Abstract
Free full text

Mouse STAT2 Restricts Early Dengue Virus Replication
Summary
Dengue virus encodes several interferon antagonists. Among these the NS5 protein binds STAT2, a necessary component of the type-I interferon signaling pathway, and targets it for degradation. We now demonstrate that the ability of dengue NS5 to associate with and degrade STAT2 is species specific. Thus, NS5 is able to bind and degrade human STAT2 but not mouse STAT2. This difference was exploited to demonstrate, absent manipulation of the viral genome, that NS5 mediated IFN antagonism is essential for efficient virus replication. Moreover, we demonstrate that differences in NS5 mediated binding and degradation between human and mouse STAT2 maps to a region within the STAT2 coiled-coil domain. By using STAT2−/− mice, we also demonstrate that mouse STAT2 restricts early dengue virus replication in vivo. These results suggest that overcoming this restriction through transgenic mouse technology may help in the development of a long-sought immune-competent mouse model of dengue virus infection.
Introduction
Dengue virus (DENV) exists in four serotypes (DENV1-DENV4) and is grouped in the flavivirus genus along with a number of additional human pathogens including Yellow Fever Virus (YFV), West Nile Virus (WNV), Japanese Encephalitis Virus (JEV) and Tick-Borne Encephalitis Virus (TBEV) (Kuno et al., 1998). These viruses have a positive strand, non-segmented genome of ~11Kb, the organization of which is highly conserved; encoding from 5′ to 3′, three structural proteins (C, M, and E) followed by seven non-structural proteins (NS1, NS2a, NS2b, NS3, NS4a, NS4b and NS5) (Cleaves and Dubin, 1979; Wengler et al., 1978). The genome is translated as a single endoplasmic reticulum (ER) bound polyprotein, which is co and post-translationally processed by both the viral (NS2b-3 protease) and cellular proteases (Coia et al., 1988; Rice et al., 1985). The virus and its arthropod vector, Aedes aegypti is endemic in over 100 countries around the world including the southern United States (Graham, 1903; Gubler, 1998).
DENV infection, when symptomatic, can result in one of three diseases; dengue fever, (DF), dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS) according to the severity of the symptoms presented (Ashburn and Craig, 2004). In the case of DF, patients suffer a mild febrile illness that includes headache and joint pain. DHF symptoms include those of DF plus signs of hemorrhaging, thrombocytopenia and plasma leakage. Without proper care, DHF can progress into potentially fatal DSS, characterized by hypovolemic shock (Kabra et al., 1999). An estimated fifty to one hundred million DENV infections occur annually, resulting in over 24,000 deaths-predominantly children under 14 years of age (Halstead, 1998).
In spite of its global health impact, there is currently no vaccine or effective anti-viral therapeutic available for DENV (Sampath and Padmanabhan, 2009; Whitehead et al., 2007). One of the primary obstacles to developing such a tool is the lack of robust animal models in which efficacy of a given vaccine or drug can be tested prior to its administration in humans (Chaturvedi et al., 2005). Mouse models have proven useful in this respect for many human viral pathogens including influenza, SARS and Ebola virus (Halfmann et al., 2009; Hu et al., 2009; van der Laan et al., 2008). In addition, mice provide a convenient system for study due to their relative small size, inexpensive maintenance costs and the extensive array of mouse specific genetic tools and reagents available.
Difficulties in developing mouse models for DENV infection result mostly from the animal’s high resistance to viral infection, manifested by a transient low viremia even after high dose challenges (reviewed in (Yauch and Shresta, 2008)). Several studies have elucidated the critical role of Type-I Interferon (IFN) in mediating this resistance. Specifically, these studies have shown that mice deficient in Type-I IFNα/β receptor (IFNAR) or in Signal Transducer and Activator of Transcription 1 (STAT1) expression are compromised in their ability to clear DENV at early time points, exhibiting detectable viral load in the serum at 24 hours post-infection (hpi) for STAT1−/− mice and up to 72hpi in the IFNAR−/− mice. Thus, the type-I IFN pathway is necessary for viral clearance at these early steps. By way of comparison, IFNGR1−/−, mice which are IFNα/β signaling competent but lack the Type-II IFNγ receptor (IFNGR) remain non-viremic upon DENV challenge. However, enhanced morbidity and mortality can be achieved by infecting mice that are deficient for both IFNAR and IFNGR (AG129 mice), indicating a greater role for the type-II IFN pathway at later stages post-infection (Shresta et al., 2004b; Shresta et al., 2005). Though valuable insight has been obtained from these mouse strains, their immune-deficiencies limit the scope of questions that can be addressed, including questions on the efficacy of vaccines and therapeutics.
