Abstract
Free full text

A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA damage response
Abstract
Repair of DNA double strand breaks is critical to genomic stability and the prevention of developmental disorders and cancer. A central pathway for this repair is homologous recombination (HR). Most knowledge of HR is derived from work in prokaryotic and eukaryotic model organisms. We performed a genome-wide siRNA-based screen in human cells. Among positive regulators of HR we identified networks of DNA damage response and pre-mRNA processing proteins, and among negative regulators we identified a phosphatase network. Three candidate proteins localized to DNA lesions including RBMX, a heterogeneous nuclear ribonucleoprotein that has a role in alternative splicing. RBMX accumulated at DNA lesions via multiple domains in a poly(ADP-ribose) polymerase 1-dependent manner and promoted HR by facilitating proper BRCA2 expression. Our screen also revealed that off-target depletion of Rad51 is a common source of RNAi false-positives, sounding a cautionary note for siRNA screens and RNAi-based studies of HR.
INTRODUCTION
The presence of double-strand breaks (DSBs) in DNA is a detrimental event, and failure to repair DSBs can cause loss of telomeric regions of chromosomes, complex translocations or cell death. In humans this can lead to severe developmental abnormalities and cancer. Organisms have evolved two major pathways for DSB repair: non-homologous end joining (NHEJ) and homologous recombination (HR). NHEJ results in the potentially error-prone religation of DSB ends. HR is an error-free repair mechanism that operates during the S or G2 phase of the cells cycle and primarily utilizes the replicated sister chromatid as a template for repair1. HR is initiated when one strand of the DSB is resected, a process mediated by CtIP, the 5’-3’ exonuclease ExoI, and possibly other nucleases2, 3. This generates a 3’ ssDNA overhang that is protected from further degradation by the ssDNA binding protein RPA. RPA is displaced by the recombinase Rad51, generating a nucleoprotein filament that coordinates the search for a homologous sequence and facilitates strand invasion of the template DNA4. In humans, BRCA2 and the Rad51 paralogs (Rad51B, Rad51C, Rad51D, XRCC2 and XRCC3) promote and maintain the nucleation of Rad51, and a host of other repair proteins modulate HR in both the positive and negative direction5.
To probe the HR pathway in human cells, we performed a genome-wide siRNA screen; and through this screen, we uncovered cellular functions required for HR and identified proteins that localize to sites of DNA damage. Screen data also revealed that Rad51 is a common off-target of siRNAs, which presents a cautionary note to those studying HR with siRNAs and highlights the vulnerability of RNAi screens to off-target effects in general. Among the candidates we identified as positive regulators of HR was RBMX, a heterogeneous nuclear ribonucleoprotein (hnRNP) that associates with the spliceosome, binds RNA and influences alternative splicing. We found that RBMX is required for resistance to DNA damage and accumulates at sites of DNA damage in a poly(ADP-ribose) polymerase (PARP1) dependent manner.
RESULTS
A genome-wide siRNA screen to identify regulators of homologous recombination
We performed an siRNA screen to identify components of the mammalian HR machinery using a well-characterized GFP-based reporter (DR-GFP) (Fig. 1a)6, 7. DR-GFP carries two mutant versions of GFP; one with two premature stop codons and an internal I-SceI endonuclease restriction site (SceGFP), the other with 3’ and 5’ end truncations (iGFP)6. Neither SceGFP nor iGFP express a functional protein; however, a gene conversion event between the mutants –generated by recombinational repair of an I-SceI-induced DSB– can reconstitute wild-type GFP. In this manner GFP expression is an accurate readout for HR. For our screen, we employed the osteosarcoma cell line DR-U2OS that has a single, stably integrated copy of DR-GFP, and we drove expression of I-SceI with an adenovirus (AdNGUS24i)7.
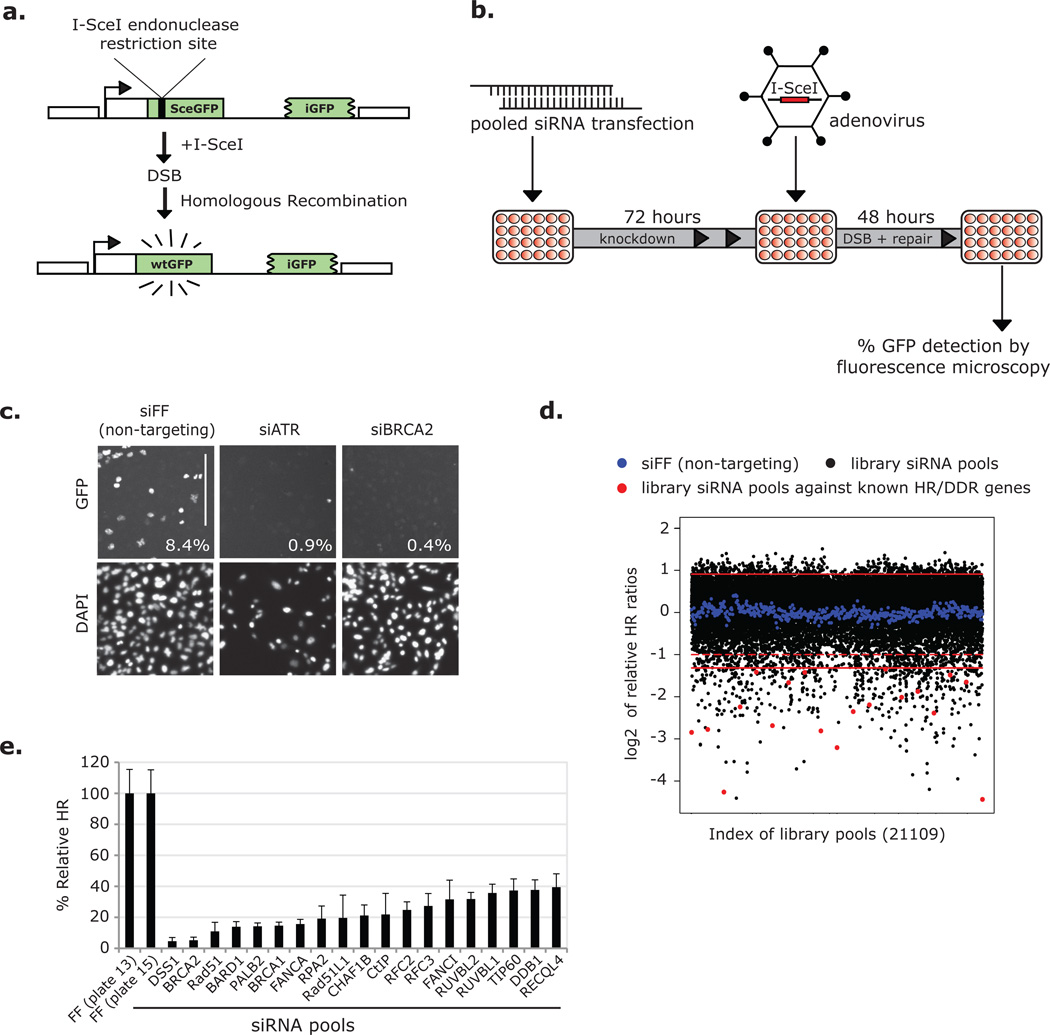
A genome-wide siRNA-based screen for homologous recombination (HR) genes. (a) Schematic of DR-GFP construct. (b) Schematic of the high-throughput (HTP) HR screen. Arrayed pools of siRNAs were reverse transfected into DR-U2OS cells in 384 well plates. Cells were infected with the I-SceI expressing adenovirus AdNGUS24i at an MOI of ~10 after 72 hours and 48 hours later were fixed and imaged. (c) DR-U2OS cells were transfected with positive (siATR and siBRCA2) and negative (siFF) control siRNAs and assayed for HR in high throughput. Images were taken on the automated screening platform and are presented in representative portions. % GFP+ cells –as calculated from full images– are included. Scale bar represents 300 µm. (d) Relative HR ratios (presented as log2 values) from 22109 library siRNA pools and 476 negative controls (siFF). Solid red lines indicate 2 s.d. from the screen-wide mean of relative HR ratios (presented as log2 values). These were used as cutoff values to determine pools scoring with increased or decreased HR. The region between the dotted red line and the lower solid red line indicates a scoring range from which most additional candidate HR mediators were selected. (e) Known regulators of HR or the DDR that scored under the 2 s.d.-based cutoff.
