Abstract
Purpose
MicroRNA (miRNA) alterations are likely to contribute to the development of pancreatic cancer and may serve as markers for the early detection of pancreatic neoplasia.Experimental design
To identify the miRNA alterations that arise during the development of pancreatic cancer, we determined the levels of 735 miRNAs in 34 pancreatic intraepithelial neoplasias (PanIN) and 15 normal pancreatic duct samples isolated by laser capture microdissection using TaqMan miRNA microarrays. Differential expression of selected miRNAs was confirmed by FISH analysis and by quantitative real-time reverse transcription PCR (qRT-PCR) analysis of selected candidate miRNAs in an independent set of PanIN and normal duct samples.Results
We identified 107 aberrantly expressed miRNAs in different PanIN grades compared with normal pancreatic duct samples and 35 aberrantly expressed miRNAs in PanIN-3 lesions compared with normal pancreatic duct samples. These differentially expressed miRNAs included those that have been previously identified as differentially expressed in pancreatic ductal adenocarcinomas (PDAC; including miR-21, miR-200a/b/c, miR-216a/b, miR-217, miR-146a, miR-155, miR-182, miR-196b, miR-203, miR-222, miR-338-3p, miR-486-3p, etc.) as well as miRNAs not previously described as differentially expressed in these lesions (miR-125b, miR-296-5p, miR-183*, miR-603, miR-625/*, miR-708, etc.). miR-196b was the most selectively differentially expressed miRNA in PanIN-3 lesions.Conclusions
Many miRNAs undergo aberrant expression in PanIN lesions and are likely to be important in the development of PDAC. The miRNAs, such as miR-196b, whose expression is limited to PanIN-3 lesions or pancreatic cancers could be useful as diagnostic markers.Free full text

MicroRNA Alterations of Pancreatic Intraepithelial Neoplasms (PanINs)
Abstract
Purpose
MicroRNA alterations are likely to contribute to the development of pancreatic cancer and may serve as markers for the early detection of pancreatic neoplasia.
Experimental Design
To identify the microRNA alterations that arise during the development of pancreatic cancer we determined the levels of 735 miRNAs in 34 pancreatic intraepithelial neoplasias (PanINs) and 15 normal pancreatic duct samples isolated by laser capture microdissection using TaqMan miRNA microarrays. Differential expression of selected miRNAs was confirmed by fluorescent in-situ hybridization analysis and by qRT-PCR analysis of selected candidate microRNAs in an independent set of PanIN and normal duct samples.
Results
We identified 107 aberrantly expressed miRNAs in different PanIN grades compared with normal pancreatic duct samples, and 35 aberrantly expressed miRNAs in PanIN-3 lesions compared with normal pancreatic duct samples. These differentially expressed miRNAs included those that have been previously identified as differentially expressed in pancreatic ductal adenocarcinomas (including miR-21, miR-200a/b/c, miR-216a/b, miR-217, miR-146a, miR-155, miR-182, miR-196b, miR-203, miR-222, miR-338-3p, miR-486-3p and others) as well as miRNAs not previously described as differentially expressed in these lesions (miR-125b, miR-296-5p, miR-183*, miR-603, miR-625/*, miR-708 and others). MiR-196b was the most selectively differentially expressed miRNA in Panin-3 lesions.
Conclusions
Many miRNAs undergo aberrant expression in PanIN lesions and are likely to be important in the development of pancreatic ductal adenocarcinoma. MicroRNAs such as miR-196b whose expression is limited to PanIN-3 lesions or pancreatic cancers could be useful as diagnostic markers.
Introduction
Pancreatic cancer is the fourth leading cause of cancer-related deaths in the United States(1). In the United States in 2010, 43,140 new patients were diagnosed, and approximately 36,800 Americans died from pancreatic cancer (1). The poor prognosis and high mortality rate result, at least in part, from the generally late presentation of the disease and the lack of effective therapies (2). Although early detection is considered the best way to cure pancreatic cancer, most early-stage pancreatic cancers do not cause symptoms. As a result there is considerable interest in pancreatic screening for individuals considered to be at significantly increased risk of developing pancreatic cancer, such as those with an inherited predisposition. Since even early-stage invasive pancreatic cancer is usually incurable, the primary goal of pancreatic screening programs has been to prevent the development of invasive pancreatic cancer by detecting and treating pancreatic precursor lesions. The commonest of these precursor lesions are PanINs (Pancreatic Intraepithelial Neoplasias) (3). Since PanINs are too small to be reliably detected by pancreatic imaging tests (4, 5), there is considerable interest in identifying markers of advanced PanIN (6, 7) to improve our ability to detect advanced PanIN during pancreatic screening. Although numerous studies have described the molecular alterations of pancreatic ductal adenocarcinomas (PDAC) (8–11), fewer studies have investigated the timing of such alterations in PanIN lesions. Understanding the molecular alterations of PanINs may not only identify markers for pancreatic screening but also to identify important biological pathways.
Changes in the expression of microRNAs (miRNAs) are important to the development of cancer. MiRNAs are small endogenous non-coding RNAs of 14 – 24 nucleotides that negatively regulate protein expression at the post-transcriptional level by inhibiting translation and/or by targeting messenger RNAs (mRNAs) for degradation (12). Furthermore, since miRNAs are stable and detectable in human plasma, they are being investigated for their utility as diagnostic serum markers (13).
Alterations in the expression of microRNAs are suspected to contribute to the development and progression of pancreatic and other cancers (14–16). Pancreatic ductal adenocarcinomas overexpress several miRNAs including miR-21, miR-34, miR-146a, miR-155, miR-196a-2, miR-200a/b (13, 17–24).
Although several studies have reported microRNA alterations of pancreatic cancer (4, 17, 20, 22, 24), the role of these alterations during early pancreatic neoplastic development is not well understood. Mendell et al. revealed that the repression of the miR-143/145 cluster by oncogenic Ras promotes pancreatic cancer development (25). But to date few miRNAs have been examined for alterations in mouse or human PanIN lesions (26) (27).
In this study we employed the TaqMan Array Human MicroRNA Cards (Sanger miRbase v16) to comprehensively profile PanIN miRNA expression relative to normal pancreatic duct cells.
Materials and Methods
PanIN specimens
Fresh pancreatic tissues were snap-frozen in liquid nitrogen, embedded in Tissue-Tek OCT compound medium (Sakura FineTek USA, Torrance, CA) and stored at −80°C. The samples were subsequently sectioned onto UV-treated PALM membrane slides (Carl Zeiss MicroImaging, Inc., Thornwood, NY) for laser capture microdissection (LCM) and stored at −80°C (5). In each case, the PanINs lesions were examined histologically and the diagnosis was confirmed by two of the authors (R. H. H and SM. H), who are expert pancreatic pathologists. PanINs were graded as PanIN-1 (low-grade), PanIN-2 (intermediate-grade), and PanIN-3 (high-grade) as previously described (28). All specimens were collected and analyzed with the approval of the Johns Hopkins Committee for Clinical Investigation (29).
