grant ID: P41 GM079575-03
Grant ID: P41 GM079575
Grant ID: P41 GM079575-01
Grant ID: P41 GM079575-02





