Abstract
Free full text

A GWAS follow-up study reveals the association of the IL12RB2 gene with systemic sclerosis in Caucasian populations
Abstract
A single-nucleotide polymorphism (SNP) at the IL12RB2 locus showed a suggestive association signal in a previously published genome-wide association study (GWAS) in systemic sclerosis (SSc). Aiming to reveal the possible implication of the IL12RB2 gene in SSc, we conducted a follow-up study of this locus in different Caucasian cohorts. We analyzed 10 GWAS-genotyped SNPs in the IL12RB2 region (2309 SSc patients and 5161 controls). We then selected three SNPs (rs3790567, rs3790566 and rs924080) based on their significance level in the GWAS, for follow-up in an independent European cohort comprising 3344 SSc and 3848 controls. The most-associated SNP (rs3790567) was further tested in an independent cohort comprising 597 SSc patients and 1139 controls from the USA. After conditional logistic regression analysis of the GWAS data, we selected rs3790567 [PMH= 1.92 × 10−5 odds ratio (OR) = 1.19] as the genetic variant with the firmest independent association observed in the analyzed GWAS peak of association. After the first follow-up phase, only the association of rs3790567 was consistent (PMH= 4.84 × 10−3 OR = 1.12). The second follow-up phase confirmed this finding (Pχ2 = 2.82 × 10−4 OR = 1.34). After performing overall pooled-analysis of all the cohorts included in the present study, the association found for the rs3790567 SNP in the IL12RB2 gene region reached GWAS-level significant association (PMH= 2.82 × 10−9 OR = 1.17). Our data clearly support the IL12RB2 genetic association with SSc, and suggest a relevant role of the interleukin 12 signaling pathway in SSc pathogenesis.
INTRODUCTION
Systemic sclerosis or scleroderma (SSc) is a rare complex connective tissue disorder characterized by extensive fibrosis of multiple organs produced by vascular damage and autoimmune dysfunction (1,2). Patients are commonly classified into two major subgroups: the limited cutaneous SSc (lcSSc) and the diffuse cutaneous (dcSSc) form of the disease (3). Positive autoantibody titers are a main feature of this disabling condition, especially anticentromere autoantibodies (ACA) and antitopoisomerase autoantibodies (ATA) (1,2). To date, a number of genes have been implicated in an increased susceptibility to SSc, confirming the genetic component of this complex disease (4,5). Some of these genes are shared with other related autoimmune diseases, supporting the idea of common pathogenic pathways underlying autoimmune imbalance (6,7).
Recently, our group published the first genome-wide association study (GWAS) conducted in Caucasian SSc patients (5). GWASs are often followed by follow-up studies focused on the regions where association peaks are observed, not only in the associations which reached the GWAS significance level, but also those which are below the GWAS level but might result in true association with the disease. In this line, a single-nucleotide polymorphism (SNP) at the IL12RB2 locus showed a suggestive association signal in the previously mentioned GWAS [PMH= 1.92 × 10−5 odds ratio (OR) = 1.19 (1.10–1.29)] (5).
Noteworthy, interleukin 12 (IL-12) binding to its receptor powerfully induces IFNγ production and promotes T helper differentiation in Th1 cells (8). In addition, several experimental and clinical studies have implicated IL-12 and IFNγ in the development of autoimmune inflammation (8,9). The IL-12 receptor (IL-12R) comprises two subunits, IL-12R β1 subunit (IL-12Rβ1) and IL-12R β2 subunit (IL-12Rβ2), which are both homologous to gp130 (a shared component of the receptors for several type I cytokines) (10).
IL12RB2 encodes IL-12Rβ2, which constitutes the transducing component of the receptor heterodimer and recruits different tyrosine kinases, signal transducers and activators of transcription (11–13). Interestingly, animal models lacking IL12Rβ2 signaling develop autoimmune events (14). Moreover, polymorphisms in the IL12RB2 gene region and upstream this locus have been related to several human autoimmune disorders, such as psoriasis (PS) (15), primary biliary cirrhosis (PBC) (16), Behçet disease (17,18) and giant cell arteritis (GCA) (19).