We screened the Dharmacon human siGENOME siRNA library in triplicate, which is arrayed as 21,121 single-target pools of 4 distinct siRNAs. Briefly, DR-U2OS cells were plated in 384 well plates, reverse transfected with siRNAs, and infected with AdNGUS24i at a multiplicity of infection (MOI) of ~10 (Fig. 1b). At this high titer changes in cell number had little effect on assay results (Supplementary Information, Fig. S1a). Cells were fixed, stained with Hoechst, and the % GFP+ cells per well were determined by fluorescence microscopy on an automated platform (Fig. 1c). The average of % GFP+ cells from each experimental triplicate was normalized to that from on-plate, non-targeting control wells transfected with an siRNA against firefly luciferase (siFF) to obtain a relative HR ratio for each library pool (Fig. 1d; Supplementary Information, Table S1). Indicative of a successful screen, we recovered 19 genes known to be involved in HR and the DDR, including Rad51, BRCA1 and BRCA2 (Fig. 1d–e).
Identification and validation of candidate HR regulators
Hits from the screen were defined as siRNA pools that decreased or increased relative HR >2 standard deviations (s.d.) from the screen-wide mean (cutoff values ~ 40% or 188% relative HR). From the corresponding genes, 510 candidate HR mediators and 484 candidate HR suppressors were identified (Supplementary Information, Table S1). We extended the list of candidate mediators by 131 genes corresponding to siRNA pools that trended in the screen (primarily with 40–50% relative HR) (Supplementary Information, Table S1–2). These additional genes had also been identified in previous DDR screens. Next we deconvolved the 641 siRNA pools against candidate mediators and the strongest 250 pools against candidate suppressors (including 1 duplicate pool) and rescreened each siRNA individually (Supplementary Information, Table S2–3). As expected, siRNAs from both candidate sets enriched for the appropriate phenotype (Fig. 2a).
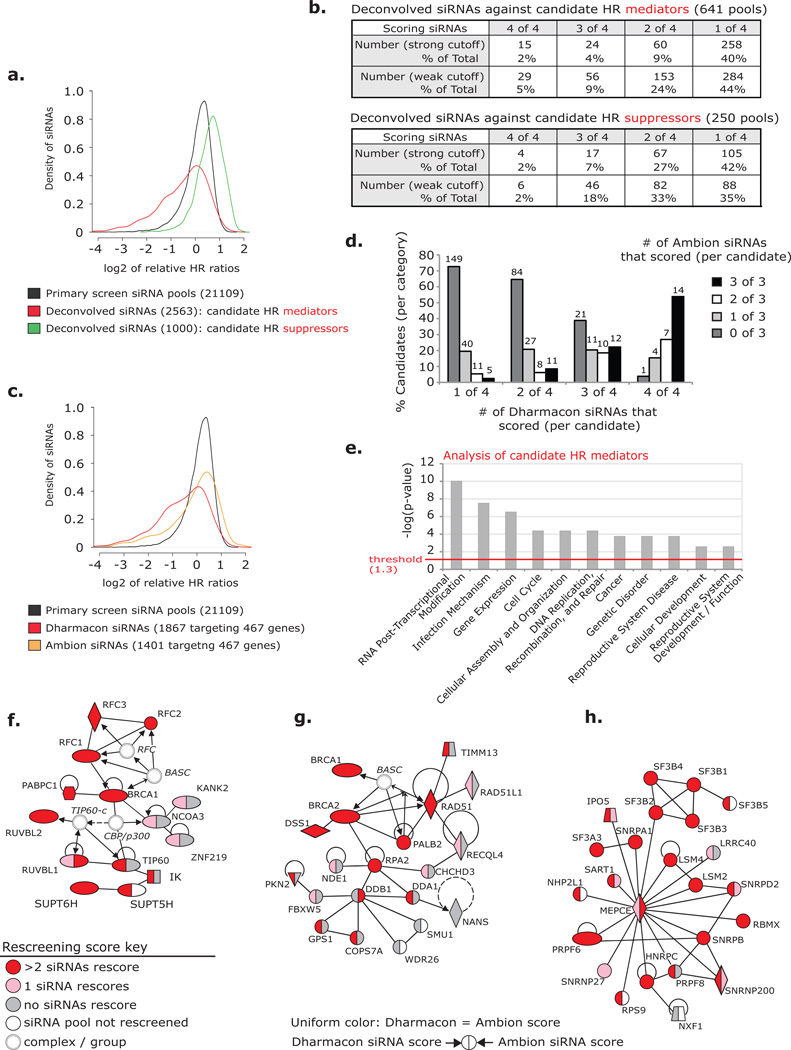
Rescreen and validation of candidate HR genes. (a) The distributions of relative HR ratios (as log2 values) for three sets of screened siRNAs: primary screen pools (black), deconvolved Dharmacon siRNAs against candidate HR mediators (red), and deconvolved Dharmacon siRNAs against candidate HR suppressors (green). (b) The number and percentage of Dharmacon siRNA pools that rescored with 1, 2, 3 or 4 siRNAs after deconvolution. Results analyzed using strong (2 s.d. from the screen-wide mean-based: 40% and 188% relative HR) or weak (1.5 s.d.-based: 59% and 169%) cutoff values. (c) The distributions of relative HR ratios (as log2 values) for primary screen pools (black: as presented in a), siRNAs from select Dharmacon pools against candidate mediators (red), and rescreened Ambion siRNAs against candidate mediators (orange). Dharmacon and Ambion siRNAs target the same 467 candidate genes. (d) Percentage of candidate HR mediator genes that scored with 0, 1, 2, or 3 Ambion siRNAs in categories of those that scored with 1, 2, 3, or 4 Dharmacon siRNAs. Weak cutoff was used for scoring. Number of candidates indicated. (e) Functional gene categories enriched among candidate HR mediators identified using Ingenuity Pathway Analysis (IPA). (f–h) Interaction networks generated from candidates that scored as HR mediators in the primary screen identified by IPA: (f) DDR network including components of the TIP60 complex and the RFC DNA clamp loader, (g) DDR / HR network including canonical HR proteins and Cul4A ubiquitin ligase associated proteins, (h) pre-mRNA processing network. Color key (representative of rescreening data): red indicates a candidate that rescored with >2 siRNAs (out of 4 Dharmacon and 3 Ambion), pink indicates that 1 siRNA rescored, gray indicates that 0 siRNAs rescored, and white indicates a candidate that was not rescreened. Line key: solid lines indicate direct interactions, dashed lines indicate indirect actions, arrows indicate the direction of interactions, and lines without arrowheads indicate binding. Shape key: ovals indicate transcription regulators, hexagons indicate translational regulators, diamonds (vertical) indicate enzymes, diamonds (horizontal) indicate peptidases, trapezoids indicate transporters, inverted triangles indicate kinases, squares indicate cytokines, circles indicate other, double circles indicate groups of proteins or complexes.
We evaluated the rescreened siRNAs with both strong and weak phenotype cutoffs (Fig. 2b). Strong siRNAs were those that rescored below (for mediator siRNAs) or above (for suppressor siRNAs) the 2 s.d.-based thresholds from the primary screen (40% and 188% relative HR, respectively). Weak cutoffs were based on 1.5 s.d. from the primary screen mean (<59% or <169% relative HR for mediators and suppressors, respectively). We considered candidate siRNA pools validated if ≥3 individual siRNAs (out of 4) rescored with at least a weak HR value (14% of pools for HR mediators and 20% of pools for HR suppressors) (Fig. 2b). The higher validation rate for siRNA pools targeting HR suppressors is a result of rescreening only the strongest 250 pools. The strongest scoring pools against candidate mediators also yielded a higher rate of validation (Supplementary Information, Fig. S1b). We evaluated siRNA toxicity throughout and observed no correlation between cell growth and relative HR levels (Supplementary Information, Fig. S1c–e and Tables S1–3; see Supplementary Results).
To further evaluate the candidate mediator list and better understand the 68% of pools that rescored with 1–2 siRNAs, we conducted a second round of rescreening using siRNAs from the Ambion Silencer Select library targeting 467 candidate mediators (3 siRNAs / gene) (Supplementary Information, Table S4). These siRNAs also enriched for reduced HR but at a level substantially less than observed among Dharmacon siRNAs targeting the same 467 genes (Fig. 2c). We reason that the independently selected Ambion siRNAs had a reduced incidence of off-targeting and were, therefore, more likely to score true positives. Importantly, candidates that validated with 3–4 (of 4) Dharmacon siRNAs had greater likelihood of scoring with 2–3 (of 3) Ambion siRNAs (over candidates that scored with fewer Dharmacon siRNAs), even when known HR / DDR mediators were not considered (Fig. 2d; Supplemental Information, Fig. S1f).