Tissue microarray construction
Tissue microarrays (TMA) were constructed from the archival formalin-fixed paraffin-embedded tissue blocks of surgically resected primary pancreatic ductal adenocarcinoma (PDAC) using a manual Tissue Puncher/Arrayer (Beecher Instruments, Silver Spring, MD) as previously described (30). A total of 94 1.4-mm cores (42 pancreatic ductal adenocarcinoma, 44 PanINs, and 8 normal pancreatic ducts) were arrayed on the recipient blocks. The PanIN lesions comprised 13 PanIN-1, 15 PanIN-2, and 16 PanIN-3 lesions.
Cells and culture conditions
The human pancreatic ductal epithelial cell line (HPDE, provided by Dr. Ming-sound Tsao at the University of Toronto) was cultured in serum free keratinocyte media supplemented with supplied growth factors according to the manufacturer’s instructions. Twenty-nine pancreatic cancer cell lines, including A38-41, A38-44, AsPC-1, BxPC-3, CAPAN-1, CAPAN-2, CFPAC-1, HPAFII, Hs766T, Mia PACA-2, Pa01C, Pa02C, Pa03C, Pa07C, Pa08C, Pa09C, Pa14C, Pa16C, Pa18C, Pa20C, Pa21C, Pa28C, PANC-1, Panc486, PK8, PK9, PL11, Su86.86 and SW1990 were utilized in this study. All cancer cell lines were maintained in DMEM (4.5 mg/ml glucose, Invitrogen) supplemented with 10% fetal bovine serum (FBS) and antibiotics (100 µg/ml streptomycin and 100 units/ml penicillin).
Laser capture microdissection
As a guide, one frozen section slide was stained with hematoxylin and eosin (H & E). Thirty-four PanIN lesions (12 PanIN-1, 11 PanIN-2, and 11 PanIN-3 lesions) and 15 samples of normal pancreatic ductal epithelial cells adjacent to PanIN lesions from patients with pancreatic ductal adenocarcinoma (n=6) or other diagnoses (IPMN or serous cystadenoma, n=9) were selectively isolated with the PALM laser microdissection platform (PALM, Carl Zeiss MicroImaging, Inc., Thornwood, NY) according to the manufacturer’s protocols (31) (Supplemental Figure 1A – D). A separate set of PanIN lesions (6 PanIN-1 lesions, 3 PanIN-2 lesions, and 2 PanIN-3 lesions) and 9 samples of normal pancreatic ductal epithelium (from patients with either pancreatic ductal adenocarcinoma (n=1) or benign neoplasms (serous cystadenoma, IPMN, n=8) were laser capture microdissected in the same fashion to validate the differential expression of candidate miRNAs. In addition, to measure the expression of candidate miRNAs in pancreatic cancer, neoplastic cells from 14 primary pancreatic ductal adenocarcinomas (PDACs) were also laser capture microdissected. We microdissected a mean of 20,000 PanIN and normal ductal epithelial cells to help ensure the detection of abundance for the microarray analysis, and a mean of 4,000 normal, PanINs and PDAC cells for the validation analysis.
RNA isolation
Total RNA was extracted using mirVana miRNA isolation kit (Ambion 1560) for cultured cells and RNAqueous-Micro kit (Ambion 1931) for microdissected cells, following the manufacturer’s protocols (total RNA isolation procedure and RNAqueous-Micro procedure for LCM, respectively). The extracted RNA was quantified by the absorbance at 260 nm and the purity of the extracted RNA was evaluated by the absorbance ratio at 260/280 nm with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Rockland, DE).
MiRNA expression profiling
Comprehensive miRNA expression profiling was performed with the TaqMan Array Human MicroRNA Cards (Cards A and B, v2.1 and v3.0, respectively; Applied Biosystems) using the 7900HT thermocycler (Applied Biosystems). These two cards are designed with 750 unique assays of human miRNAs from Sanger miRbase v14, of which we identified 735 human miRNAs from Sanger miRbase v16 (Supplemental Table 1). MicroRNAs were amplified after specific reverse transcription and preamplification using Megaplex Assay Performance (Megaplex RT Primer Pools and Megaplex PreAmp pools, both from Applied Biosystems) according to manufacturer’s instructions (Applied Biosystems) and normalized against RNU6B (U6 snRNA, an endogenous control assay designed in both cards). Relative expression was determined using the ΔΔ Ct method and a ≥ 32 Ct value was interpreted as amplification too low to quantify.
Individual miRNA expression detection
MiRNAs that were candidates for being differentially expressed were analyzed using the TaqMan Small RNA Assay (Applied Biosystems), a two-step quantitative real-time reverse-transcription polymerase chain reaction (qPCR). The 7900HT thermocycler was used to measure the abundance of individual candidate differentially expressed miRNAs. Candidate microRNAs were amplified after specific reverse transcription and preamplification using Megaplex Assay Performance for LCM samples, or after specific reverse transcription using TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) for cultured cell samples, according to manufacturer’s instructions (Applied Biosystems) and normalized against RNU6B. All candidate miRNAs’ and RNU6B’s primers were provided from Applied Biosystems. Each sample was run in triplicate. Relative expression was determined using the ΔΔ Ct method and a ≥ 32 Ct value was interpreted as amplification too low to quantify.
Locked nucleic acid fluorescent in situ hybridization (LNA-FISH)
LNA-FISH was performed on TMA slides using LNA oligonucleotide probes against miR-196b and U6, both labeled with fluorescein at the 5’-end (Exiqon, Vedbaek, Denmark), according to the protocol described in de Planell-Saguer’s paper (32). LNA-U6 was used as a positive measure of probe specificity. Briefly, after deparaffinization, slides were prehybridized for 30 min at 50°C (30°C below the RNA Tm of LNA-miR-196b probe which is 80°C) in a humid chamber with prehybridization buffer, then incubated with hybridization buffer containing LNA-miR-196b probe (1:1000) for 1 hour at 50°C in a hybridization oven. After several washes, the slides were incubated in TNB blocking buffer (PerkinElmer, Waltham, MA) containing goat anti-fluorescein antibody peroxidase conjugated (FITC/HRP; Rockland, Gilbertsville, PA) (1:1000) for 30 min in a humid chamber at room temperature (RT). Then the signals were amplified using tyramide signal amplification (TSA; PerkinElmer, Waltham, MA) for 10min in a humid chamber at RT. After incubation in DAPI staining solution for 5min at RT, slides were mounted with Prolong Gold anti-fade reagent (Invitrogen, Carlsbad, CA) and incubated overnight at 4°C and signals visualized using a fluorescent microscope. All steps beginning with hybridization were performed in the dark. The LNA-miR-196b-FISH results were quantified at a single-cell level by counting expression spots per cell as previously described (33).