Hence, with the aim of investigating the possible role of the IL12RB2 gene in SSc, we conducted a GWAS follow-up study in different European and US Caucasian cohorts.
RESULTS
IL12RB2 region analysis in the GWAS set
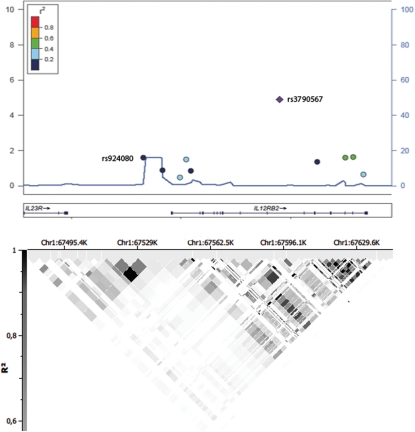
Ten SNPs in the IL12RB2 region were included in the initial GWAS analysis set, six of them were found to be significantly associated with SSc, but only four remained significant after GC correction (Table 1). However, conditioned logistic regression revealed that among the initially observed associations, only the rs3790567 association was independent from the others (Table 1). HapMap linkage disequilibrium patterns defined rs3790566 (not included in the GWAS phase) as the unique tag-SNP for rs3790567. Hence, both the most-associated SNP (rs3790567) and this tag-SNP (rs3790566) were selected for replication.
Table 1.
Pooled logistic regression of IL12RB2 genetic variants in the GWAS cohort (2309 SSc patients and 5161 controls)
| SNP | Chr: 1 position (bp) | Minor allele | Plog | OR | PGC | P-value: add to rs3790567 | OR and rs3790567 | P-value rs3790567 add to SNP | OR rs3790567 and to SNP | r2 with rs3790567 |
|---|---|---|---|---|---|---|---|---|---|---|
| rs924080 | 67,532,728 | G | 2.93 × 10−2 | 1.08 | 3.91 × 10−2 | 0.12 | 1.06 | 2.16 × 10−5 | 1.19 | 0.02 |
| rs12131065 | 67,541,594 | A | 0.16 | 0.94 | 0.18 | 0.20 | 0.95 | 7.72 × 10−6 | 1.19 | 0.001 |
| rs3790558 | 67,549,609 | C | 0.31 | 1.04 | 0.34 | 0.19 | 0.95 | 4.74 × 10−6 | 1.23 | 0.23 |
| rs10489627 | 67,552,264 | G | 4.88 × 10−2 | 1.08 | 0.06 | 0.83 | 0.99 | 4.98 × 10−5 | 1.20 | 0.23 |
| rs2066445 | 67,554,563 | A | 0.09 | 0.93 | 0.11 | 0.11 | 0.93 | 7.62 × 10−6 | 1.20 | 0.0005 |
| rs3790567 | 67,594,965 | A | 6.36 × 10−6 | 1.20 | 1.92 × 10−5 | NA | NA | NA | NA | NA |
| rs3828069 | 67,612,161 | G | 4.24 × 10−2 | 0.91 | 0.05 | 0.44 | 0.96 | 4.28 × 10−5 | 1.19 | 0.08 |
| rs4297265 | 67,624,923 | G | 1.71 × 10−2 | 1.09 | 2.41 × 10−2 | 0.39 | 0.96 | 9.86 × 10−5 | 1.23 | 0.44 |
| rs2270614 | 67,628,609 | A | 1.66 × 10−2 | 1.09 | 2.34 × 10−2 | 0.41 | 0.96 | 1.04 × 10−4 | 1.23 | 0.44 |
| rs7555183 | 67,633,215 | A | 0.24 | 1.05 | 0.27 | 0.92 | 1.00 | 1.31 × 10−5 | 1.20 | 0.08 |
Chr, chromosome; Plog, logistic regression P-value; OR, odds ratio; PGC, GC corrected P-value. Last columns, single locus test P-value when SNP added to rs3790567, single locus test OR when SNP added to rs3790567, single locus test P-value when rs3790567 added to SNP in logistic regression analyses, single locus test OR when rs3790567 added to SNP in logistic regression analyses and pairwise r2 of SNP with rs3790567. NA, not applicable.