In vertebrates, the Type-I IFN pathway is a critical component of the antiviral response. Cellular proteins that contain Pattern Recognition Receptors (PRRs) bind to virus specific components termed Pathogen Associated Molecular Patterns (PAMPs). This results in activation of IFNα/β production and eventual IFNα/β secretion from the PAMP containing cell (Kawai and Akira, 2007). The secreted IFN then binds to the IFNAR in a paracrine and autocrine fashion, thus activating the IFN signaling pathway (Cleary et al., 1994; Novick et al., 1994). Receptor binding stimulates activation of the Janus Kinases Jak1 and Tyk2 which associate with the cytoplasmic tail of the IFNAR receptor (Colamonici et al., 1995; Domanski et al., 1997). These kinases in turn phosphorylate the STAT1 and STAT2 proteins (Greenlund et al., 1995; Gupta et al., 1996; Qureshi et al., 1995; Shuai et al., 1994; Shuai et al., 1993). Phosphorylated STAT1 and STAT2 form a heterodimer and when subsequently bound to Interferon Regulatory Factor 9 (IRF9) form the transcription factor complex Interferon Stimulated Gene Factor 3 (ISGF3) (Fu et al., 1990; Kessler et al., 1988). ISGF3 then translocates into the nucleus where it binds to promoter regions termed Interferon Stimulated Response Elements (ISREs) and induces transcription of IFN Stimulated Genes or ISGs. There are over a hundred ISGs identified thus far, which function to create an antiviral state within a cell (Aebi et al., 1989; Clemens and Williams, 1978; Pavlovic et al., 1990; Samuel, 1981).
DENV encodes several proteins which block the IFN production or IFN signaling capabilities of the infected cell. These include the NS2b-3 protease which limits IFN production (Rodriguez-Madoz JR et al., 2010) and the NS2a, NS4a and NS4b proteins which can independently inhibit the IFN signaling pathway to some extent, with NS4b being sufficient to block STAT1 phosphorylation when over-expressed (Munoz-Jordan et al., 2003). In addition, the NS5 protein was demonstrated to bind to STAT2 and target it for degradation through as yet unidentified mechanisms (Ashour et al., 2009; Mazzon et al., 2009). This loss of STAT2 protein recapitulates what is observed during dengue infection (Ashour et al., 2009; Jones et al., 2005). Curiously, the degradation activity but not the binding function of NS5 requires that NS5 be expressed within the context of a polyprotein which undergoes proteolytic processing for its maturation. This manner of expression mirrors closely how NS5 is translated and matured during infection, though it remains unclear why it results in a gain of function for this viral protein.
The STAT protein family consists of seven members. Of the seven, only STAT1 and STAT2 are critical for the IFN pathway. STAT domain organization is conserved and includes an N-terminal domain, a coiled-coil domain, an DNA binding domain, an SH2 domain, and a trans-activation domain. With the exception of STAT2, the protein sequence of the STATs is highly conserved between mouse and human, ranging from 85% identity for STAT6 to 98% for STAT3. In comparison, STAT2 sequence identity between these two species is only 68% (Park et al., 1999; Paulson et al., 1999). STAT2 is frequently targeted for degradation by viral proteins including the V protein of hPIV2 and the NS1 protein of RSV (Andrejeva et al., 2002; Lo et al., 2005; Parisien et al., 2001).
In this study we show that DENV NS5 is able to bind and degrade human STAT2 (hSTAT2) in both human and mouse cells, but is unable to bind or degrade mouse STAT2 (mSTAT2) in either cell type. Furthermore, we show that DENV is sensitive to the action of IFNα/β in mouse cells, but not in human cells when treatment occurs after infection, and that this phenotype correlates with the species origin of STAT2. Thus, without any genetic manipulation of the viral genome, we have shown the biological relevance of NS5 mediated IFN antagonism. Finally, we map the ability of NS5 to bind and degrade STAT2 to a region within the hSTAT2 coiled-coil domain. Taken together, we conclude that mSTAT2 is a restriction factor for DENV and that mutating this protein through transgenic technology may result in an improved mouse model for DENV infection.