Network analysis
Next we evaluated candidate genes for enrichment of functional categories and interaction networks using Ingenuity Pathway Analysis (IPA, Ingenuity Systems, http://www.ingenuity.com; see Methods section for a description of the gene lists submitted to IPA). Both mediators and suppressors were enriched for genes functionally categorized as DNA replication, recombination and repair, which we expected for mediators of HR, but not necessarily for suppressors as little is known about what activities limit recombination (Fig. 2e; Supplementary Information, Fig. S2a). Among candidate mediators two gene networks with known HR genes were identified (Fig. 2f–g). These highlight roles for the RFC DNA clamp loader and the TIP60 histone acetylase complex in HR, and suggest a role for factors associated with DDB1 and the Cul4A ubiquitin ligase. Nine components of the TIP60 complex scored or trended in the primary screen: TIP60, RUVBL1, RUVBL2, DMAP1, Brd8, p400, ING3, MRGBP and MRG15. The most significantly enriched category among candidate mediators was RNA post-transcriptional modification, which also produced a strong interaction network (Fig. 2e, h). Although the involvement of RNA processing proteins in the DDR is poorly understood, several large-scale genetic and proteomic analyses of the DDR have shown similar enrichments8–10. A role in promoting HR could explain the enrichment in each screen. Among HR suppressors a small network of phosphatases emerged, which may act to limit the activity of kinases that promote HR (Supplementary Information; Fig. S2b).
IPA also identified categories of candidate genes that relate to the design of the screen but not HR. The DR-GFP based HR assay depends on infection of an adenovirus, expression of I-SceI and GFP, and normal cell cycle progression; and it is, therefore unlikely that candidates functionally categorized under infection mechanism, gene expression and cell cycle regulation represent biological true positives (Fig. 2e; Supplementary Information, Fig. 2a). Specifically, a network of RNA polymerase II (RNAP2) and mediator subunits was identified among candidate HR mediators (Supplementary Information, Fig. S2c).
Three candidate HR regulators localize to sites of DNA damage: HIRIP3, RBMX, DDX17
We made GFP fusions of 22 candidates and evaluated each for relocalization after DNA damage (Supplementary Information, Table S5). Two candidate mediators, HIRIP3 and RBMX, and one suppressor, DDX17, accumulated at regions of DNA damaged by microirradiation (Fig. 3a). RBMX is an RNA-binding protein that associates with the spliceosome and plays a role in alternative splicing, and DDX17 is a DEAD-box RNA helicase that is phosphorylated in response to IR8, 11, 12. HIRIP3 interacts with the histone chaperone HIRA and binds histones H2B and H313. Interestingly, HIRA and two additional HIRA-associated proteins (UBN1 and CAIN) also localized to DNA damage after microirradiation (Supplementary Information, Fig. S3a).
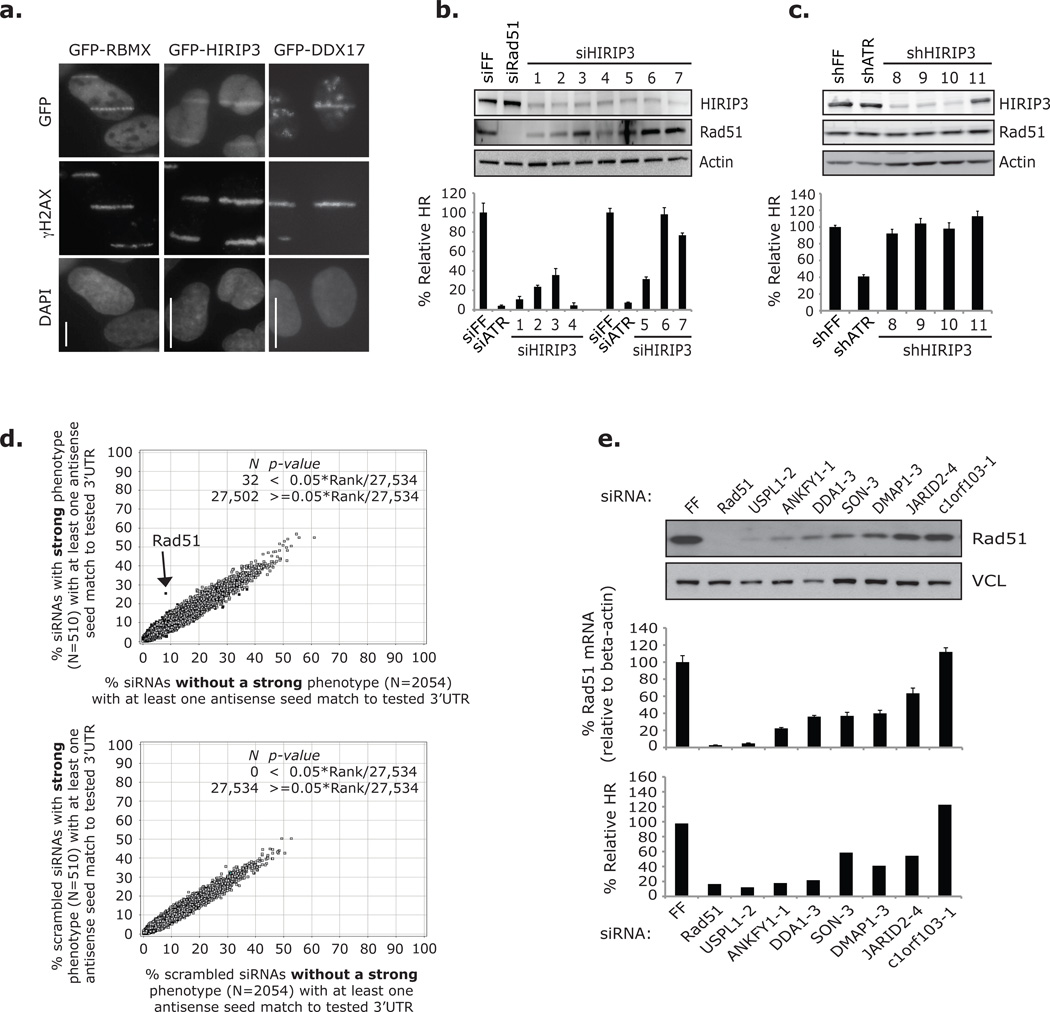
Off-target Rad51-depletion was a major source of false positives among Dharmacon siRNAs identified by the primary screen. (a) Cells expressing GFP fusions of RBMX, HIRIP3 or DDX17 were microirradiated and prepared for imaging after 0–5 (RBMX and DDX17) and 15–20 minutes (HIRIP3) with an antibody against γH2AX. GFP was observed directly. Nuclei were stained with DAPI. Scale bars indicate 10 µm. Images were prepared from three separate experiments and are not intended for comparison. (b) HR assay results from DR-U2OS and HIRIP3 / Rad51 western blot analysis from U2OS cells transfected with siRNAs against HIRIP3 in three separate experiments: one to acquire western blot data and two for HR analysis. Experimental data is grouped with corresponding controls. Error bars represent ± s.d. across three replicates. (c) HR assay results and corresponding HIRIP3 / Rad51 western blot analysis from DR-U2OS cells transduced with shRNAs against HIRIP3. Error bars represent ± s.d. across three replicates. (d) GESS analysis of Dharmacon siRNAs against candidate mediator genes. Scatter plots represent the percentage of siRNAs in 2 groups that have at least one 7-nucleotide antisense seed sequence match to 27,534 human 3’UTRs. Upper plot: compares siRNAs that individually rescored with a strong phenotype (y-axis) to those that did not (x-axis). Lower plot: compares the same 2 groups of siRNAs after scrambling the seed sequences and serves as a control. 3’UTRs that significantly enriched for seed matches in either siRNA group are black. Arrow illustrates the 3’UTR of Rad51. (e) HR assay results and Rad51 mRNA / protein levels from cells transfected with seven siRNAs predicted to off-target Rad51 by a 7-nucleotide antisense seed region match to the Rad51 3’UTR. Western blot and RT-qPCR results are from the same experiment in U2OS cells. Primers used against Rad51 mRNA recognize four transcript variants. Error bars in RT-qPCR analysis represent ± s.e.m. across three replicates. HR data is from the HTP Dharmacon rescreening analysis in DR-U2OS cells and represent the average of two replicates. Full scans of blots in b–c, e are shown in Supplementary Information, Fig. S13.
The HIRIP3-targeting siRNA pool gave a strong HR defect in our primary screen, but while seven siRNAs and three shRNAs were individually shown to deplete HIRIP3, only five of these caused substantial HR defects; and none of three screened HIRIP3 Ambion siRNAs scored (Fig. 3b–c; Supplementary Information, Table S4). Indicative of an off-target effect, expression of siRNA-resistant HIRIP3 did not rescue the siHIRIP3-2 HR defect. Because these HIRIP3 siRNAs (as well as siRNAs against UBN1 and HIRA) caused defective HR without correlation to on-target depletion, we suspected HR might be particularly sensitive to off-target effects (Supplementary Information, Fig. S3b–f; see Supplementary Results).