Statistical analysis
Principal components analysis (PCA) mapping of comprehensive miRNA expression profiling was performed by using Partek Genomics Suite software (Partek Incorporated, St. Louis, MO). Differences in median expression were determined using the Mann-Whitney U test, differences in proportions of expressing samples were determined with Pearson’s chi-square X2 test, and differences in LNA-FISH abnormalities between categorical variables were determined with Student’s t-test. Statistical significance was defined as a value of P < 0.05. To adjust for multiple comparisons, we calculated the false discovery rate (FDR) from the subset of significantly deregulated miRNAs with P < 0.05 (34). All statistical analyses were performed using the SPSS Statistics 19.0 software.
Results
MiRNA expression differs in PanIN Lesions and normal pancreatic ducts
The microRNA profiles of 34 laser capture microdissected PanIN lesions were compared with 15 LCM samples of normal pancreatic duct using TaqMan Array Human MicroRNA Cards containing 735 human miRNA assays. MicroRNA concentrations were normalized against RNU6B. We first used principal component analysis (PCA) to compare the global miRNAs expression profile of PanIN lesions with that of samples of normal pancreatic ductal epithelial cells (Supplemental Figure 1E) using the data from TaqMan Array Human MicroRNA Cards. This analysis revealed that miRNAs effectively separated PanIN lesions (all PanINs or each group of PanINs, PanIN-1, PanIN-2, and PanIN-3) and normal pancreatic ducts, while PanIN lesions of different grades overlapped in their miRNA profiles (Supplemental Figure 1E).
Numerous miRNAs are aberrantly expressed in PanIN lesions
To identify differentially expressed miRNAs among PanINs overall relative to normal pancreatic duct samples, we calculated the median fold-change of each miRNA between PanIN and normal duct samples analyzed by TaqMan microarray using the ΔΔ Ct method (Supplemental Table 2). This analysis identified 65 significantly differentially expressed miRNAs in PanIN lesions and normal pancreatic duct samples (fold change > 2 or < 0.5, respectively; P < 0.05, corrected for FDR; Table 1).
Table 1
Significantly dysexpressed miRNAs in PanIN lesions
| Assay | Normal | PanINs (fold) | Up/Down | P value | FDR |
|---|---|---|---|---|---|
| hsa-miR-486-3p | 1 | 37.5 | Up | 0.000 | 0.001 |
| hsa-miR-875-5p | 1 | 16.0 | Up | 0.021 | 0.039 |
| hsa-miR-422a | 1 | 15.2 | Up | 0.002 | 0.008 |
| hsa-miR-21 | 1 | 15.2 | Up | 0.003 | 0.016 |
| hsa-miR-182 | 1 | 13.4 | Up | 0.000 | 0.002 |
| hsa-miR-146a | 1 | 10.5 | Up | 0.001 | 0.005 |
| hsa-miR-183 | 1 | 10.4 | Up | 0.002 | 0.009 |
| hsa-miR-18a | 1 | 9.9 | Up | 0.006 | 0.023 |
| hsa-miR-494 | 1 | 9.9 | Up | 0.021 | 0.040 |
| hsa-miR-603 | 1 | 9.6 | Up | 0.002 | 0.010 |
| hsa-miR-183* | 1 | 9.1 | Up | 0.000 | 0.002 |
| hsa-miR-18b | 1 | 8.4 | Up | 0.008 | 0.025 |
| hsa-miR-625 | 1 | 8.3 | Up | 0.000 | 0.003 |
| hsa-miR-708 | 1 | 7.9 | Up | 0.003 | 0.017 |
| hsa-miR-135b | 1 | 7.9 | Up | 0.013 | 0.035 |
| hsa-miR-625* | 1 | 7.8 | Up | 0.002 | 0.011 |
| hsa-miR-203 | 1 | 7.8 | Up | 0.003 | 0.018 |
| hsa-miR-338-3p | 1 | 7.8 | Up | 0.013 | 0.035 |
| hsa-miR-429 | 1 | 7.3 | Up | 0.002 | 0.012 |
| hsa-miR-103 | 1 | 7.0 | Up | 0.02 | 0.038 |
| hsa-miR-200a | 1 | 6.8 | Up | 0.011 | 0.028 |
| hsa-miR-425 | 1 | 6.7 | Up | 0.004 | 0.020 |
| hsa-miR-29c | 1 | 6.7 | Up | 0.016 | 0.038 |
| hsa-miR-29b | 1 | 6.3 | Up | 0.002 | 0.012 |
| hsa-miR-15b | 1 | 6.3 | Up | 0.037 | 0.048 |
| hsa-miR-22* | 1 | 6.0 | Up | 0.005 | 0.022 |
| hsa-miR-101 | 1 | 5.8 | Up | 0.022 | 0.042 |
| hsa-miR-874 | 1 | 5.7 | Up | 0.000 | 0.004 |