Despite the loss of the observed association after correction for multiple testing (Table 1), we also included rs924080 in the first follow-up phase. This genetic variant was located in the intergenic region between IL12RB2 and IL23R, and it was the last GWAS SNP contained in the IL23R haplotype block (Fig. 1). In addition, this polymorphism mapped in a recombination hotspot identified in the HapMap Project (Phase II, Caucasian and Asian populations; http://www.hapmap.org) and previous reports (17).
European follow-up phase
Table 2 shows the pooled analysis of seven independent white European cohorts of the three SNPs analyzed in the first follow-up phase. No evidence of association was observed for rs924080. Despite an initial association of rs3790566 and rs3790567, after performing Bonferroni multiple test correction only the association of rs3790567 remained significant (Table 2). The pooled analysis of this genetic variant in the GWAS cohort and the independent follow-up set reached a notable statistically significant association [PMH= 5.19 × 10−7 OR = 1.16 (1.09–1.22), Table 3].
Table 2.
Genotype and allele distribution of IL12RB2 genetic variants in the European SSc patients and controls follow-up study (3344 SSc/3848 controls)
| SNP | 1/2 | CTRL | SSc | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1/1 (n) | 1/2 (n) | 2/2 (n) | MAF | 1/1 (n) | 1/2 (n) | 2/2 (n) | MAF | PMH | OR | 95% CI | PBonf | PBD | ||
| rs924080 | C/T | 0.22 (827) | 0.48 (1807) | 0.29 (1094) | 0.46 | 0.22 (687) | 0.49 (1545) | 0.30 (934) | 0.46 | 0.96 | 1.00 | 0.93–1.07 | 1 | NS |
| rs3790566 | T/C | 0.08 (280) | 0.37 (1334) | 0.55 (1978) | 0.26 | 0.09 (273) | 0.39 (1196) | 0.52 (1617) | 0.28 | 3.35 × 10−2 | 1.09 | 1.01–1.18 | 0.10 | NS |
| rs3790567 | A/G | 0.08 (241) | 0.37 (1169) | 0.56 (1773) | 0.26 | 0.09 (282) | 0.38 (1187) | 0.52 (1616) | 0.28 | 4.84 × 10−3 | 1.12 | 1.04–1.22 | 0.01 | NS |
SSc, systemic sclerosis patients; CTRL, healthy controls; 1/2, minor allele/major allele; MAF, minor allele frequency; PMH, allelic Mantel–Haenszel fixed effects model P-value; OR, odds ratio; 95% CI, 95% confidence interval; PBD, Breslow–Day test P-value; NS, not statistically significant.
Table 3.
Genotype and allele distribution of IL12RB2 rs3790567 genetic variant in SSc patients and controls in a three-step association study
| CTRL | SSc | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Population (CTRL/SSc) | AA (n) | AG (n) | GG (n) | MAF | AA (n) | AG (n) | GG (n) | MAF | PMH | OR | 95% CI | PBD |
| GWAS cohort (5161/2309) | 0.06 (332) | 0.37 (1911) | 0.57 (2918) | 0.25 | 0.08 (196) | 0.40 (919) | 0.52 (1194) | 0.28 | 1.92 × 10−5 | 1.19 | 1.10–1.29 | NS |
| European follow-up (3183/3085) | 0.08 (241) | 0.37 (1169) | 0.56 (1773) | 0.26 | 0.09 (282) | 0.38 (1187) | 0.52 (1161) | 0.28 | 4.84 × 10−3 | 1.12 | 1.04–1.22 | NS |
| GWAS + European follow-up (8344/5394) | 0.07 (573) | 0.37 (3080) | 0.56 (4691) | 0.25 | 0.09 (478) | 0.39 (2106) | 0.52 (2810) | 0.28 | 5.19 × 10−7 | 1.16 | 1.09–1.22 | NS |
| US follow-up (1139/597) | 0.05 (60) | 0.37 (417) | 0.58 (662) | 0.24 | 0.10 (59) | 0.39 (231) | 0.51 (307) | 0.29 | a2.82 × 10−4 | 1.34 | 1.14–1.57 | NA |
| GWAS + European + US follow-up (9483/5991) | 0.07 (633) | 0.37 (3497) | 0.56 (5353) | 0.25 | 0.09 (537) | 0.39 (2337) | 0.52 (3117) | 0.28 | 2.82 × 10−9 | 1.17 | 1.11–1.24 | NS |
Controls are used as reference for all comparisons. CTRL, healthy controls; SSc, systemic sclerosis; MAF, minor allele (A) frequency; PMH, allelic Mantel–Haenszel fixed effects model P-value; aallelic Chi-square uncorrected P-value; OR, odds ratio; 95% CI, 95% confidence interval; PBD, Breslow–Day test P-value; NS, not statistically significant; NA, not applicable.