Off-target Rad51 depletion contributes to screen false positives
To search for off-target effects in our screen data set we employed Genome-wide Enrichment of Seed Sequences (GESS) analysis, which identifies 3’UTRs with enriched sequence complementary to the seed regions of siRNAs that score in genome-wide date sets (over non-scoring siRNAs)14. siRNA seed regions are nucleotides 2–8 of either RNA strand (sense or antisense), and seed sequence complementarity to the 3’UTR of a gene transcript is thought to elicit repression by pathways endogenously engaged by microRNA-containing RISC complexes15, 16. GESS analysis of our Dharmacon rescreen data revealed that siRNAs against candidate mediators with strong HR defects (40% relative HR cutoff) were 3-fold enriched for antisense seed sequences matches of 7-nucleotides to the 3’UTR of Rad51, compared to siRNAs that did not rescore with a strong phenotype (an increase from 8% to 25%, Fisher’s Exact Test p-value = 4.65×10−23) (Fig. 3d). The sense strands of strongly scoring Dharmacon siRNAs, however, gave no enrichment for seed matches to the Rad51 3’UTR (Fisher’s Exact Test p-value = 0.5986), and no enrichment for seed region complimentary (from both strands) to the Rad51 coding region (CDS) was observed (Fisher’s Exact Test p-value = 0.8886) (Supplementary Information, Fig. S4a). From these data, we predict a 17% false positive rate due to off-target Rad51 depletion among our strong scoring Dharmacon siRNAs. Importantly though, the strong scoring Ambion siRNAs against candidate mediators were not enriched for antisense (or sense) seed complimentarily to the Rad51 3’UTR (Fisher’s Exact Test p-value = 0.3526 antisense and 0.7485 sense), suggesting that this off-target effect does not confound the data from those reagents (Supplementary Information, Fig. S4b). Interestingly, GESS analysis of Dharmacon siRNAs that scored for increased HR identified enrichment of three 3’UTRs belonging to genes that have yet to be implicated in HR: ITGB1BP3, FAM153C and EDC3 (Supplementary Information, Fig. S4c).
Predicted off-target Rad51 depletion was confirmed for 6 of 7 screened Dharmacon siRNAs with a 7-nucleotide antisense seed region match to the 3’UTR of Rad51, and both Rad51 mRNA and protein depletion by these siRNAs correlated with the % Relative HR determined for each during rescreening analysis (Fig. 3e). The HR defects caused by four HIRIP3 siRNAs (including three from the primary screen pool) also correlated with off-target Rad51 depletion (Fig. 3b–c; Supplementary Information, Fig. S4d). However, of these only siHIRIP3-5 had a 7-nucleotide seed match to the Rad51 3’UTR, indicating that Rad51 off-targeting also occurs through mechanisms that are independent of complete seed matches and suggesting that the incidence of Rad51 off-targeting in our screen is underrepresented by the GESS estimation of false positives. Consequently, we refined our list of validated candidate HR mediators to 121 that scored with at least 3 of 7 combined Ambion and Dharmacon siRNAs after eliminating Dharmacon siRNAs predicted to deplete Rad51 by a 7-nucleotide antisense seed region match to the Rad51 3’UTR (Supplementary Information, Table S6).
Candidate HR effector RBMX accumulates at regions of DNA damage in a PARP-dependent manner
Next we focused on RBMX, a second candidate that localized to DNA damage. RBMX is a nuclear hnRNP protein that regulates alternative splicing in at least two possible ways: one through RNA binding by a RNA Recognition Motif (RRM) and one independent of the RRM11. X-linked RBMX and a Y chromosome-encoded paralog (RBMY) are found in the pseudoautosomal region of their respective sex chromosomes and are conserved among mammals17. In humans, there are several intron-less retrogenes of RBMX present on various autosomes18. Expression of RBMX and at least one of its retrogenes (RBMXL1) is ubiquitous throughout tissue types, while expression of RBMY is restricted to male germ cells indicating a role in spermatogenesis and possibly meiosis17–19. RBMX has also been proposed to be a tumor suppressor20, 21.
GFP-RBMX localization to DNA damage was apparent in ~20–40% of U2OS cells, and both endogenous and Flag/Ha (FHA)-tagged RBMX microirradiation tracks (or “stripes”) could be observed with antibodies after Triton-X pre-extraction (Fig. 4a–c; Supplementary Information, Fig. S5a–b). The percentage of cells with GFP-RBMX tracks increased after depletion of endogenous RBMX suggesting that background chromatin binding obscures detection (Fig 4a). GFP-RBMX localization to DNA damage was transient, occurring between 0 and 10 minutes after microirradiation (longer at room temperature); and following this initial recruitment, GFP-RBMX was removed from damaged DNA causing a localization pattern we refer to as an “anti-stripe” (Fig. 4b–c; Supplementary Information, Fig. S5c). HA-tagged and endogenous RBMX formed anti-stripes as well (Supplementary Information, Fig. S5a–b). We failed to observe RBMX accumulation at ionizing radiation-induced foci, although this may be a consequence of low signal-to-noise ratio or the transient nature of RBMX recruitment. We previously showed that RNAP2 forms anti-stripes after microirradiation in a manner correlated with transcriptional repression at sites of active DNA repair22. Consistent with this interpretation, we also observed anti-stripe localization of other hnRNP proteins after microirradiation (Supplementary Information, Fig. S5d).
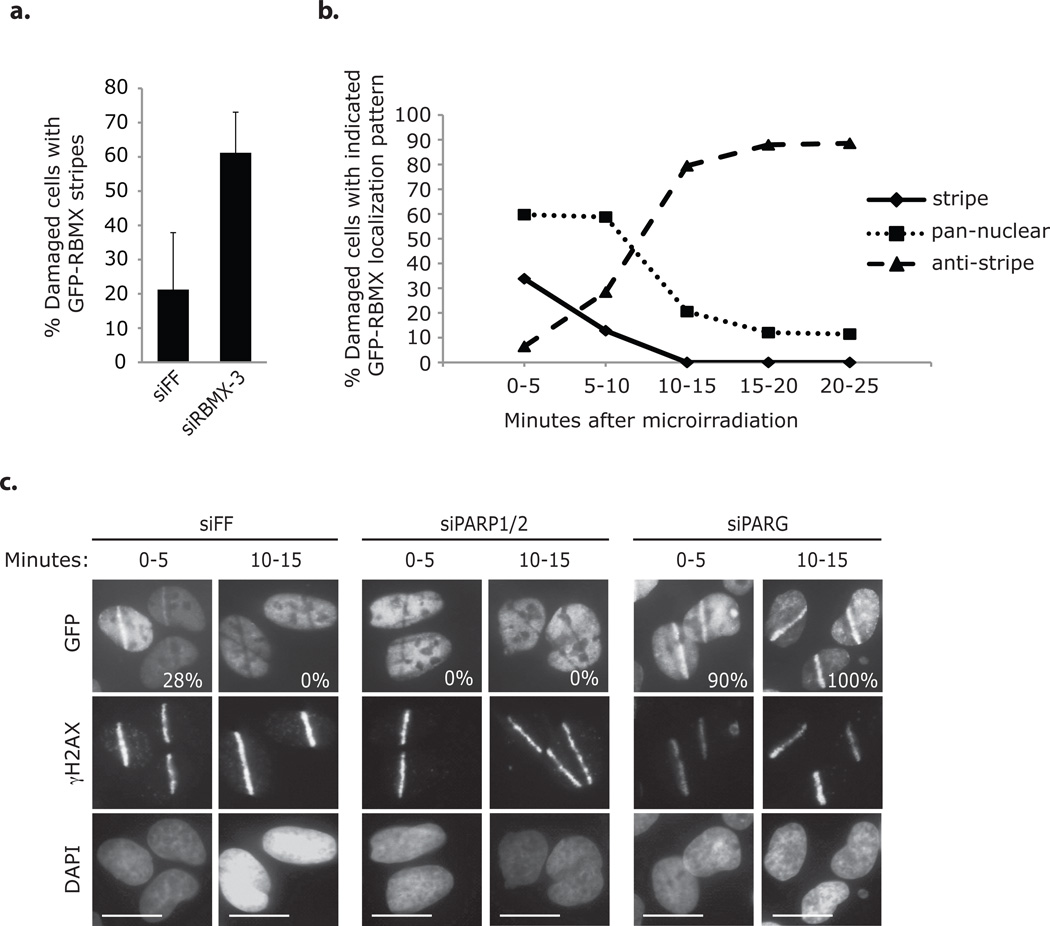
RBMX accumulates transiently at sites of DNA damage in a PARP-dependent manner. (a) U2OS cells expressing siRBMX-3 resistant GFP-RBMX and transfected with the indicated siRNAs were microirradiated (one at a time for 5 minutes) and then immediately processed for immunofluorescence with an antibody against γH2AX. Cells were evaluated for GFP-RBMX accumulation at γH2AX stained laser tracks (approx. 60–190 cells / condition). Error bars represent ± s.d. across three replicates. (b) U2OS cells expressing GFP-RBMX were microirradiated (one at a time for 5 minutes) and processed for immunofluorescence with an antibody against γH2AX at the indicated times. Cells were evaluated for GFP-RBMX accumulation at γH2AX stained laser tracks (approx. 130–190 cells / condition). Data represent the mean of two replicates. (c) U2OS cells expressing GFP-RBMX and transfected with the indicated siRNAs were microirradiated (one at a time for 5 minutes) and processed for imaging with an antibody against γH2AX at the indicated times. GFP was observed directly. Nuclei were stained with DAPI. The percentages of cells with GFP-RBMX accumulation at γH2AX stained laser tracks are indicated (approx. 110–160 cells / condition, n=1). siPARP1/2 indicates two pools of 4 siRNAs targeting PARP1 and PARP2. siPARG indicates a pool of 4 siRNAs targeting PARG. Scale bars indicate 10 µm.