| hsa-miR-148b | 1 | 5.7 | Up | 0.008 | 0.025 |
| hsa-miR-29a | 1 | 5.7 | Up | 0.012 | 0.032 |
| hsa-miR-130b | 1 | 5.6 | Up | 0.011 | 0.029 |
| hsa-miR-190 | 1 | 5.4 | Up | 0.013 | 0.036 |
| hsa-miR-222 | 1 | 5.3 | Up | 0.002 | 0.013 |
| hsa-miR-106b | 1 | 5.0 | Up | 0.011 | 0.030 |
| hsa-miR-31 | 1 | 4.7 | Up | 0.022 | 0.042 |
| hsa-miR-652 | 1 | 4.5 | Up | 0.003 | 0.018 |
| hsa-miR-93 | 1 | 4.4 | Up | 0.003 | 0.019 |
| hsa-miR-664 | 1 | 4.3 | Up | 0.032 | 0.045 |
| hsa-miR-378 | 1 | 4.0 | Up | 0.01 | 0.027 |
| hsa-miR-193a-3p | 1 | 4.0 | Up | 0.011 | 0.031 |
| hsa-miR-320b | 1 | 3.9 | Up | 0.001 | 0.005 |
| hsa-miR-155 | 1 | 3.9 | Up | 0.012 | 0.033 |
| hsa-miR-423-5p | 1 | 3.8 | Up | 0.011 | 0.032 |
| hsa-miR-331-5p | 1 | 3.8 | Up | 0.046 | 0.049 |
| hsa-miR-1274b | 1 | 3.7 | Up | 0.03 | 0.045 |
| hsa-miR-194 | 1 | 3.4 | Up | 0.035 | 0.047 |
| hsa-miR-200a* | 1 | 3.3 | Up | 0.001 | 0.006 |
| hsa-miR-193b | 1 | 3.3 | Up | 0.002 | 0.014 |
| hsa-miR-192* | 1 | 3.3 | Up | 0.006 | 0.024 |
| hsa-let-7f | 1 | 3.3 | Up | 0.022 | 0.043 |
| hsa-miR-135b* | 1 | 3.2 | Up | 0.002 | 0.015 |
| hsa-miR-331-3p | 1 | 3.2 | Up | 0.009 | 0.026 |
| hsa-let-7g | 1 | 3.2 | Up | 0.041 | 0.048 |
| hsa-miR-151-5P | 1 | 3.1 | Up | 0.021 | 0.041 |
| hsa-miR-185 | 1 | 3.1 | Up | 0.027 | 0.044 |
| hsa-miR-200b* | 1 | 3.0 | Up | 0.002 | 0.015 |
| hsa-miR-34c-5p | 1 | 2.7 | Up | 0.005 | 0.022 |
| hsa-miR-330-3p | 1 | 2.6 | Up | 0.001 | 0.007 |
| hsa-miR-200c | 1 | 2.5 | Up | 0.01 | 0.028 |
| hsa-miR-17* | 1 | 2.5 | Up | 0.047 | 0.050 |
| hsa-miR-95 | 1 | 2.3 | Up | 0.004 | 0.021 |
| hsa-miR-550a | 1 | 2.3 | Up | 0.012 | 0.034 |
| hsa-miR-200b | 1 | 2.1 | Up | 0.032 | 0.046 |
| hsa-miR-629 | 1 | 2.0 | Up | 0.015 | 0.037 |
| hsa-miR-296-5p | 1 | 0.3 | Down | 0.001 | 0.008 |
To independently validate the differential expression of miRNAs identified by microarray analysis, we chose 13 candidate overexpressed miRNAs (miR-146a, miR-182, miR-193a-3p, miR-193b, miR-200a, miR-200b, miR-200c, miR-21, miR-29b, miR-425, miR-486-3p, miR-708, and miR-874) and one candidate underexpressed miRNA (miR-296-5p) for qPCR analysis in an independent set of 20 samples, including 9 normal pancreatic duct samples and 11 PanIN lesions isolated by laser capture microdissection. We confirmed the differential expression of 13 of these 14 miRNAs (all except miR-296-5p). These results are shown in Figure 1, Supplemental Figure 2 and Supplemental Table 3.
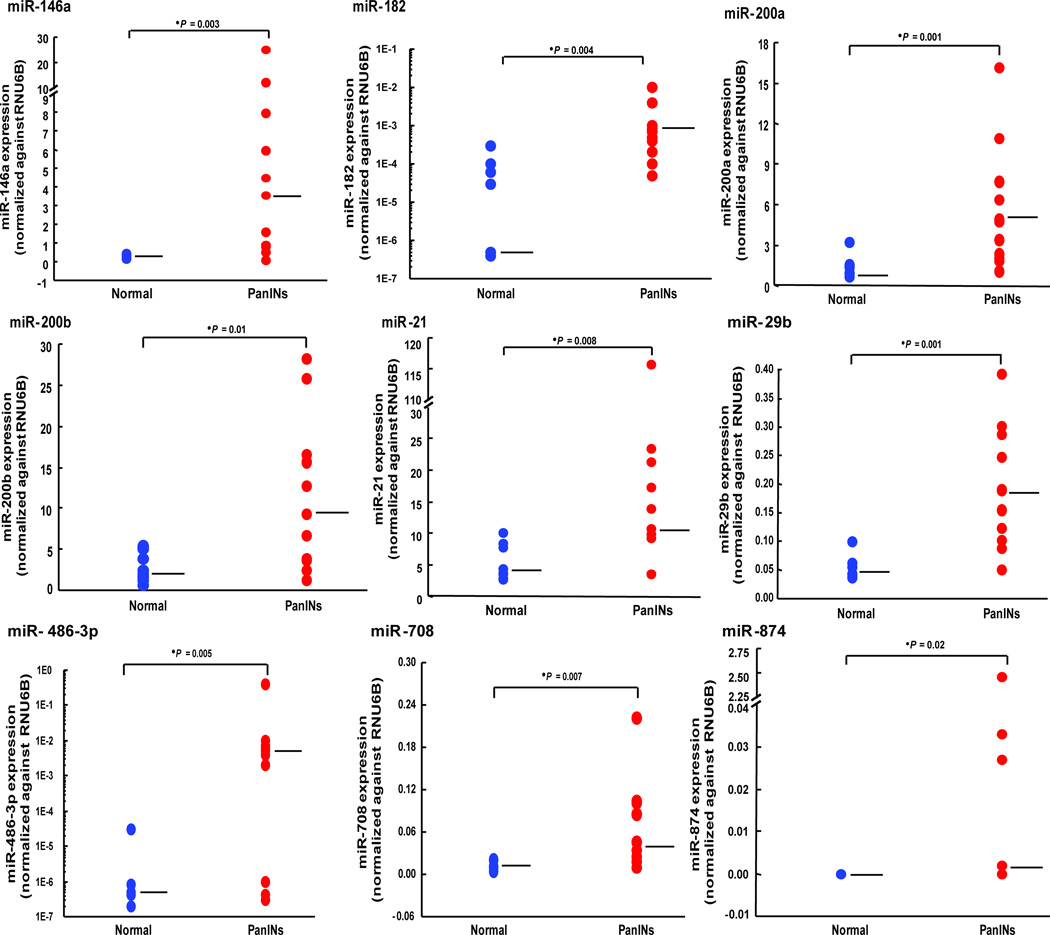
The levels of candidate miR-146a, miR-182, miR-200a/b, miR-21, miR-29b, miR-486-3p, miR-708, and miR-874 expressions were measured in 9 normal pancreatic ducts (blue) and in 11 PanIN lesions (red). Each sample was run in triplicate. Horizontal bars represent medians.
MiRNAs alterations in different grades of PanINs
Since PanINs undergo a series of histological and molecular changes as they progress towards invasive adenocarcinoma, we next compared TaqMan Array microRNA profiles of intermediate-to-high-grade PanIN lesions (PanIN-2/-3) and those of low-grade PanIN lesions (PanIN-1) with normal pancreatic ducts. This comparison yielded 10 significantly overexpressed and 8 significantly underexpressed miRNAs in PanIN-2/-3 samples compared with PanIN-1/Normal-Duct samples (fold change > 2 or < 0.5, respectively; Supplementary Table 4), of which 4 of the overexpressed miRNAs (miR-18b, miR-21, miR-338-3p, and miR-874) and 6 of the underexpressed miRNAs in the higher grade lesions (miR-139-3p, miR-214, miR-216b, miR-296-5p, miR-370, and miR-622) reached significance after adjusting for FDR. Several of these miRNAs were among the differentially expressed miRNAs between PanINs and normal pancreata (Table 1).