The subgroup and autoantibody titer stratified pooled analyses comprising the GWAS and the European follow-up cohorts showed firm statistically significant risk association signals in all the subgroups of the disease considered (Supplementary Material, Tables S1–S2).
US follow-up phase
In order to confirm the rs3790567 signal, an independent US cohort was included (597 SSc and 1139 controls). Case–control frequency analysis revealed a strong association [Pχ2= 2.82 × 10−4 OR = 1.34 (1.14–1.57), Table 3]. After stratification, only lcSSc subgroup reached statistical significance, probably due to a lack of power since the other subgroups are relatively smaller (Supplementary Material, Tables S1–S2).
The overall pooled analysis of rs3790567 comprising the GWAS set and both the European and the US follow-up sets reached GWAS-level statistically significant association in the whole set of SSc patients [PMH= 2.82 × 10−9 OR = 1.17 (1.11–1.24)] and remained significant after stratification in all the subgroups (Table 3 and Supplementary Material, Tables S1–S2). Hence, we suggest that the association found in rs3790567 most likely belonged to the whole SSc set of patients rather than any of its subgroups. The rs3790567 individual population allele distributions and association tests are shown in Supplementary Material, Tables S3–S5.
IL23R locus dependence analysis
Aiming to further confirm the independence of the reported IL12RB2 signal from the IL23R locus, we analyzed the association of the SNPs in the IL23R region which were included in the GWAS initial phase and their effect on the IL12RB2 rs3790567 association. The IL23R region comprised 27 SNPs and only 4 of them showed some marginal association with SSc, considering uncorrected P-values (Supplementary Material, Table S7). Nevertheless, the association observed in rs3790567 was found independent of these weak signals (Supplementary Material, Table S7).
DISCUSSION
Our data clearly support an association of IL12RB2 rs3790567 with SSc. The risk effect of the IL12RB2 rs3790567 minor allele is consistent in all the analyzed cohorts with the exception of the Italian population. In contrast to other cohorts, the minor allele rs3790567*A is over-represented in controls compared with SSc patients in the Italian sample set. The Italian control group showed the highest minor allele frequency among all the included populations, and the linkage disequilibrium between rs3790567 and rs3790566 in the Italian cohort was considerably lower (r2= 0.70) than in the other European populations (r2> 0.90). In addition, this over-representation of the rs3790567*A minor allele is also observed in the TSI (Tuscan in Italy) population in the HapMap Project (Phase III) (MAFTSI= 0.30) when compared with the CEU population (MAFCEU= 0.26). However, the linkage disequilibrium observed between rs3790567 and rs3790566 in the Hapmap TSI population compared with the CEU population decreased very slightly (r2TSI= 0.97, r2CEU= 1). Hence, it is likely that the observed discrepancies in the Italian set were due to ethnic differences in linkage disequilibrium patterns. Supporting this notion, BD test revealed significant heterogeneity in the lcSSc overall pooled analysis caused by the Italian patients (PBD with the Italian population = 0.04; PBD without the Italian population = 0.45). Although cases and controls were geographically matched, the potential effect of population substructure in the replication cohorts could not be controlled by deriving principal components on a population-specific basis, as it was performed for the GWAS cohorts, due to the lack of high-throughput genotype information for these individuals. Considering the reported heterogeneous genetic background for Italian populations (20), the influence of this factor on the deviation observed in our Italian subset cannot be ignored.