RBMX recruitment to DNA damage was independent of ATM signaling and H2AX but dependent on poly(ADP-ribose) polymerase 1 (PARP1) (Fig. 4c; Supplementary Information, Fig. S6a–c). PARP1 mediates recruitment of repair proteins to DNA damage via the transient polymerization of branched poly(ADP-ribose) (PAR) structures; and it is likely that RBMX localization is similarly facilitated1, 22, 23. First, GFP-RBMX track formation occurred coincident with PAR formation at breaks (Supplementary Information, Fig. S6d). Second, inhibition of PARP1 –via the chemical inhibitor KU0058948 or by siRNA-mediated depletion– prevented GFP-RBMX track formation and caused early formation of anti-stripes (Fig. 4c; Supplementary Information, Fig. S6a–b). Third, transfection of cells with siRNAs against PARG, the PAR disassembly enzyme, increased the percentage of cells with GFP-RBMX tracks after microirradiation and prolonged localization at damage (Fig. 4c).
RBMX promotes DNA repair by homologous recombination
The RBMX siRNA pool in our primary screen decreased HR to 7% of controls, comparable to the effect of depleting BRCA2 and Rad51 (5% and 11%, respectively). All four siRNAs from this pool (siRBMX-1 through -4), as well as two Ambion siRNAs (siRBMX-5 and -6) and three independently selected shRNAs (shRBMX-7, -9 and -10), caused defective HR in a manner correlating with RBMX depletion (Fig. 5a–c; Supplementary Information, Fig. S7a–b, and Table S4). We ruled out obvious off-target effects for these RNAi reagents, and found that expression of siRNA-resistant RBMX rescued the siRBMX-3 associated HR defect (Fig. 5d–e; Supplementary Information, Fig. S7b–g). There were no confounding effects on the cell cycle distribution in the rescue assay (Supplementary Information, Fig. S8a–b).
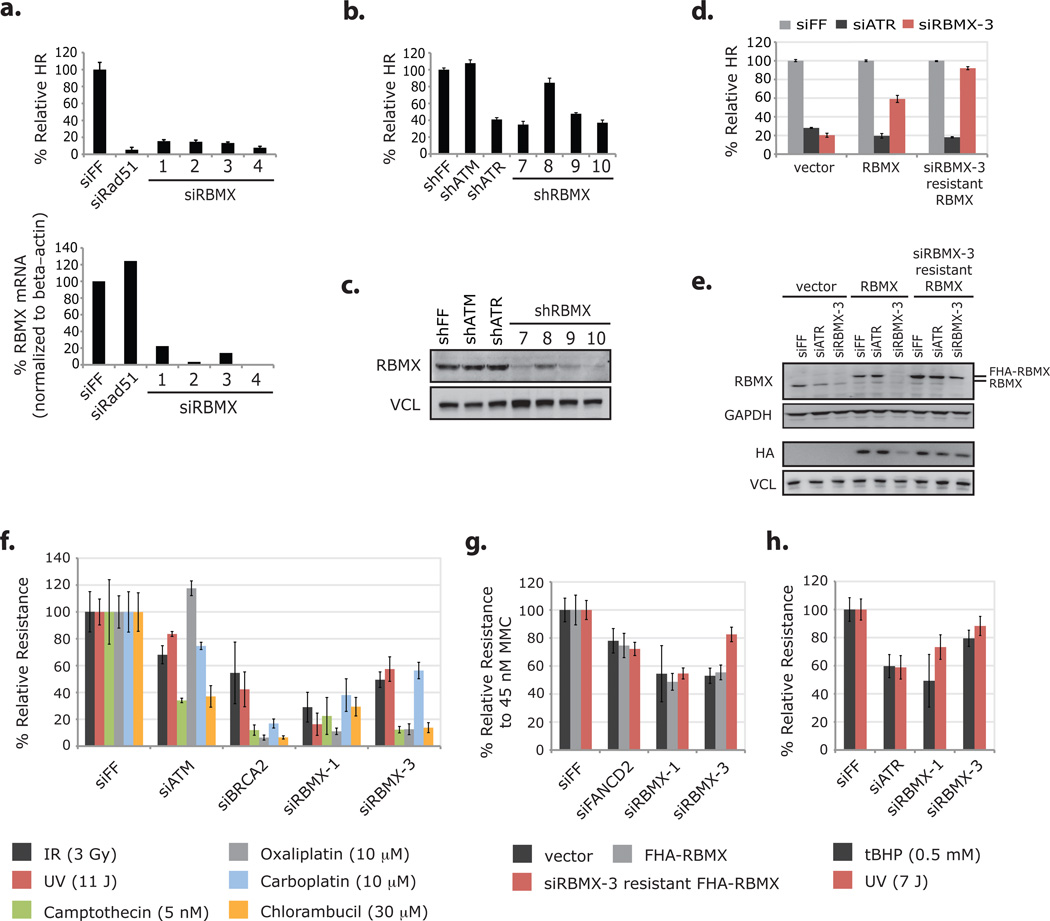
RBMX promotes homologous recombination and resistance to DNA damaging agents. (a) HR assay results from DR-U2OS cells transfected with RBMX-targeting siRNAs and corresponding levels of RBMX mRNA as measured by RT-qPCR (normalized to beta-actin mRNA). Error bars in HR analysis represent ± s.d. across three replicates and data in RT-qPCR analysis represent the mean of two replicates. (b) HR assay results from DR-U2OS cells transduced with RBMX-targeting shRNAs. Error bars represent ± s.d. across three replicates. (c) Western blot analysis of RBMX levels from cells analyzed in b. (d) DR-U2OS cells transduced with control vector, FHA-RBMX and siRBMX-3 resistant FHA-RBMX cDNAs and then transfected with indicated siRNAs were assayed for HR efficiency. Data from cells with the same cDNA were normalized to the siFF condition. Error bars represent ± s.d. across three replicates. (e) Whole-cell extracts from cells in d were immunoblotted with the indicated antibodies. Two western blots of the same extracts are presented and panels are grouped accordingly. (f) Multicolor competition assay for resistance to DNA damaging agents31. Briefly, U2OS cells expressing GFP and transfected with the indicated siRNAs were mixed in a 1:1 ratio with dsRed U2OS cells transfected with siFF. Cell mixtures were treated with the indicated DNA damaging agents, and after 8 days, the relative survival of GFP to dsRed cells was determined by FACS analysis. GFP / dsRed ratios were normalized to those from undamaged pools to control for the relative growth effects of the siRNAs. Error bars represent ± s.d. across three replicates. (g) DR-U2OS cells transduced with the indicated cDNAs and then transfected with the indicated siRNAs were treated with 45 nM MMC, and after 7 days relative resistance was determined by the CellTiter-Glo Luminescent Cell Viability Assay. Error bars represent ± s.d. across three replicates. (h) U2OS cells transfected with the indicated siRNAs were treated with tBHP (for 1 hour) or UV, and after 5 days relative resistance was determined by the CellTiter-Glo Luminescent Cell Viability Assay. Error bars represent ± s.d. across three replicates. Full scans of blots in c and e are shown in Supplementary Information, Fig. S13.
RBMX-targeting siRNAs also sensitized cells to DNA damaging agents that engage the HR machinery for repair, including DSB-inducing irradiation (IR), replication stress-inducing camptothecin, and several crosslinking agents (mitomycin C, chlorambucil, oxaliplatin, and carboplatin) (Fig. 5f–g). The sensitivity to mitomycin C caused by siRBMX-3 was significantly attenuated by expression of siRNA-resistant FHA-RBMX (Fig. 5g). Interestingly, RBMX depletion also caused sensitivity to ultraviolent light (UV) and tert-butyl hydroperoxide (tBHP), both of which cause DNA lesions not primarily repaired by HR (Fig. 5f, h). Because PARP is important for the repair of single strand DNA breaks induced by tBHP, sensitization of cells to tBHP by RBMX depletion is consistent with a role for RBMX in PARP-mediated repair.