Results
NS5 of DENV mediates human STAT2 but not mouse STAT2 degradation
Previous data has indicated that DENV is able to degrade hSTAT2 and STAT2 from non-human primates (using Vero cells) (Ashour et al., 2009; Jones et al., 2005). As mice are resistant to DENV infection, and this resistance is dependent on a functioning IFN system, we were interested in determining whether DENV could degrade mouse STAT2 (mSTAT2). To address this question we generated clonal U6A cells (human cells deficient in STAT2 expression derived from the non deficient 2FTGH cell line) stably expressing FLAG tagged versions of either hSTAT2 or mSTAT2. These two cell lines expressed similar levels of FLAG-tagged STAT2 protein and recovered their ability to respond to IFN treatment (relative to the parental U6A cells) as measured by a bio-assay (Park et al., 2003b) utilizing the IFN sensitive Newcastle Disease Virus expressing GFP (NDV-GFP), (Fig. 1A left panels, Figure S1A and S1B). We next infected both these cell lines with DENV and compared the levels of FLAG-tagged STAT2 protein 24hpi (Fig.1B). Virus infection of hSTAT2-FLAG containing cells showed significant reduction in hSTAT2-FLAG levels as compared to hSTAT2-FLAG uninfected cells (Fig. 1B lanes 3 and 4). However, infected mSTAT2-FLAG expressing cells showed no decrease in FLAG-tagged STAT protein indicating DENV is unable to degrade the mouse form of STAT2 (Fig. 1B lanes 5 and 6). In order to rule out this was not a result of cell type differences generated during clonal selection we utilized an NS5 transfection based assay to demonstrate STAT2 degradation. We have shown previously that transfection of NS5 in a form that undergoes proteolytic processing is capable of decreasing levels of over-expressed hSTAT2 in a manner sensitive to proteasome inhibition suggesting this is an active degradation process (Ashour et al., 2009). In this case we utilized an HA-tagged NS5 gene expressed downstream of the viral protease cleavage site which itself is fused to the DENV E protein (E-NS5-HA). This construct is co-transfected with the DENV protease (NS2b-3) to mediate proper proteolytic processing of NS5. U6A cells were co-transfected with E-NS5-HA (or empty plasmid control), NS2b-3-HA, STAT1-GFP, and either hSTAT2-FLAG or mSTAT-FLAG. 24 hours post-transfection we measured the amount of FLAG tagged STAT2 that was present by western blot in cells expressing E-NS5 as compared to the empty plasmid control. As expected, hSTAT2-FLAG transfected cells showed a loss of hSTAT2-FLAG as compared to the empty plasmid control lane (Fig. 1C lane 2 versus lane 1). This decrease was specific for the hSTAT2-FLAG as a similar decrease was not observed in the STAT1-GFP protein which serves as an internal negative control. Cells transfected with mSTAT2-FLAG however showed no similar decrease in mSTAT2-FLAG levels in the presence of E-NS5-HA, as compared to the negative control lane (Fig. 1C lane 4 versus lane 3) consistent with the virus infection data. We therefore conclude that DENV NS5 protein is unable to target mSTAT2-FLAG for degradation.

NS5 mediated STAT2 degradation is species specific and does not require additional species specific host factors. A. 2FTGH, U6A, U6A (hSTAT2-FLAG), U6A (mSTAT2-FLAG), wtMEF’s, STAT2−/− MEF, STAT2−/− (hSTAT2-FLAG) and STAT2−/− (mSTAT2-FLAG) were treated with 100u/mL Type-I IFN. 24hpt, cells were challenged with NDV-GFP. Virus replication levels were recorded 12hpi by live microscopy. B. U6A cells or U6A-hSTAT2-FLAG or U6A-mSTAT2-FLAG cells were infected with DENV at an MOI of 10. 24hpi, cells were lysed and levels of protein analyzed by SDS-PAGE and immune-blotting using antibodies against FLAG, NS5 and Tubulin. C. U6A cells were co-transfected with the given constructs. 24hpt, cells were lysed and levels of protein analyzed by SDS-PAGE and immune-blotting using antibodies against FLAG, GFP, HA and Tubulin. D. wtMEF’s, STAT2−/−, STAT2−/− (-hSTAT2-FLAG) or STAT2−/− (-mSTAT2-FLAG) cells were infected with DENV at an MOI of 10. 24hpi, cells were lysed and levels of protein analyzed by SDS-PAGE and immune-blotting using antibodies against FLAG, NS5, mSTAT2 and Tubulin. E. BHK21 cells were co-transfected with the given constructs. 24hpt, cells were lysed and levels of protein analyzed by SDS-PAGE and immune-blotting using antibodies against FLAG, GFP, HA and Tubulin. F. 2FTGH cells or 2FTGH derivatives (U6A, U3A and U2A) were infected with DENV at an MOI of 10. 24hpi cells were lysed and analyzed by SDS-PAGE and immune-blotting using antibodies against STAT2, NS5 and Tubulin. Asterisk denotes residual signal from immune-blot against STAT2. All western blots and microscopy data are representative of experiments performed several times. Upper arrows indicate FLAG-tagged mSTAT2 expected migration. Lower arrows indicate FLAG-tagged hSTAT2 expected migration. See also Fig. S1.