As stated above, different IL12RB2 genetics variants have been associated with multiple autoimmune disorders (15–19). However, the fact that the same IL12RB2 variant, rs3790567, has been associated with increased susceptibility to both PBC and GCA (16,19), together with the lack of association in our data of a nearby highly linked variant (rs3790566), suggest that rs3790567 intronic SNP may be tagging a functional variant or even has a yet unknown functional implication itself.
The IL12RB2 gene maps close to the IL-23R coding gene (IL23R), which are located <50 kb from each other. IL-23R binds IL-12Rβ1 chain constituting the heterodimeric receptor for IL-23 (21). Although IL23R polymorphisms have been associated with different autoimmune diseases (22–28), its implication in SSc is not clear (29–31). In this report, conditional regression analyses showed that the association of IL12RB2 rs3790567 with SSc is independent from all the studied IL23R genetic polymorphisms, even from IL23R rs11209026 (Arg281Gln) missense variant. Hence, we suggest that the reported association of the IL12RB2 gene with SSc susceptibility does not rely on the IL23R locus. Nevertheless, further studies will be necessary to investigate the possible effect of IL12RB2 genetic variants on IL23R gene expression.
IL-12 levels are increased in the serum of SSc patients as well as in the alveolar lavage fluid (BAL-f) from patients with SSc-associated interstitial lung disease (ILD) (32,33). Although IL-12 classical implication in immune imbalance has been mainly related to a pro-inflammatory cell-mediated immunity and Th1 response (9) and increased levels of IL-12 correlate with renal vascular damage (32), the role of IL-12 in SSc pathogenesis should be considered cautiously. Indeed, SSc patients and especially those with ILD have a Th2-polarized response (34). Additionally, it has been suggested that IL-12 drives a drift from a Th2 to Th1 response which improves skin score in SSc patients (35). Moreover, IL-12 is known to have anti-fibrotic effects in fibroblasts (36), and the administration of IL-12 coding plasmid to the tight skin SSc mouse model prevents collagen accumulation in the skin (37). On the other hand, the implication IL-12Rβ2 in autoimmune events seems to be complex as well. For instance, IL12rb2 knock-out mice do not display IL-12-mediated NK cytotoxicity (38) and the IL-12/IL-12Rβ2 axis is known to be critical for the generation of Th1 autoreactive cells (39), but, despite this, these mice develop spontaneous autoimmune pathology (immune-complex glomerulonephritis) and B-cell tumors by a strong IL-6 up-regulation (14,40). In addition, IL-12R signals predominantly through the STAT pathway, especially STAT4 (37,40). In this regard, it should be noted that polymorphisms in the STAT4 gene are well-established risk factors for SSc (4). Hence, it is likely that genetically predisposed individuals may present subtle differences in IL-12 signaling pathway regulation that could influence the prognosis of SSc.
To date, only a few SSc-related loci have reached a GWAS-level significance (i.e. P-value < 5.00 × 10−8), both in the previously mentioned GWAS and recent studies: the HLA region, STAT4, TNPO3-IRF5, CD247, PSORS1C1, TNIP1 and IRF8 (5,41,42). Hence, we consider that the reported GWAS-level significant association may firmly contribute to the genetic knowledge of the disease.
In conclusion, we report for the first time the association of an IL12RB2 genetic variant with SSc. Our data together with previous reports identify IL12RB2 as a common genetic risk factor for autoimmunity.