RBMX influences HR by facilitating proper expression of BRCA2
Next we evaluated the effect of RBMX depletion on known HR events, specifically Rad51 nucleation onto resected ssDNA and the coordinated upstream signaling. RBMX depletion caused defective formation of IR-induced Rad51 foci, which was attenuated by expression of siRNA-resistant FHA-RBMX (Fig 6a–c; Supplementary Information, Fig. S8c–d). Although the siRNAs used in these experiments (siRBMX-1 and -3) caused slight reductions to Rad51 protein levels (but not Rad51 mRNA), the effect caused by siRBMX-3 (unlike HR) was not rescued by siRNA-resistant FHA-RBMX indicating that there is a negligible, RBMX-independent effect of the siRNAs on Rad51 levels (Supplementary Information, Fig. S7b, e, S8a). Depletion of RBMX had no effect on RPA2 or Chk1 phosphorylation after IR or camptothecin treatment suggesting that, in the absence of RBMX resection at breaks proceeds properly (Supplementary Information, Fig. S9a–c).
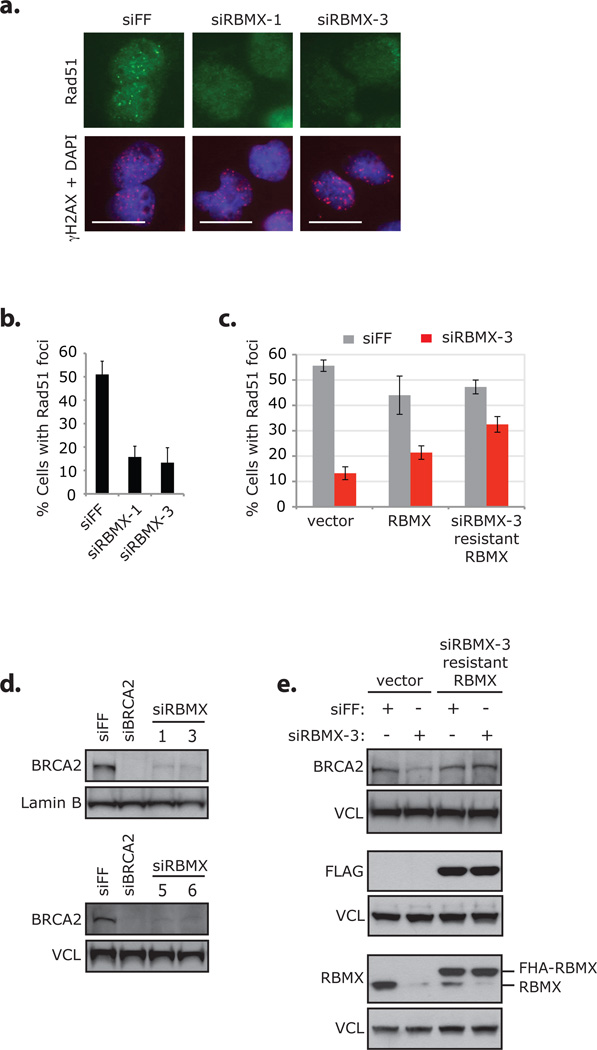
RBMX promotes formation of IR-induced Rad51 foci by facilitating proper expression of BRCA2. (a) DR-U2OS cells were transfected with the indicated siRNAs, treated with 10 Gy IR, and processed for imaging with antibodies against Rad51 and γH2AX after 4 hours. Nuclei were stained with DAPI. Scale bars indicate 20 µm. Data from the same experiment is presented in Supplementary Information, Fig. S7d–g, S8c. Individual adjustment of color channels in γH2AX + DAPI was required to illustrate foci; identical adjustment parameters were used. (b) DR-U2OS cells transfected with the indicated siRNAs were damaged and processed for immunofluorescence in same manner as described in a. Cells with Rad51 foci were counted by eye; because transfection with siRNAs against RBMX causes some changes to nuclear morphology, only normal shaped nuclei were counted (approx. 100–130 cells / condition). Error bars represent ± s.d. across four replicates. Data from the same siRNA transfected cells are presented in Supplementary Information, Fig S9b–c. (c) DR-U2OS cells transduced with control vector, FHA-RBMX or siRBMX-3 resistant FHA-RBMX cDNAs were transfected with indicated siRNAs and treated with 10 Gy IR. After 8 hours cells were processed for immunofluorescence with antibodies against Rad51 and γH2AX and counted (200 cells / condition). Error bars represent ± s.d. across four replicates. Representative images of cells are presented in Supplementary Information, Fig. S8d. (d) Whole-cell extracts from DR-U2OS cells transfected with the indicated siRNAs were immunoblotted with antibodies against BRCA2 or Lamin B / vinculin (VCL). Extracts analyzed in the upper blot were evaluated for protein levels of additional HR and DDR proteins (Supplementary Information, Fig. S12a); and additional data from this blot are presented in Supplementary Information, Fig. S12b (the Lamin B panel is reproduced as a control in that figure). Extracts analyzed in the lower blot were also used in Supplementary Information, Fig. S7b / S12d. (e) Whole-cell extracts from undamaged DR-U2OS cells evaluated in c were immunoblotted with the indicated antibodies. Three western blots of the same extracts are presented and panels are grouped accordingly. Full scans of blots in d–e are shown in Supplementary Information, Fig. S13.
To determine the region(s) of RBMX responsible for promoting HR and facilitating localization to DNA damage, we tested a series of GFP- and FHA-tagged RBMX mutants (Supplementary Information, Fig. S10a–d; see Supplementary Results). Deletion and point mutation of the conserved nucleotide-interacting residues in the RRM rendered the siRNA-resistant FHA-RBMX unable to rescue HR comparable to FHA-RBMX with a wild-type RRM at the indicated levels of expression; however, the RRM was dispensable for DNA damage localization. Next, we asked if RBMX localization to DNA damage is required for HR; and surprisingly, we found that HR efficiency was not decreased under PARP inhibition or depletion conditions that prevented RBMX accumulation at DNA damage (Supplementary Information, Fig. S11a–b). PARP1 depletion also did not substantially alter HR in an RBMX-independent manner (Supplementary Information, Fig. S11c).
Because the RNA binding region of RBMX was important for HR but rapid recruitment of RBMX to DNA lesions was not, we reasoned that RBMX might promote HR by influencing protein expression through pre-requisite splicing events. While RBMX depletion had no effect on many repair proteins we tested, including PALB2, BRCA1 and RPA2 (and no effect on BRCA1 foci formation), we found that BRCA2 (and to some extent ATR) levels were decreased by RBMX siRNAs in a manner that could be rescued by siRNA-resistant FHA-RBMX (Fig. 6d–e; Supplementary Information, Fig. S12a–e). Because of these results, we evaluated siRNAs against seven additional pre-mRNA processing genes identified as candidate HR mediators by our screen and found that some of these also had an effect on BRCA2 expression (Supplementary Information, Fig. S12f–j).
DISCUSSION
Here, we describe an unbiased screen to identify regulators of the mammalian HR machinery that yielded two candidate gene sets (510 positive and 484 negative regulators). We present these lists as a well-curated resource. Of 510 candidate mediators, we have validated 121 with ≥3 individual siRNAs. This validated list contains known HR proteins, including the recently characterized 9-1-1 complex-interacting protein RHINO, as well as uncharacterized HR proteins24. Among these is c4orf21, which was recently shown to influence crosslink repair in a genome-wide screen for sensitivity to mitomycin C and contains a predicted DNA binding domain and helicase-like region25. Both candidate mediators and suppressors will aid future characterization of HR regulation; but, as it is well established that NHEJ proteins suppress HR, the HR suppressor set may also yield positive regulators of NHEJ26. This suppressor set may also contribute to genetic engineering studies of gene targeting in mammalian cells.
Our initial candidate characterization work (primarily of HIRIP3) led to the insight that siRNA-based HR studies are plagued by off-target effects and using a method for the identification off-targets in genome-wide datasets (GESS), we determined that in our screen Dharmacon siRNAs with strong HR defects were 3-fold enriched for antisense seed sequence complementarity to the Rad51 3’UTR (Fig. 3d). We confirmed off-target depletion of Rad51 mRNA and protein by 6 screened Dharmacon siRNAs with such complementarity (of 7 tested) (Fig. 3e). Interestingly though, 3 siRNAs identified by the screen to cause defective HR (siHIRIP3-1, siHIRIP3-2 and siHIRIP3-4) depleted Rad51 without full seed complementarity to the 3’UTR; and 4 independently selected siRNAs (of the 14 siRNAs used to evaluate HIRIP3, HIRA and the HIRA-associated candidate proteins) were observed to deplete Rad51 protein, but only 1 of these had a full antisense seed match (Fig. 3b; Supplementary Information, Fig. S3e–f, S4d). Therefore additional mechanisms of Rad51 off-targeting by siRNA may be common, and Rad51 hypersensitivity to RNAi may confound study results even when RNAi reagents are chosen without selective pressure.