In order to determine whether STAT2 is the only species specific factor responsible for DENV mediated degradation, we repeated the DENV infections in a murine cell system containing hSTAT2-FLAG or mSTAT2-FLAG. Mouse embryonic fibroblasts (MEFs) derived from STAT2 KO mice were generated to stably express hSTAT2-FLAG or mSTAT2-FLAG in a clonal fashion. Both clonal populations expressed similar levels of STAT2-FLAG, and expression of STAT2-FLAG rescued the ability of these cells to respond to IFN as measured by the NDV-GFP bioassay (Fig. 1A, right panels and Fig. S1A). We next infected these two populations of cells, in addition to wt and STAT2 KO MEFs, with DENV and measured the level of STAT2 and NS5 24hpi. As can be seen in Fig. 1D, levels of endogenous mSTAT2 in the wtMEFs are increased in the presence of DENV (lane 2 versus lane 1). STAT2 is an ISG, and therefore the most likely explanation for this increase is that IFN produced by DENV infection is activating the IFN pathway resulting in increased levels of ISG’s including endogenous mSTAT2. By contrast, levels of hSTAT2-FLAG in MEFs infected with DENV were reduced compared to uninfected cells (Fig. 1D lane 6 versus lane 5), indicating that NS5 mediated degradation of hSTAT2-FLAG does not require additional human factors. Cells expressing mSTAT2-FLAG, upon infection showed levels of mSTAT2-FLAG similar to the uninfected control samples (Fig. 1D lanes 7 and 8). Thus the species specificity responsible for STAT2 degradation observed occurs at the level of STAT2 and is not due to additional host factors. We repeated the transfection based STAT2 degradation assay, this time in the hamster cell line BHK21, in order to determine whether NS5 alone was able to degrade STAT2 without additional human host factors. To mimic expression of NS5 as a precursor (absent the need of NS4b), HA-tagged NS5 was expressed in these experiments as a full length protein fused downstream of ubiquitin (Ub-NS5-HA). This fusion protein was then placed downstream of the signal-peptide containing DENV E protein (E-Ub-NS5-HA) which results in initial ER localization of the polypeptide. De-ubiquitinating proteins (DUBs) cleave at the C-terminus of the ubiquitin sequence, releasing NS5 into the cytosol (Ashour et al., 2009). As a negative control for this experiment we replaced full length NS5 with NS5 containing an N-terminal truncation of 277 residues that results in a loss of hSTAT2-FLAG binding (E-Ub-NS5-HA 278-900) (Ashour et al., 2009). Akin to the previous results, NS5 was able to mediate the loss of over-expressed hSTAT2-FLAG but not of mSTAT2-FLAG (Fig 1E). Similar results were obtained when we co-transfected E-NS5-HA and NS2b-3-HA with either hSTAT2-FLAG pr mSTAT2-FLAG in STAT2−/− MEFs (Fig. S1C).
Finally, we infected human U2A cells (IRF9 deficient) and U3A cells (STAT1 deficient) to determine whether additional factors of the ISGF3 complex were required for DENV mediated hSTAT2 degradation, as is the case with some paramyxoviruses that degrade STAT proteins (Horvath, 2004). Fig. 1F demonstrates that hSTAT2 degradation occurs in the absence of either of these proteins indicating they are not essential for this process.
NS5 associates with hSTAT2 but not mSTAT2
As NS5 associates with hSTAT2 and this association is most likely required for hSTAT2 degradation during infection, we performed immune-precipitation experiments to determine whether NS5-HA was able to associate with mSTAT2-FLAG. We transfected HA-tagged NS5 1-900 and 278-900 constructs into BHK21 cells along with STAT1-FLAG (used as an internal negative control) and either mSTAT2-FLAG or hSTAT2-FLAG. Immune-precipitation against the HA tag of full length NS5 pulled down hSTAT2-FLAG but not mSTAT2-FLAG (Fig. 2A lanes 3 and 4). Of note, levels of hSTAT2-FLAG in the whole cell extract (WCE) of samples co-transfected with E-Ub-NS5-HA 1-900 are lower as compared to samples co-transfected with E-Ub-NS5-HA 278-900, most likely the result of active degradation (Fig. 2A, panel 3, lane 3 versus 1). We confirmed the immune-precipitation result, this time performing the reciprocal immune-precipitation within the context of viral infection. 293T cells were transfected with FLAG tagged STAT1, hSTAT2, mSTAT2 or IRF9. 24hpt, these samples were infected with DENV, and 24hpi we lysed the cells and performed immune-precipitation for the FLAG epitope. As seen in Fig. 2B, NS5 co-precipitated only in the hSTAT2-FLAG transfected samples (lane 2). Of note, levels of NS5 in the WCE of the hSTAT2-FLAG sample are significantly lower than NS5 levels in the remaining three WCE lanes (panel three, lane 2). We speculate that this could be due to the fact that high levels of hSTAT2-FLAG in the cell most likely titrates NS5 away from the replication complex resulting in lower levels of DENV infection. In any case, these results demonstrate that NS5 associates with hSTAT2-FLAG and not mSTAT2-FLAG.