MATERIALS AND METHODS
Subjects
The GWAS cohort was comprised of 2309 SSc patients and 5161 controls of Caucasian ancestry from Spain, Germany, The Netherlands and USA from a previously published study (5). The first follow-up phase consisted of 3085 SSc patients and 3183 controls from seven European Caucasian cohorts (Spain, Germany, The Netherlands, Italy, Sweden, UK and Norway). The second follow-up step comprised 1736 additional USA Caucasian individuals (597 SSc and 1139 controls). All the patients fulfilled the 1980 American College of Rheumatology (ACR) classification criteria for SSc (43) or the criteria proposed for early-SSc (44). In addition, patients were classified as having lcSSc or dcSSc as described in LeRoy et al. (3).
The following clinical data were collected for the ascertainment of the clinical phenotype of SSc patients: age, gender and presence of SSc-specific autoantibodies (Ab) ATA and ACA (Supplementary Material, Table S6). The control population consisted of unrelated healthy individuals recruited in the same geographical regions as SSc patients and matched by age, sex and ethnicity with the SSc patients groups.
The study was approved by local ethical committees from all the participating centers. Both patients and controls were included in the study after written informed consent. DNA from patients and controls were obtained using standard methods.
SNP selection
In the screening GWAS phase, we included a 116 kb region spanning the IL12RB2 region and ~13 kb upstream and downstream from this locus, from base pair 67 530 000 to 67 646 000 in chromosome 1, in the GWAS cohorts. After QC filtering as described in Radstake et al. (5), genotyping data for 10 SNPs over this region on chromosome 1 were available. The same procedure was applied for the analysis of the IL23R region, which comprised 163 kb and 27 SNPs.
TaqMan SNP genotyping of the follow-up cohorts was performed in a 7900HT Real-Time Polymerase Chain Reaction (PCR) System from Applied Biosystems following the manufacturer's suggestions (Foster City, CA, USA).
Statistical analysis
Significance was calculated using 2 × 2 contingency tables and Fisher's exact test or χ2 when necessary, to obtain P-values, OR and 95% confidence intervals using PLINK (v1.07) software (http://pngu.mgh.harvard.edu/purcell/plink/). P-values below 0.05 were considered statistically significant. Bonferroni correction and GC as described in Radstake et al. (5) were applied. The Hardy–Weinberg equilibrium (HWE) was tested for all the SNPs comparing the observed genotype distribution in controls with the expected genotype distribution under HWE by means of Fisher's exact test or χ2 when necessary as described in Radstake et al. (5). The logistic regression and conditioned logistic regression analyses (considering the different cohorts as covariables) were performed using PLINK software. Linkage disequilibrium patterns across the region in the HapMap Project Phase I and II (CEU population) defined the haplotype-tagging SNPs using Haploview (v.4.2) software (http://www.broadinstitute.org/haploview/haploview). The SNPs included in the GWAS phase were forced-included in the list of SNPs. Over this region on chromosome 1, the recombination rate was estimated from HapMap public database using LocusZoom (v.1.1) software (http://csg.sph.umich.edu/locuszoom/) (45). SNP & Variation Suite Version 7.5.1 (Golden Helix Inc.) and LocusZoom software were used for the composition of Figure 1. Cochran–Mantel–Haenszel meta-analysis was performed to control for the differences among populations as implemented in PLINK software. In addition, the Breslow–Day test (BD test) was performed as implemented in PLINK in each meta-analysis to assess the homogeneity of the association among populations. The power of the whole set of SSc patients and controls reached 100%. Power was calculated using the software Power Calculator for Genetic Studies 2006 (46) and assuming an additive model (P-value = 0.01 OR = 1.20). The genotyping success call rate in the GWAS cohort was of 99.8%, while in the replication set was over 95%.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by the following grants: J.M. was funded by GEN-FER from the Spanish Society of Rheumatology, SAF2009-11110 from the Spanish Ministry of Science, CTS-4977 from Junta de Andalucía, Spain, in part by Redes Temáticas de Investigación Cooperativa Sanitaria Program, RD08/0075 (RIER) from Instituto de Salud Carlos III (ISCIII), Spain and by Fondo Europeo de Desarrollo Regional (FEDER). T.R.D.J.R. was funded by the VIDI laureate from the Dutch Association of Research (NWO) and Dutch Arthritis Foundation (National Reumafonds). J.M. and T.R.D.J.R. were sponsored by the Orphan Disease Program grant from the European League Against Rheumatism (EULAR). B.P.C.K. is supported by the Dutch Diabetes Research Foundation (grant 2008.40.001) and the Dutch Arthritis Foundation (Reumafonds, grant NR 09-1-408). T.W. was granted by DFG WI 1031/6.1 and DFG KFO 250 TP03. N.O. was funded by PI-0590-2010, Consejería de Salud, Junta de Andalucía, Spain. The USA studies were supported by NIH/NIAMS Scleroderma Registry and DNA Repository (N01-AR-0-2251), NIH/NIAMS-RO1- AR055258 and NIH/NIAMS Center of Research Translation in Scleroderma (1P50AR054144) and the Department of Defense Congressionally Directed Medical Research Programs (W81XWH-07-01-0111).