Since PanIN-3 lesions are an important lesion to identify in patients undergoing pancreatic screening, we next compared miRNA profiles of PanIN-3 with those of normal pancreatic duct samples. This comparison yielded 26 significantly overexpressed and 9 significantly underexpressed miRNAs in PanIN-3 lesions compared with normal pancreatic duct samples (fold change > 2 or < 0.5, respectively; corrected for FDR; Table 2 and Supplemental Table 5). Only one of these miRNAs, (miR-196b) was selectively overexpressed in PanIN-3 lesions relative to other PanINs (Table 1). In addition, 8 miRNAs (miR-125b, miR-126, miR-127-3p, miR139-3p/5p, miR-216b, miR-218, and miR-452) were selectively underexpressed in PanIN-3 lesions relative to other PanINs (Table 2).
Table 2
Aberrantly expressed miRNAs in PanIN-3 lesions
| Assay | Normal | PanIN-3 (fold) | Up/Down | PanINsa | P value | FDR |
|---|---|---|---|---|---|---|
| hsa-miR-196b | 1 | 64.9 | Up | − | 0.002 | 0.010 |
| hsa-miR-486-3p | 1 | 49.8 | Up | Up | 0.005 | 0.013 |
| hsa-miR-21 | 1 | 23.2 | Up | Up | 0.017 | 0.024 |
| hsa-miR-338-3p | 1 | 14.9 | Up | Up | 0.024 | 0.033 |
| hsa-miR-18a | 1 | 14.7 | Up | Up | 0.039 | 0.041 |
| hsa-miR-183* | 1 | 14.5 | Up | Up | 0.001 | 0.003 |
| hsa-miR-182 | 1 | 12.8 | Up | Up | 0.002 | 0.009 |
| hsa-miR-18b | 1 | 11.9 | Up | Up | 0.014 | 0.020 |
| hsa-miR-183 | 1 | 10.6 | Up | Up | 0.017 | 0.026 |
| hsa-miR-422a | 1 | 10.2 | Up | Up | 0.039 | 0.043 |
| hsa-miR-603 | 1 | 9.6 | Up | Up | 0.036 | 0.040 |
| hsa-miR-190 | 1 | 7.1 | Up | Up | 0.031 | 0.039 |
| hsa-miR-29b | 1 | 6.8 | Up | Up | 0.039 | 0.044 |
| hsa-miR-93 | 1 | 6.7 | Up | Up | 0.024 | 0.034 |
| hsa-miR-425 | 1 | 6.7 | Up | Up | 0.017 | 0.027 |
| hsa-miR-146a | 1 | 6.6 | Up | Up | 0.011 | 0.016 |
| hsa-miR-874 | 1 | 5.6 | Up | Up | 0.017 | 0.029 |
| hsa-miR-101 | 1 | 5.3 | Up | Up | 0.039 | 0.046 |
| hsa-miR-652 | 1 | 5.1 | Up | Up | 0.012 | 0.017 |
| hsa-miR-193a-3p | 1 | 4.8 | Up | Up | 0.039 | 0.047 |
| hsa-miR-625 | 1 | 4.5 | Up | Up | 0.014 | 0.021 |
| hsa-miR-135b | 1 | 3.4 | Up | Up | 0.045 | 0.050 |
| hsa-miR-320b | 1 | 3.1 | Up | Up | 0.024 | 0.036 |
| hsa-miR-135b* | 1 | 2.8 | Up | Up | 0.021 | 0.031 |
| hsa-miR-222 | 1 | 2.6 | Up | Up | 0.039 | 0.049 |
| hsa-miR-106b | 1 | 2.4 | Up | Up | 0.014 | 0.023 |
| hsa-miR-452 | 1 | 0.4 | Down | − | 0.002 | 0.011 |
| hsa-miR-126 | 1 | 0.3 | Down | − | 0.018 | 0.030 |
| hsa-miR-218 | 1 | 0.3 | Down | − | 0.006 | 0.014 |
| hsa-miR-125b | 1 | 0.3 | Down | − | 0.012 | 0.019 |
| hsa-miR-127-3p | 1 | 0.2 | Down | − | 0.024 | 0.037 |
| hsa-miR-139-3p | 1 | 0.2 | Down | − | 0.001 | 0.007 |
| hsa-miR-139-5p | 1 | 0.2 | Down | − | 0.001 | 0.004 |
| hsa-miR-216b | 1 | 0.1 | Down | − | 0.000 | 0.001 |
| hsa-miR-296-5p | 1 | 0.1 | Down | Down | 0.001 | 0.006 |
Silencing and induction of miRNA expression in PanIN lesions
Analysis of differences in the median expression of miRNAs may fail to identify differences in miRNA expression that occur as a result of either aberrant silencing or aberrant induction of expression in a subset of PanIN lesions. We therefore classified the miRNA expression profiles in PanIN and normal duct samples into expressing and non-expressing and compared them to identify changes in the proportion of expressing samples among the groups. For each miRNA, we classified expressing samples as those with a qRT-PCR Ct value <32. This classification yielded 2 significantly upregulated miRNAs (miR-516a-3p and miR-659) and 16 significantly downregulated miRNAs (miR-107, miR129-3p, miR-181c, miR-184, miR-198, miR-216a/b, miR-217, miR-370, miR-372, miR-379, miR-433, miR-539, miR-543, miR-618 and miR-758) in PanIN lesions compared with normal pancreatic duct samples, after adjusting for FDR (Supplementary Table 6). MiR-196b was significantly more likely to be expressed and miR-216b more likely to be silenced in PanIN-3 lesions compared with normal pancreatic ductal samples (Table 2 and Supplemental Table 6). All of the miRNAs identified as differentially expressed were also identified as differentially expressed in PanINs vs. normal pancreatic duct samples or between PanIN-2/3 lesions vs. PanIN-1/normal duct samples (Supplemental Tables 4 and 6).
We next analyzed the expression of three of the candidate overexpressed miRNAs (miR-182, miR-196b, and miR-486-3p) and three candidate underexpressed miRNAs (miR125b, miR216b, and miR-296-5p) in additional PanIN-3 lesions by qPCR. Combined analysis of these samples with PanIN-3 samples and normal pancreatic duct samples including samples from the original set of PanIN-3 and normal pancreatic duct samples (24 normal pancreatic ducts and 13 PanIN-3 samples in total), confirmed the differential expression of all 6 miRNAs after adjusting for FDR (Figure 2 and Supplementary Figure 3).