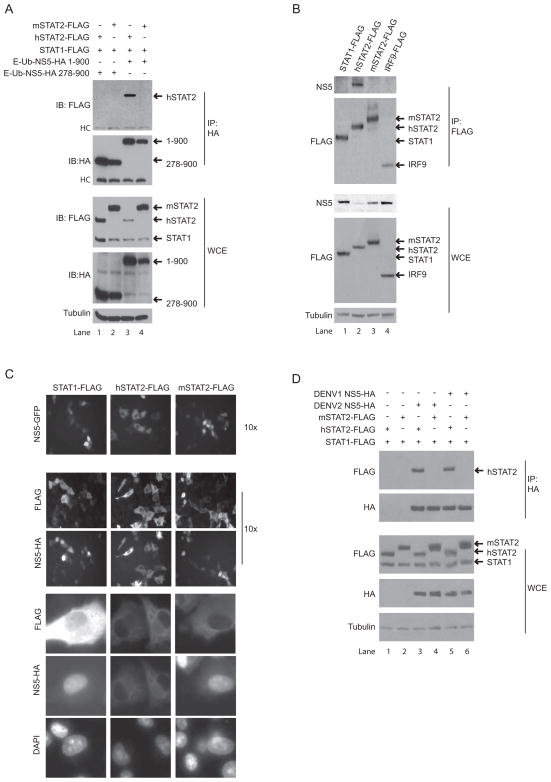
NS5 associates with hSTAT2 but not mSTAT2. A. BHK21 cells were transfected with plasmids encoding for FLAG tagged STAT1 and STAT2, and HA tagged NS5 proteins. Cells were lysed 24hpt and immune-precipitation was performed against the HA epitope. Immune-precipitated samples and the total cell extracts were analyzed by SDS-PAGE and immune-blotting using antibody against FLAG, HA and Tubulin. B. 293T cells were transfected with plasmids encoding for FLAG tagged STAT1, STAT2 and IRF9 proteins. 24hpt, cells were infected with DENV at an MOI of 10. 24hpi, cells were lysed and immune-precipitation was performed against the FLAG epitope. Immune-precipitated samples and the total cell extract were analyzed b SDS-PAGE and immune-blotting using antibodies against FLAG, NS5 and Tubulin. C. Plasmids encoding for STAT1, hSTAT2, mSTAT2 and NS5 proteins were transfected into BHK21 cells. 24hpt, cells were live imaged for GFP fluorescence (top panels) and subsequently fixed for immune-staining using antibody against HA (secondary FITC fluorophore) and FLAG (secondary Texas Red fluorophore) epitopes as well as DAPI stain to visualize DNA. Panels 2 and 3 are 10X images of the cells after fixing and staining for NS5 and the STAT protein. The lower three panels are from the same experiment (though not necessarily from the same field as panels 2 and 3) and have been enlarged for easy visualization of the cytoplasmic and nuclear compartments. D. Same as A. All western blots and microscopy data are representative of experiments performed several times.
Additional evidence of the species specific difference in association between NS5 and STAT2 was observed through analysis of NS5 localization in the presence of over-expressed hSTAT2 or mSTAT2 in BHK cells. In the presence of endogenous levels of hSTAT2, DENV NS5 is predominantly nuclear when over-expressed and during infection (Brooks et al., 2002; Forwood et al., 1999; Kapoor et al., 1995). Given the nuclear localization of NS5 and the predominantly cytoplasmic localization of unstimulated STAT2, we hypothesized that high levels of STAT2 (capable of associating with NS5) may interfere with NS5 nuclear localization. We therefore tested whether differential localization was observed when we co-expressed either hSTAT2 or mSTAT2 with NS5. Fig. 2C demonstrates that whereas NS5-GFP and NS5-HA localize to the nucleus when co-expressed with mSTAT2-FLAG, NS5-GFP and NS5-HA co-expressed with hSTAT2-FLAG localize to the cytoplasm, suggesting that hSTAT2 association with NS5 retains NS5 in the cytoplasm in an over-expression system. STAT1-FLAG was used as a negative control for these experiments as it has been demonstrated to not associate with NS5. Thus, through the use of two independent assays, we conclude that NS5 association with STAT2 is a species specific event.
It is possible that species specificity of NS5 antagonism is unique to DENV2 or to the 16681 virus strain used in the above studies. We therefore examined the ability of NS5 derived from DENV1 (Western Pacific strain) to associate with hSTAT2-FLAG and mSTAT2-FLAG. DENV1 NS5-HA was co-transfected with either hSTAT2-FLAG or mSTAT2-FLAG into BHK21 cells and immune-precipitation was subsequently performed against the HA epitope. Similar to results obtained with DENV2 NS5 (16681) (Fig. 2D lanes 3 and 4), DENV1 NS5 co-precipitated hSTAT2-FLAG but not mSTAT2-FLAG (Fig. 2D lanes 5 and 6). Thus, NS5 species specificity can be extended to at least one other serotype and is not a feature unique to DENV2 16681.
NS5 is unable to block IFN signaling in the presence of mSTAT2-FLAG
Since NS5 is unable to associate with or degrade mSTAT2, we wanted to determine if mSTAT2 expression would restore IFN signaling in NS5 expressing cells. To test this, we performed IFN signaling reporter assays in 293T cells. Increasing amounts of FLAG tagged STAT1, IRF9 and either hSTAT2 (hISGF3) or mSTAT2 (mISGF3) were co-transfected with E-Ub-NS5-HA and the IFN-driven chloramphenicol-acetyltransferase (CAT) reporter plasmid ISRE-54-CAT-GFP. As a negative control, we replaced full length NS5 with E-Ub-NS5-HA 278-900. In addition, we included a truncated form of NS5 (E-Ub-NS5-HA 10-900) that is deficient in degrading hSTAT2 but retains hSTAT2 binding and IFN antagonism activity (Ashour et al., 2009).