ACKNOWLEDGEMENTS
We thank Sofia Vargas, Sonia García and Gema Robledo for their excellent technical assistance and all the patients and control donors for their essential collaboration. We thank Banco Nacional de ADN (University of Salamanca, Spain) and the Norwegian Bone Marrow Donor Registry who supplied part of the control DNA samples. We are also thankful to EUSTAR (The EULAR Scleroderma Trials and Research group) and the German Network of Systemic Sclerosis for the facilitation of this project.
APPENDIX
Spanish Scleroderma Group: Norberto Ortego-Centeno and Raquel Ríos, Unidad de Enfermedades Sistémicas Autoinmunes, Servicio de Medicina Interna, Hospital Clínico Universitario San Cecilio, Granada; Nuria Navarrete, Servicio de Medicina Interna, Hospital Virgen de las Nieves, Granada; Rosa García Portales, Servicio de Reumatología, Hospital Virgen de la Victoria, Málaga; María Teresa Camps, Servicio de Medicina Interna, Hospital Carlos Haya, Málaga; Antonio Fernández-Nebro, Servicio de Reumatología, Hospital Carlos Haya, Málaga; María F. González-Escribano, Servicio de Inmunología, Hospital Virgen del Rocío, Sevilla; Julio Sánchez-Román and Mª Jesús Castillo, Servicio de Medicina Interna, Hospital Virgen del Rocío, Sevilla; Mª Ángeles Aguirre and Inmaculada Gómez-Gracia, Servicio de Reumatología, Hospital Reina Sofía, Córdoba; Benjamín Fernández-Gutiérrez and Luis Rodríguez-Rodríguez, Servicio de Reumatología, Hospital Clínico San Carlos, Madrid; Esther Vicente, Servicio de Reumatología, Hospital La Princesa, Madrid; José Luis Andreu, Servicio de Reumatología, Hospital Puerta del Hierro, Madrid; Paloma García de la Peña, Servicio de Reumatología, Hospital Madrid Norte Sanchinarro, Madrid; Francisco Javier López-Longo and Lina Martínez, Servicio de Reumatología, Hospital General Universitario Gregorio Marañón, Madrid; Vicente Fonollosa, Servicio de Medicina Interna, Hospital Valle de Hebrón, Barcelona; Gerard Espinosa, Servicio de Medicina Interna, Hospital Clinic, Barcelona; Carlos Tolosa, Servicio de Medicina Interna, Hospital Parc Tauli, Sabadell; Anna Pros, Servicio de Reumatología, Hospital Del Mar, Barcelona; Mónica Rodríguez Carballeira, Servicio de Medicina Interna, Hospital Universitari Mútua Terrasa, Barcelona; Francisco Javier Narváez, Servicio de Reumatología, Hospital Universitari de Bellvitge, Barcelona; Miguel Ángel González-Gay, Servicio de Reumatología, Hospital Universitario Marqués de Valdecilla, Santander; Bernardino Díaz, Luis Trapiella and María Gallego, Servicio de Medicina Interna, Hospital Central de Asturias, Oviedo; María del Carmen Freire and Inés Vaqueiro, Unidad de Trombosis y Vasculitis, Servicio de Medicina Interna, Hospital Xeral-Complexo Hospitalario Universitario de Vigo, Vigo; María Victoria Egurbide, Servicio de Medicina Interna, Hospital de Cruces, Barakaldo; Luis Sáez-Comet, Unidad de Enfermedades Autoinmunes Sistémicas, Servicio de Medicina Interna, Hospital Universitario Miguel Servet, Zaragoza; Federico Díaz and Vanesa Hernández, Servicio de Reumatología, Hospital Universitario de Canarias, Tenerife; Emma Beltrán and Servicio de Reumatología, Hospital del Doctor Peset Aleixandre, Valencia; José Andrés Román-Ivorra, Servicio de Reumatología, Hospital Universitari i Politecnic La Fe, Valencia.