Candidate miRNA expression profiles in pancreatic cancers
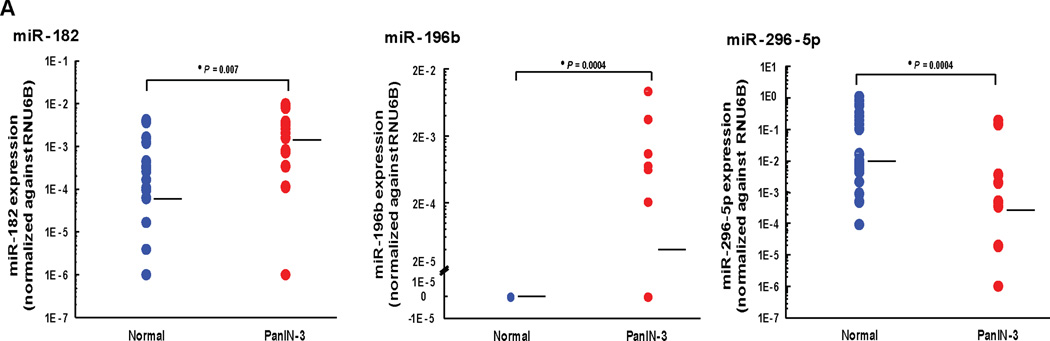
We next examined pancreatic cancers for miRNA expression of miRNAs that have not been described as differentially expressed in invasive pancreatic cancers previously including 2 miRNAs that were overexpressed in PanINs (miR-182 and miR-196b) and one that was underexpressed (miR-296-5p). Expression of these miRNAs was examined in 29 pancreatic cancer cell lines and in the non-neoplastic pancreatic epithelial line, HPDE. As shown in Supplemental Figure 4, only miR-296-5p showed differential expression in pancreatic cancer cell lines relative to HPDE. To confirm that miRNA expression patterns of pancreatic cancer cell lines were the same in primary pancreatic cancer cells, we examined the expression of these three candidate miRNAs in 14 samples of laser capture microdissected PDAC cells compared with 8 samples of microdissected normal pancreatic ductal cells. As shown in Supplemental Figure 4, the expression of these 3 miRNAs was significantly different in primary PDAC samples relative to normal pancreatic ducts for all three miRNAs, including miR-182 (P value < 0.001), miR-196b P value < 0.001) and miR-296-5p (P value = 0.002), respectively.
Expression of miRNA-196b in PanIN-3 lesions by LNA-FISH
Since miRNA-196b is the most differentially expressed miRNA in PanIN-3 lesions by miRNA microarray analysis, we further examined its expression in PanIN lesions using LNA-FISH. We found that the mean expression of miR-196b in 16 PanIN-3 lesions was 3.1 spots/cell (95% CI, 2.0 – 4.3) vs. 0.5, spots/cell (95% CI; 0.2 – 0.7) in 8 normal pancreatic duct cells (P = 0.027). MiRNA-196b levels were also significantly higher in the PanIN-3 lesions than in 13 PanIN-1 (0.4 spots/cell, 0.2 – 0.7); and 15 PanIN-2 lesions 0.6 spots/cell, 0.1 – 1.0) (P=0.010, and 0.011, respectively) (Figure 3 and Supplementary Figure 5). MiRNA levels were significantly higher in 42 pancreatic ductal adenocarcinomas (7.1 spots/cell, 6.0 – 8.3) than in PanIN-3 lesions, lower grade PanINs and normal pancreatic duct cells (Figure 3).
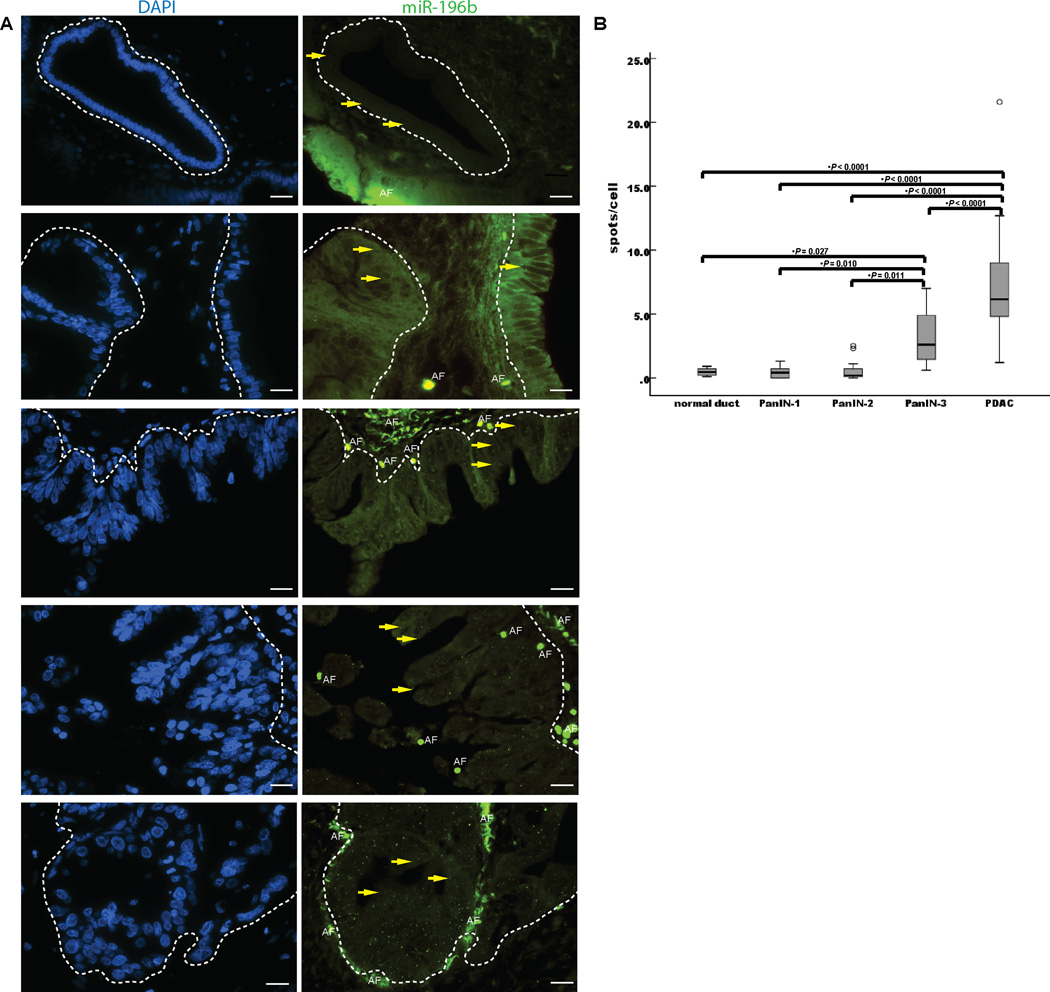
A, yellow arrows indicate LNA-miR-196b-FISH probe signals (green spots) for normal pancreatic ducts, PanIN-1 lesions, PanIN-2 lesions, PanIN-3 lesions, and PDAC lesions (from upper to lower, respectively), which are circled with white dashed lines. DAPI (blue) indicates nucleic acid staining. B, quantitative analysis of LNA-miR-196b-FISH probe with spots/cell in TMAs. The lower and upper borders of the boxes mark the 95% confidence interval of the mean. The center horizontal line is drawn at the sample mean. The vertical lines drawn from the boxes extend to the minimum and the maximum. AE, autofluorescence. Original magnification, 40×.