ISGF3 dose dependent increases in GFP (Fig. 3A) and CAT (Fig. 3B) activity were observed in both hISGF3 and mISGF3 transfected 293T cells in the negative control E-Ub-NS5-HA 278-900 containing samples. The values on the Y-axis in Fig. 3B represent the reporter activity in ISGF3-FLAG transfected cells treated with IFN divided by the same activity in non ISGF3-FLAG transfected cells (which express endogenous IRF9, STAT1 and STAT2). Fold induction at both the 0.2ng, and 2.0ng amounts were comparable between hISGF3-FLAG and mISGF3-FLAG containing samples indicating that these two complexes were equally efficient in activating ISG transcription. E-Ub-NS5-HA containing samples showed significant inhibition of hISGF3-FLAG activity when levels of hISGF3-FLAG plasmids were co-transfected relative to the E-Ub-NS5-HA 278-900 negative control. E-Ub-NS5-HA 10-900 also significantly inhibited hISGF3-FLAG activity at the 0.2ng 2.0ng and 20ng amounts, although to a lesser extent than full length NS5, indicating that binding to STAT2 inhibits ISGF3 but that degradation further enhances this inhibition. In contrast to the hISGF3-FLAG results, 293T cells transfected with mISGF3-FLAG showed intact signaling activity at all three doses in E-Ub-NS5-HA 10-900 containing samples. Of note, E-Ub-NS5-HA 1-900 co-transfected with the lowest amount of mISGF3 showed some inhibition of reporter activity. This may be due to a present but low affinity of NS5 for mSTAT2 or it may represent the loss of transcriptional activity contributed by the endogenous hSTAT2 (which would be targeted for degradation in the presence of full length NS5). Nevertheless, we conclude that NS5 mediated signaling antagonism is compromised in cells expressing mSTAT2. FLAG tagged STAT1, STAT2 and IRF9 protein levels were undetectable at the 0.2 and 2.0ng level across all samples (Fig. 3C top panels). Notably, we observed transfected-ISGF3-FLAG dose dependent increases in the protein levels of both E-Ub-NS5-HA 10-900 and E-Ub-NS5-HA 1-900 in the hISGF3-FLAG samples (Fig. 3C, second panel, lanes 6–11). In contrast, E-Ub-NS5-HA 1-900 and E-Ub-NS5-HA 10-900 protein levels were decreased in a dose dependent manner in the mISGF3-FLAG transfected samples (Fig. 3C, second panel, lanes 17–22). These opposing effects on expression level were observed only in samples containing an NS5 protein capable of hSTAT2 association since E-Ub-NS5-HA 278-900 protein levels in the hISGF3-FLAG transfections were similar to the mISGF3-FLAG transfections (Fig. 3C, second panel lanes 3–5 vs. 14–17). This may reflect differences in the stability of STAT2 associated vs. non-associated NS5 and/or NS5 stability during IFN signaling activation and will be followed up on in future studies.
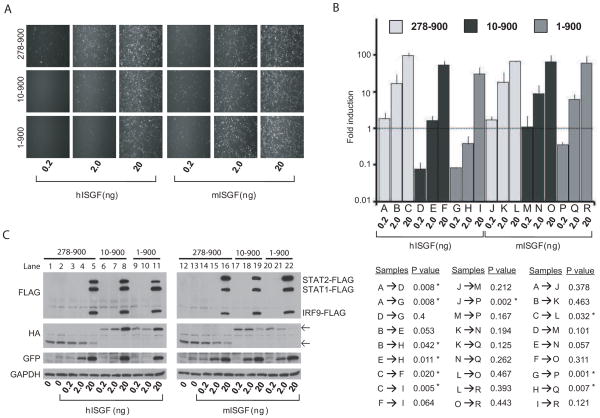
NS5 inhibits hSTAT2 mediated signaling but not mSTAT2 mediated signaling. A. 293T cells were co-transfected with increasing amounts of hISGF3-FLAG (hSTAT2-FLAG, STAT1-FLAG and IRF9-FLAG) or mISGF3-FLAG (mSTAT2-FLAG, STAT1-FLAG and IRF9-FLAG) plus ISRE-CAT-GFP, pCAGGS-Firefly luciferase and either E-Ub-NS5-HA 278-900, E-Ub-NS5-HA 10-900 or E-Ub-NS5-HA 1-900. 24hpt cells were treated with 100u/mL Type-I IFN. Reporter activity (measured by GFP signal) was visualized 48hpt by live microscopy. Results are representative of two independent experiments. B. Cells were transfected and IFN treated same as A. 48hpt, cells were lysed and CAT activity was measured. Fold induction of the ISGF3-FLAG transfected cells is calculated relative to the fold induction observed with ISRE-54-GFP-CAT transfected IFN treated cells versus untreated cells, both in the absence of over-expressed ISGF3 components. This value is set as 1 and symbolized by the dashed horizontal line. Data was obtained from one experiment performed in triplicate using independent sources for plasmids and cells. Bars indicate standard deviation of the samples. P values of the statistical differences calculated between each sample (using unpaired, one-tailed, Student’s T-test), are provided in the bottom panel. Those differences that were found to be significant (p-value ≤ 0.05) between two samples are signified by an asterisk. C) Western blot analysis of one representative experiment. Lanes 1 and 12 represent the control samples that do not over-express ISGF3 components or receive IFN treatment. Lanes 2 and 13 represent the control samples that do not over-express ISGF3 components and receive IFN treatment. Bands corresponding to the predicted molecular weights of NS5 1-900 and 10-900 are indicated by the upper arrow in the second panel. Bands corresponding to the predicted molecular weight of NS5 278-900 are indicated by the lower arrow of the second panel. Antibodies against FLAG, HA, GFP and GAPDH were used.