REFERENCES
Articles from Human Molecular Genetics are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/hmg/ddr522
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/hmg/article-pdf/21/4/926/17257701/ddr522.pdf
Citations & impact
Impact metrics
Citations of article over time
Article citations
Immunogenetics of Systemic Sclerosis.
Genes (Basel), 15(5):586, 05 May 2024
Cited by: 0 articles | PMID: 38790215 | PMCID: PMC11121022
Review Free full text in Europe PMC
TYK2: an emerging therapeutic target in rheumatic disease.
Nat Rev Rheumatol, 20(4):232-240, 11 Mar 2024
Cited by: 2 articles | PMID: 38467779
Review
The Immunogenetics of Systemic Sclerosis.
Adv Exp Med Biol, 1367:259-298, 01 Jan 2022
Cited by: 4 articles | PMID: 35286699
Updates on genetics in systemic sclerosis.
Inflamm Regen, 41(1):17, 15 Jun 2021
Cited by: 14 articles | PMID: 34130729 | PMCID: PMC8204536
Review Free full text in Europe PMC
Pathogenesis and Function of Interleukin-35 in Rheumatoid Arthritis.
Front Pharmacol, 12:655114, 13 May 2021
Cited by: 10 articles | PMID: 34054534 | PMCID: PMC8155723
Review Free full text in Europe PMC
Go to all (57) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
SNPs (Showing 10 of 10)
- (16 citations) dbSNP - rs3790567
- (3 citations) dbSNP - rs3790566
- (2 citations) dbSNP - rs924080
- (1 citation) dbSNP - rs10489627
- (1 citation) dbSNP - rs3828069
- (1 citation) dbSNP - rs3790558
- (1 citation) dbSNP - rs7555183
- (1 citation) dbSNP - rs11209026
- (1 citation) dbSNP - rs2066445
- (1 citation) dbSNP - rs2270614
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Behçet's disease risk association fine-mapped on the IL23R-IL12RB2 intergenic region in Koreans.
Arthritis Res Ther, 19(1):227, 10 Oct 2017
Cited by: 12 articles | PMID: 29017598 | PMCID: PMC5633897
Identification of CSK as a systemic sclerosis genetic risk factor through Genome Wide Association Study follow-up.
Hum Mol Genet, 21(12):2825-2835, 09 Mar 2012
Cited by: 62 articles | PMID: 22407130 | PMCID: PMC3368627
Association study of ITGAM, ITGAX, and CD58 autoimmune risk loci in systemic sclerosis: results from 2 large European Caucasian cohorts.
J Rheumatol, 38(6):1033-1038, 01 Mar 2011
Cited by: 20 articles | PMID: 21362770 | PMCID: PMC3404507
Transethnic meta-analysis identifies GSDMA and PRDM1 as susceptibility genes to systemic sclerosis.
Ann Rheum Dis, 76(6):1150-1158, 17 Mar 2017
Cited by: 54 articles | PMID: 28314753 | PMCID: PMC6733404
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NIAMS NIH HHS (6)
Grant ID: R01-AR055258
Grant ID: R01 AR055258
Grant ID: 1P50AR054144
Grant ID: N01 AR002251-013
Grant ID: P50 AR054144
Grant ID: N01-AR-0-2251