Numerous miRNAs are aberrantly expressed during pancreatic carcinogenesis
We further compared miRNAs’ expression in each PanIN grade with those in normal pancreatic duct cells by the timing of aberrant PanIN expression (fold change > 2 or < 0.5, Figure 4 and Supplemental Table 7). This comparison yielded 107 miRNAs significantly aberrantly expressed during one or more PanIN stages, including 37 miRNAs not previously identified as aberrantly expressed in pancreatic cancer (13, 17, 19, 20, 24, 26, 35–40). Almost all of these miRNAs were identified by comparing the miRNA levels of PanINs as a group to normal pancreatic ducts. One exception is miR-494 which was significantly elevated in PanINs as an overall group, (Table 1), but not in any one PanIN subgroup. Among the miRNAs not previously reported to be aberrantly expressed includes overexpressed miRNAs: miR-135b*, miR-18b, miR-183*, miR-516a-3p, miR-603, miR-625, miR-629, miR-652, miR-659, miR-708, miR-874, and underexpressed miRNAs: miR-10a*/b*, miR136*, miR-146b-3p, miR-223*, miR-433, miR-452, miR-539, miR-543, miR-551b, miR-618, miR-744*, miR-758 and miR-885-5p, and others. In addition, some of the miRNAs we identified as differentially expressed miRNAs have been previously found to show the same differential expression in pancreatic cancers. These include miRNAs overexpressed in PanINs: miR-129-3p, miR-130b, miR-133a, miR-135b, miR-148a/b, miR-151-5p, miR-183, miR-185, miR-193a-3p, miR-200c, miR-320b, miR-330-3p, miR-331-3p/5p, miR-34c-5p, miR-378, miR-422a, miR-423-5p and miR-425, and miRNAs underexpressed in PanINs including miR-125b, miR-126, miR-127-3p, miR-137, miR-181c, miR-184, miR-198, miR-205, miR-32, miR-370, miR-372 and miR-379 (17, 20, 22, 24, 37, 40). Furthermore, several miRNAs were identified as significantly overexpressed only in low-grade PanINs (only in PanIN-1 or PanIN-1/ PanIN-2 lesions) including miR-129-3p, miR-130b, miR-133a, miR-148a/b, miR-151-5p, miR-185, miR-200c, miR-330-3p, miR-331-3p/5p, miR-34c-5p, miR-378 and miR-423-5p.
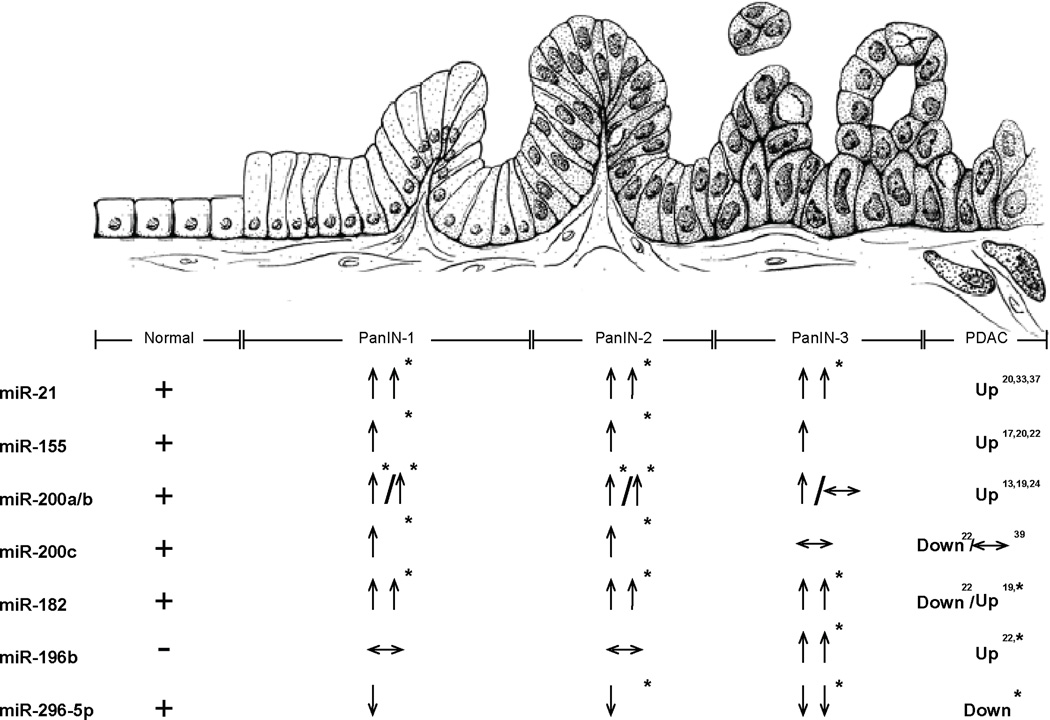
Normal duct cells (far left), PanIN lesions (center), and invasive PDAC (far right). “+” and “−”: indicates presence or absence of miRNA expression in normal ducts, respectively. “↑” and “↓”, indicates changes in level of expression (2–10 fold and 0.1–0.5 fold, respectively). “↑↑” and “↓↓”, indicates changes in level of expression (>10-fold and <0.1-fold, respectively). * indicates P value < 0.05.
Discussion
In this study we have comprehensively profiled the expression of miRNAs in PanINs and normal pancreatic duct samples. Many of the differentially expressed miRNAs we identified in PanIN lesions have been also identified as differentially expressed in invasive pancreatic cancers (Supplemental Table 7). These differentially expressed miRNAs include overexpression of let-7f/g, miR-101, miR-103, miR-106b, miR-146a, miR-15b, miR-155, miR-18a, miR-182, miR-190, miR-193b, miR-194, miR-196b, miR-200a/b, miR-203, miR-21, miR-222, miR-29a/b/c, miR-31, miR-338-3p, miR-429, miR-486-3p, miR-93 and miR95, and underexpression of miR-107, miR-139-3p/5p, miR-216a/b, miR-217, miR-218 and miR-483-5p (13, 17, 20, 22, 37). Several of these miRNAs have been found to have oncogenic or tumor suppressive functions. These include miR-107, miR-146a, miR-155, miR-200a/b, miR-203, miR-21, miR-216a/b and miR-217(13, 22, 38, 41–43).
The overexpression of let-7a, miR-200 and miR-21 we observed in PanIN lesions confirm the results of Du Rieu et al who found overexpression of these miRNAs in mouse PanIN lesions, and found that the miR-21 upregulation preceded phenotypic changes in pancreatic duct cells (26). In contrast, LaConti et al found overexpression of miR-10, miR-16, miR-21, miR-100 and miR-155 in mouse PanINs, whereas we identified overexpression of only miR-21 and miR-155 in human PanINs (27).