mSTAT2 expression inhibits virus production in an IFN dependent manner
As NS5 antagonist function is abrogated in cells expressing mSTAT2, we next determined whether mSTAT2 expression would have a deleterious effect on DENV production. For this purpose, we measured virus titers in the supernatant of IFN treated hSTAT2 or mSTAT2 stable expressing cells. These experiments were performed in both a human background (using 2FTGH cells and the STAT2 deficient U6A derivatives) and a mouse background (using WT MEFs and the STAT2 KO derivatives). Infected cells (MOI of 0.1) were treated 12hpi with universal IFN and supernatant was collected 36hpi. We chose these conditions to reflect mainly the antiviral effects of IFN in cells already infected with DENV. The use of universal IFN allowed us to treat both mouse and human cells with the same IFN. IFN treatment of 2FTGH cells resulted in less than 10% inhibition in virus titer as compared to the untreated 2FTGH cells (Fig. 4A, see also Fig. S2 for the virus titers obtained in these experiments). A similar low level of IFN mediated inhibition was observed in the hSTAT2-FLAG stable expressing U6A cells. This is in contrast to cells expressing mSTAT2-FLAG which show an 85% decrease in virus titer after IFN treatment (Fig. 4A). We repeated these experiments using wt MEFs, STAT2 KO MEFs, or MEFs stably expressing hSTAT2-FLAG or mSTAT2-FLAG. Whereas wtMEF cells and mSTAT2-FLAG expressing cells demonstrated 95% and 67% inhibition of virus titer after IFN treatment respectively, inhibition was not observed in the STAT KO cells and hSTAT2-FLAG expressing cells (Fig. 4B). Taken together, these results demonstrate that mSTAT2 but not hSTAT2 inhibits virus replication in an IFN dependent manner. Furthermore, these results suggest that STAT2 degradation is an important step in the virus life cycle and is necessary for optimal virus infection in the presence of the IFN response.
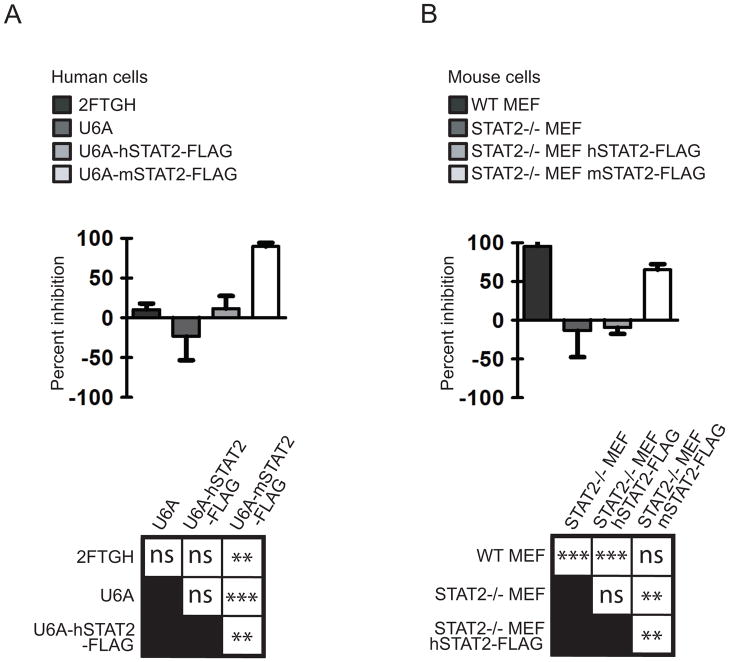
DENV is sensitive to mSTAT2 expression in an IFN dependent manner. A. 2FTGH, U6A, U6A (hSTAT2-FLAG) and U6A (mSTAT2-FLAG) were infected with DENV at an MOI of 0.1. 12hpi, cells were treated with 100u/mL of Type-I IFN. 36hpi, supernatant was collected and levels of virus were measured by titration in Vero cells. Percent inhibition is measured relative to the levels achieved in the same cell line not treated with IFN (see Material and Methods). No virus was detected at the zero hour time point. Data was obtained from three independent experiments and bars indicate standard deviation of the samples. ANOVA results (Bonferroni multiple comparison test) are presented on the lower panel. ns= no significance. ***= extremely significant (p-value <0.001). **= very significant (p-value 0.001–0.01). B. wtMEF’s, STAT2−/− MEF, STAT2−/− (hSTAT2-FLAG) and STAT2−/− (mSTAT2-FLAG) were infected with DENV at an MOI of 0.1 and processed same as in B. No virus was detected at the zero hour time point. Data was obtained from three independent experiments and bars indicate standard deviation of the samples. Statistical analysis same as A. See also Fig. S2.