It is tempting to speculate that some as yet unidentified structural element within the Rad51 mRNA hypersensitizes this particular transcript to siRNA-mediated depletion, a miRNA enhancer of sorts. The cell cycle regulated expression of Rad51 may aid its hypersensitivity to RNAi, as it would presumably require less time to deplete than a stable protein. Undoubtedly, however, a key aspect of this hypersensitivity must be the critical dependence of the HR process on Rad51 levels, whose highly cooperative filament assembly on ssDNA should be exquisitely sensitive to protein levels. Supporting this, we observe that a slight decrease of Rad51 protein does not affect HR but reduction below a certain threshold correlates with a strong phenotype (Fig. 3e).
We have successfully identified the RNA binding protein RBMX as a regulator of HR, and in characterizing RBMX we have begun to define a relationship between RNA processing and DNA repair. The enrichment of RNA processing factors in our set of candidate HR regulators and the identification of this network in other large-scale studies of the DDR, have made it difficult to ignore the notion that RNA processing and DNA repair functionally intersect8–10. Roles for a few RNA binding proteins (RBPs) in the DDR have already been elucidated. The protein hnRNP K is induced after DNA damage and acts together with p53 to promote damage-induced transcriptional programs, and hnRNP A0 binds to and stabilizes Gadd45α mRNA in response to DNA damage27, 28. The authors of the hnRNP A0 study hypothesize that RBPs generally influence the DDR by globally affecting mRNA stability after DNA damage28, 29.
Our study extends the body of work connecting specific RBPs directly to the DDR by establishing that RBMX localizes to sites of DNA damage. The biochemical consequences of transiently accumulating RBMX to sites of damage remains to be elucidated. It is possible that the ability of RBMX to localize at these sites through two independent interaction domains could help to bundle PAR structures and hold breaks together to facilitate DNA repair. It is also possible that RBMX localizes a specific noncoding RNA to sites of breaks to facilitate repair. Importantly, however, the role for RBMX in HR is likely through indirect regulation of BRCA2, possibly due to a pre-requisite splicing event. siRNAs against other pre-mRNA processing genes have some effect on BRCA2 (albeit less substantially), and so we hypothesize that depletion of splicing factors in general may cause defective DNA repair through mis-splicing or alternative splicing of key effectors. BRCA2 and ATR are large genes that contain a considerable number of exons (26 and 47 respectively), which may render them particularly susceptible to misregulated splicing. In this way, the pre-mRNA processing genes on our candidate list likely represent indirect, but biologically significant, components of DNA repair.
In this work, we provide a set of genes involved in HR as a foundation upon which future studies can build. In addition to the biological insights herein, we have shown that siRNA screens can be plagued by off target effects in a significant manner. This provides a note of caution in interpreting siRNA screen results and argues strongly that each screen be subject to a search for significant off-targets.
METHODS
Cell culture
Human U2OS and DR-U2OS osteosarcoma cells and mouse embryonic fibroblasts were grown in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 units / ml of penicillin, and 0.1 mg / ml streptomycin (Invitrogen). For HTP screening, DR-U2OS cells were grown in McCoy’s 5A media with 10% FBS. DR-U2OS cells were provided by Maria Jasin7.
Antibodies and Inhibitors
Primary antibodies are listed in Supplementary Information, Table S7. Secondary antibodies for immunofluorescence were Alexa Fluor® 488 and 594 conjugated (Invitrogen). Secondary antibodies for western blots were from Jackson Laboratory. The PARP inhibitor (KU-0058948) was used at 1 µM (from KuDOS Pharmaceuticals Ltd., provided Simon Boulton). The ATM inhibitor (KU-55933) was used at 10 µM (Sigma).
Plasmids, shRNAs, siRNAs and RT-qPCR
RBMX, HIRIP3, DDX17, RBMY, hnRNP-K, hnRNP-C, Histone H3 cDNAs were from hORFeome V5.1. Full-length RBMX, HIRIP3 and H3 were verified by sequencing. The 5’ ends of DDX17, RBMY, hnRNP-K, hnRNP-C were verified by sequencing. Mutants with point mutations and internal deletions were generated using QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies). Truncation fragments were generated by PCR and cloned into the pENTR™/D-TOPO vector using pENTR™ Directional TOPO® Cloning Kit (Invitrogen). Mutants and truncations were verified by sequencing. cDNAs were cloned into pMSCV-N-HA-Flag-GAW-IRES-PURO or pMSCV-N-EGFP-GAW-PGK-PURO using the Gateway recombination system. shRNAs were used in the pSMP-MSCV-PURO vector (Open Biosystems). siRNAs were transfected into cells at 20–50 µM using either Oligofectamine™ or Lipofectamine™ RNAiMAX Transfection Reagents (Invitrogen) according to manufacturer instructions and cells were processed for indicated experiments 2–3 days later. shRNA and siRNA sequences are listed in Supplementary Information, Table S2–4, 8–9. RNA was isolated from cells using the RNAeasy Plus kit (Qiagen) and reverse transcribed into cDNA using SuperScript III Reverse Transcriptase (Invitrogen #18080-044) according to the manufacturer instructions. Quantitative RT-PCR (RT-qPCR) was performed using Platinum Cybergreen Super Mix with Rox dye (Invitrogen #11733-046) on an Applied Biosystems 7500 Fast PCR machine. RT-qPCR primers can be found in Supplementary Information, Table S10.
HR Assay and high-throughput (HTP) screening protocol
The primary screen was performed using 21,121 siRNA pools from the Dharmacon human siGENOME siRNA library (G-005000-05) at 50 nM. Dharmacon and Ambion rescreens were conducted at 20 nM. HTP screening: DR-U2OS cells were plated on 384 well plates at 700 cells / well and reverse transfected with siRNAs using Oligofectamine™ Transfection Reagent. Positive (siATR and siBRCA2) and negative (siFF) controls were added to each plate. After 72 hours, cells were infected with the I-SceI carrying adenovirus AdNGUS24i (provided by Frank Graham, McMaster University) at an estimated MOI of 10; 48 hours after infection, cells were fixed in 3.7% formaldehyde and stained with Hoechst 33342 at a dilution of 1:5000 (Invitrogen). Changes were made to this protocol for rescreening Ambion siRNAs: (1) 500 DR-U2OS cells / well were plated (to adjust for reduced toxicity of Ambion siRNAs observed in controls), (2) the Lipofectamine™ RNAiMAX Transfection Reagent was used, and (3) AdNGUS24i was used at an MOI of ~15 (to adjust for differences between viral preparations). Automated imaging of screen plates (2–4 images per well in 2 channels) was conducted on an Image Express Micro microscope (Molecular Devices) at 4X magnification (488 nm and 350 nm wavelengths were used to detect GFP expression and Hoechst 33342 stained DNA, respectively). Automated counting of GFP+ and Hoechst stained nuclei was performed for each image with Metamorph Cell Scoring software (Molecular Devices Inc.), and a ratio of GFP+ to Hoechst stained (total) nuclei was calculated for each well using all corresponding images. Primary screen pools, rescreened Ambion siRNAs and deconvolved Dharmacon siRNAs against candidate suppressors were evaluated in triplicate; Dharmacon siRNAs against candidate mediators were evaluated in duplicate. To normalize day-to-day variability, each triplicate (or duplicate) average of % GFP+ was normalized to the average % GFP+ from on-plate negative control wells (siFF). These normalized values are the relative HR ratios, and standard deviation was calculated and propagated for each. Select images from the primary screen and Dharmacon rescreen of candidate mediators yielding high standard deviations were visually inspected, and data from images that were found to contain an irregularity (for example: were out of focus) were deleted. A cell number / well value was calculated for each well as the sum of Hoechst 33342 stained nuclei from all corresponding images, and a relative cell growth ratio for each siRNA pool was calculated by normalizing the average cell number / well of corresponding experimental wells to that of on-plate negative control wells (siFF). The number of images taken of experimental and corresponding control wells was the same.
The procedure for low-throughput HR assays was similar to the HTP protocol (above), except: (1) DR-U2OS cells were either forward or reverse transfected in 6 well plates, (2) AdNGUS24i was used at an exact MOI of 10, (3) GFP+ ratios were determined ~36–48 hour post infection by FACS analysis on a BD LSRII Flow Cytometer (BD Biosciences).