We also identified numerous differentially expressed miRNAs that have not been identified as differentially expressed in pancreatic cancers (Supplemental Table 7). For example, miR-183, a member of miR-182/183/96 family (44), targets EGR1 and PTEN, and promotes tumor cell migration (45). We also identified several differentially expressed miRNAs that had differential expression only in low-grade PanINs (PanIN-1 or PanIN-1/ PanIN-2 lesions). Levels of one such candidate, miR-200c has been reported to influence E-cadherin expression, inhibit invasion of pancreatic cancer cells and correlate with patient survival (40). Such a pattern of expression is perhaps not surprising given the different phenotypic and molecular profiles of low-grade and high-grade PanINs. Several miRNAs described as elevated in pancreatic cancers including miR-143/145 and miR-34a did not display significantly different expression patterns to normal pancreatic duct samples (Supplemental Tables 2 and 5). None of the miRNAs identified as overexpressed are located at chromosomal loci that are known targets of chromosomal amplification.
The identification of markers of high-grade dysplasia and invasive pancreatic ductal adenocarcinoma such as miR-196b could have implications for pancreatic screening. We and others have been screening individuals with extensive family histories of pancreatic cancer (4, 5, 46–48). Pancreatic screening by endoscopic ultrasound typically identifies subtle non-specific changes in the pancreas and when such pancreata are resected PanIN lesions are often found in association with lobulocentric atrophy (49). However, these imaging alterations are non-specific and are also detected in patients without an increased risk of pancreatic cancer as well as among older individuals and patients with chronic pancreatitis (50). One important goal of pancreatic screening is to identify high-grade lesions so that patients with significant lesions can be offered pancreatic resection prior to the development of invasive cancer. Markers are needed that are specific for high-grade PanIN-3 lesions and invasive pancreatic ductal adenocarcinoma among patients undergoing pancreatic screening. Such markers could potentially be detected in pancreatic fluid samples obtained after secretin infusion at the time of upper endoscopy. Our results indicate that miR-196b has such specific expression patterns and that miR-196b deserves further investigation to determine if it can identify PanIN-3 among patients undergoing screening.
In summary, we have performed a quantitative real-time PCR analysis of over 700 miRNAs in PanIN lesions to identify aberrantly expressed miRNAs at each PanIN stage. These aberrant miRNA expression patterns may provide insights into pancreatic neoplastic development. The almost exclusive expression of miR-196b in PanIN-3 lesions and pancreatic cancers compared to lower-grade PanINs and normal pancreata suggest that its detection could be useful for identifying PanIN-3 lesions in patients undergoing pancreatic screening.
Supplementary Material
1
7
8
9
10
11
12
2
3
4
5
6
Acknowledgements
Grant Support: This work was supported by NIH grants (P50-CA62924, R01-CA120432, RC2CA148376), and the Michael Rolfe Foundation.
Abbreviations used in this paper
| FDR | false discovery rate |
| LCM | laser capture microdissection |
| LNA-FISH | locked nucleic acid fluorescent in situ hybridization |
| miRNA | microRNA |
| PanIN | pancreatic intraepithelial neoplasias |
| PDAC | pancreatic ductal adenocarcinoma |
| PCA | principal component analysis |
| qRT-PCR | quantitative real time reverse-transcription polymerase chain reaction |
| TMA | tissue microarrays |
| TSA | tyramide signal amplification |
Footnotes
Conflicts of interest: The authors disclose no conflicts
Author Contributions
Conceived and designed the experiments: JY MG. Performed the experiments: JY AL SMH. Analyzed the data: JY AL MG. Wrote the paper: JY RHH MG. Agree with the manuscript’s results and conclusions: JY AL SMH RHH MG. Wrote the draft of the paper: JY MG.References
Full text links
Read article at publisher's site: https://doi.org/10.1158/1078-0432.ccr-11-2347
Read article for free, from open access legal sources, via Unpaywall:
http://clincancerres.aacrjournals.org/content/18/4/981.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1158/1078-0432.ccr-11-2347
Article citations
MicroRNA-155 and its exosomal form: Small pieces in the gastrointestinal cancers puzzle.
Cell Biol Toxicol, 40(1):77, 16 Sep 2024
Cited by: 0 articles | PMID: 39283408 | PMCID: PMC11405467
Review Free full text in Europe PMC
MicroRNA-486-3p affects cisplatin resistance in high-grade serous ovarian cancer by regulating TMIGD2: An experimental study.
Heliyon, 10(15):e34978, 19 Jul 2024
Cited by: 0 articles | PMID: 39145009 | PMCID: PMC11320304
Cancer cell plasticity: from cellular, molecular, and genetic mechanisms to tumor heterogeneity and drug resistance.
Cancer Metastasis Rev, 43(1):197-228, 08 Feb 2024
Cited by: 9 articles | PMID: 38329598 | PMCID: PMC11016008
Review Free full text in Europe PMC
MicroRNA-372 acts as a double-edged sword in human cancers.
Heliyon, 9(5):e15991, 09 May 2023
Cited by: 3 articles | PMID: 37251909 | PMCID: PMC10208947
Review Free full text in Europe PMC
Alteration in Levels of Specific miRNAs and Their Potential Protein Targets between Human Pancreatic Cancer Samples, Adjacent Normal Tissue, and Xenografts Derived from These Tumors.
Life (Basel), 13(3):608, 22 Feb 2023
Cited by: 1 article | PMID: 36983764 | PMCID: PMC10057657
Go to all (150) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
MicroRNAs as diagnostic markers for pancreatic ductal adenocarcinoma and its precursor, pancreatic intraepithelial neoplasm.
Cancer Genet, 206(6):217-221, 01 Jun 2013
Cited by: 54 articles | PMID: 23933230
Gene expression profiles in pancreatic intraepithelial neoplasia reflect the effects of Hedgehog signaling on pancreatic ductal epithelial cells.
Cancer Res, 65(5):1619-1626, 01 Mar 2005
Cited by: 154 articles | PMID: 15753353
Aberrant MicroRNA-155 expression is an early event in the multistep progression of pancreatic adenocarcinoma.
Pancreatology, 10(1):66-73, 20 Mar 2010
Cited by: 91 articles | PMID: 20332664 | PMCID: PMC2865485
Update on the pathology and genetics of exocrine pancreatic tumors with ductal phenotype: precursor lesions and new tumor entities.
Dig Dis, 19(1):15-23, 01 Jan 2001
Cited by: 16 articles | PMID: 11385247
Review
Funding
Funders who supported this work.
NCI NIH HHS (9)
Grant ID: P50 CA062924
Grant ID: P50 CA062924-18
Grant ID: P50-CA62924
Grant ID: R01-CA120432
Grant ID: R01 CA120432-05
Grant ID: RC2 CA148346-02
Grant ID: RC2 CA148346
Grant ID: RC2CA148376
Grant ID: R01 CA120432