Candidate selection
Primary screen data was collected and processed as described above in “HR Assay and high-throughput (HTP) screening protocol.” All relative HR ratios from the primary screen were compiled, and from this, 519 pools that decreased relative HR >2 s.d. from the screen-wide mean and 486 pools that increased relative HR >2 s.d. (including 2 duplicate pools against SMAD1 and TIAM2) were identified (screen mean = 1.14, s.d. = 0.37) (Supplementary Information, Table S1). siRNA pools that were unavailable for validation, corresponded to discontinued gene entries in the NCBI database, or (as stated above) were determined by visual analysis to be based on poor quality imaging were eliminated. The 510 and a selection of the 486 siRNA pools remaining (against 510 and 484 HR mediator and suppressor candidates, respectively) were deconvolved into individual duplexes and rescreened (Supplementary Information, Table S2–3). For this 131 pools (against 131 genes) were added to the 510 candidate pools against HR mediators (for 641 candidates, 2564 siRNAs total). Selection of the 131 additional candidates is described in the Results section. Only the 250 pools (including 1 duplicate pool against SMAD1) that most strongly increased HR were rescreened. siRNAs from the Ambion Silencer Select library targeting 467 (of 641) candidate HR mediators were also screened (3 siRNAs / gene, 1401 siRNAs total). Selection of these 467 candidates was based on data from our primary screen and Dharmacon rescreen analysis, as well as data from related DNA damage screens, GESS analysis and published information about each gene.
Ingenuity Pathway Analysis (IPA) was conducted on the candidate HR mediator and suppressor sets generated by applying a 2 s.d. cutoff to the primary screen data and prior to the expansion / editing of the candidate lists outlined above (519 mediators and 484 suppressors) (Supplementary Information, Table S1). These gene sets were uploaded into IPA software and scanned for functional enrichment and interaction networks based on information in the Ingenuity Knowledge Base. Select functional enrichment categories are displayed in Fig. 2e and Supplementary Information, Fig. S2a. Enrichment p-values were determined using the Fisher’s Exact Test. Select protein networks are displayed in Fig. 2f–h and Supplementary Information, Fig. S2b–c. Network nodes are colored according to the number of siRNAs that scored using a weak cutoff value based on 1.5 s.d. from the primary screen mean.
Genome-wide Enrichment of Seed Sequences (GESS) off-target analysis
GESS off-target analysis was conducted as previously described14. The 2564 Dharmacon siRNAs (from 641 pools) and 1401 Ambion siRNAs rescreened against candidate HR mediators, as well as the 1000 Dharmacon siRNAs (from 250 pools, including 1 duplicate pool) rescreened against candidate HR suppressors, were each submitted to GESS analysis. Significance p-values were determined as follows: the Yates’ Chi Square statistic and associated one-tailed p-value were calculated for each database sequence evaluated (3’UTR or CDS) if all siRNA categories (active siRNAs with or without matching, inactive siRNAs with or without matching) had more than 20 seed match events. Otherwise, a two-sided p-value was calculated from the Fisher’s Exact Test. The transcript sequences were ranked from lowest to highest p-value and the statistical significance was determined by comparing the p-value to a p-value threshold (0.05) corrected for multiple hypothesis testing using the Benjamini and Hochberg (Simes') method.
UV laser / IR induced DNA damage and immunofluorescence
UV laser-induced damage was generated as previously described30. Cells were sensitized to UV-A laser (λ = 355 nM) by 24 hour pre-treatment with 10 µM BrdU and microirradiated using a PALM MicroBeam with fluorescence illumination (Zeiss) at 40–45% laser power. Ionizing irradiation (IR)-induced damage was generated by timed exposure to a Cesium-137 source. After damage, cells were allowed to recover for the indicated times at either room temperature (RT) or 37°C and then fixed in 3.7% formaldehyde for 10 minutes at RT. Fixed cells were washed twice with PBS, permeabilized in 0.5% NP-40, washed twice with PBS, and blocked with PBG (0.2% [w/v] cold fish gelatin, 0.5% [w/v] BSA in PBS) for 30 minutes prior to immunostaining with the indicated antibodies diluted in PBG. DNA was stained with DAPI by addition of Vectashield Mounting Medium (Vector Laboratories). GFP was observed directly. Images were collected on an Axioplan2 Zeiss microscope with a AxioCam MRM Zeiss digital camera and Axiovision 4.5–4.8 software. Images intended for comparison were prepared from the same experiment with the same exposure times, and were processed for brightness and contrast in an identical manner. Those not intended for comparison and not prepared in this way are indicated.
Cell cycle analysis
Cells were prepared for cell cycle analysis using the BD Pharmingen™ APC BrdU Flow Kit according to manufacturer instructions. Cell cycle profiles were obtained by FACS analysis on a BD LSRII Flow Cytometer (BD Biosciences).
Sensitivity assays
Multicolor competition assays were performed as previously described31. CellTiter-Glo Luminescent Cell Viability Assay (Promega): Cells were transfected with the indicated siRNAs and treated as indicated after 2–3 days. After recovery from treatment, the media was changed and CellTiter-Glo reagent was added (1:17.5 dilution). The resulting luminescent signal (proportional to the amount of ATP) was read on a VICTOR X5 Multilabel Plate Reader (PerkinElmer). The signal from each treatment condition was normalized to an untreated control to adjust for the relative growth effects of the siRNAs. Data are presented normalized to that from control transfected cells (siFF).
Supplementary Material
1
18
19
2
20
21
22
23
24
25
26
10
27
28
3
4
5
6
7
8
9
11
12
13
14
15
16
17
ACKNOWLEDGEMENTS
We thank Dr. Maria Jasin for DR-U2OS cells, Dr. Philip Ng and Dr. Frank Graham for the AdNGUS24i, Dr. Peter Adams for antibodies, Dr. Simon Boulton for the PARP inhibitor, and Cecilia Cotta-Ramusino and members of the Elledge lab for advice and discussion. We thank the Institute of Chemistry and Cell Biology (ICCB)-Longwood screening facility, including Caroline Shamu, Stewart Rudnicki, Sean M. Johnston and Tiao Xie. This work was supported by a grant from the National Institutes of Health to S.J.E. A.S. was supported by T32CA09216 to the Pathology Department at the Massachusetts General Hospital and by Burroughs Wellcome Fund Career Award for Medical Scientists and is a Rita Allen Foundation and an Irma T. Hirschl scholar. S.J.E. is an investigator with the Howard Hughes Medical Institute.
Footnotes
AUTHOR CONTRIBUTIONS
B.A., A.S. and S.J.E. conceived experimental design and conducted data analysis. F.D.S. and R.W.K. performed GESS analysis. The manuscript was prepared by B.A. and S.J.E. and edited by A.S., F.D.S., and R.W.K.COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
Full text links
Read article at publisher's site: https://doi.org/10.1038/ncb2426
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3290715?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/ncb2426
Article citations
An anciently diverged family of RNA binding proteins maintain correct splicing of a class of ultra-long exons through cryptic splice site repression.
Elife, 12:RP89705, 02 Oct 2024
Cited by: 1 article | PMID: 39356106 | PMCID: PMC11446547
DSS1 restrains BRCA2's engagement with dsDNA for homologous recombination, replication fork protection, and R-loop homeostasis.
Nat Commun, 15(1):7081, 17 Aug 2024
Cited by: 0 articles | PMID: 39152168 | PMCID: PMC11329725
Involvement of the splicing factor SART1 in the BRCA1-dependent homologous recombination repair of DNA double-strand breaks.
Sci Rep, 14(1):18455, 08 Aug 2024
Cited by: 0 articles | PMID: 39117746 | PMCID: PMC11310344
hnRNPs: roles in neurodevelopment and implication for brain disorders.
Front Mol Neurosci, 17:1411639, 17 Jul 2024
Cited by: 0 articles | PMID: 39086926 | PMCID: PMC11288931
Review Free full text in Europe PMC
Therapy-induced secretion of spliceosomal components mediates pro-survival crosstalk between ovarian cancer cells.
Nat Commun, 15(1):5237, 19 Jun 2024
Cited by: 0 articles | PMID: 38898005 | PMCID: PMC11187153
Go to all (300) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Sensitivity to poly(ADP-ribose) polymerase (PARP) inhibition identifies ubiquitin-specific peptidase 11 (USP11) as a regulator of DNA double-strand break repair.
J Biol Chem, 285(19):14565-14571, 15 Mar 2010
Cited by: 99 articles | PMID: 20233726 | PMCID: PMC2863164
Requirement of heterogeneous nuclear ribonucleoprotein C for BRCA gene expression and homologous recombination.
PLoS One, 8(4):e61368, 09 Apr 2013
Cited by: 46 articles | PMID: 23585894 | PMCID: PMC3621867
Identification of regulators of poly-ADP-ribose polymerase inhibitor response through complementary CRISPR knockout and activation screens.
Nat Commun, 11(1):6118, 30 Nov 2020
Cited by: 33 articles | PMID: 33257658 | PMCID: PMC7704667
Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins.
Cold Spring Harb Perspect Biol, 7(4):a016600, 01 Apr 2015
Cited by: 474 articles | PMID: 25833843 | PMCID: PMC4382744
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Howard Hughes Medical Institute
NCI NIH HHS (2)
Grant ID: T32 CA009216
Grant ID: T32CA09216
NIGMS NIH HHS (3)
Grant ID: R01 GM044664
Grant ID: R37 GM044664
Grant ID: R37 GM044664-24