mSTAT2 is required for early control of DENV replication in vivo and in mouse macrophages
The importance of both the Type-1 IFN receptor and STAT1 in controlling DENV infection in mice has been shown previously (Shresta et al., 2004b; Shresta et al., 2005). In order to determine whether mSTAT2 was also required, we monitored replication of a mouse adapted DENV, D2S10 (Shresta et al., 2006), in infected WT and STAT2−/− mice. Infections were performed by intravenous (Fig. 5A and and5B)5B) or intracranial (Fig. 5C) injection. In the case of intravenous injection, viral RNA was detectable in the lymph node (Fig. 5A) and spleen (Fig. 5B) of WT mice at 8 hours post-infection, but was undetectable by 18 hours post-infection. This in contrast to results obtained in the STAT2−/− mice where we could detect viral RNA in both organs up to 32 hours post-infection. Notably, at 60 hours post-infection, no virus was detected in the spleen or lymph node of the WT or STAT2−/− mice. This suggests that while STAT2 is important for control of virus replication early in infection, it is not required at later time points for clearance of dengue virus from these tissues. Similar results were seen when we infected mice intracranially. Levels of virus in WT mice were low at 24 hours post-infection and were undetectable by 48 hours post-infection. However, we were still able to detect DENV in the STAT2−/− mouse at up to 72 hours post-infection (Fig. 5C). In addition, no death was observed when we monitored survival past seven days of one STAT2-deficient mouse infected through the intravenous route and one STAT2-deficient mouse infected intracranially (data not shown). Finally, we wanted to know whether macrophages, a primary in vivo target cell for DENV (Jessie et al., 2004), also required mSTAT2 for controlling DENV replication. To test this, we infected primary mouse monocyte-derived macrophages from the bone marrow of WT (WT mBMDMs) or STAT2−/− (KO mBMDMs) mice. No virus could be detected in the WT mBMDM samples (Fig. 5D squares). This is in contrast to DENV in the STAT2−/− mBMDMs which reached detectable levels of replication at 24 and 48 hours post-infection (Fig. 5D triangles). These results indicate that mSTAT2 is necessary for early control of DENV replication in vivo and also in macrophages.

mSTAT2 is required for control of DENV production in vivo and in mouse monocyte-derived macrophages. (A) WT and STAT2−/− mice were infected intravenously with 1 x 106 pfu of mouse-adapted DENV2 DS210, 24 hours after an intra-peritoneal injection of enhancing anti-DENV1 antibodies. Lymph nodes were collected at 8, 18, 32 and 60 hours post-infection and viral load determined by qPCR for NS5. Results are from a single experiment (n=3 for each time point) and error bars indicate standard deviation. The P value for the effect of genotype was 0.0001. Limit of detection in this assay was 0.03 copies/mg represented by the dashed line. B) Same as A. Lymph nodes were collected at 8, 18, 32 and 60 hours post-infection and viral load determined by qPCR for NS5. Results are from a single experiment (n=3 for each time point) and error bars indicate standard deviation. The P value for the effect of genotype was 0.0001. Limit of detection in this assay was 0.03 copies/mg represented by the dashed line. (C) WT and STAT2−/− mice were infected intracranially with 2x105 pfu of DS210. Brain tissue was collected at 24, 48 and 72 hours post-infection and viral load determined by plaque assay in BHK21 cells. The limit of detection was 0.05 PFU/mg. Results are from a single experiment (n=5 for 24hpi and 72hpi, n=6 for 48hpi) and error bars indicate standard deviation. The P value for the effect of genotype was 0.0001. D) Mouse bone marrow monocyte-derived macrophages (mBMDMs) from WT and STAT2−/− mice were infected with DENV2 16681 at an MOI of 1. Supernatant was removed at 0, 24 and 48hpi and virus titer was determined. Limit of detection in this assay is 4 Fluorescing Foci Units/mL (FFU/mL) represented by the dashed line. No virus was detected at the zero hour time point. Data was obtained from one infection performed in triplicate and bars indicate standard deviation of the samples. Statistically significant difference (p≤0.05) was determined using Student’s T-test (unpaired, one-tailed). ND= not detected.
NS5 interacts with the coiled coil domain of hSTAT2
To determine which specific region(s) of hSTAT2 is/are targeted by NS5, we first used a functional assay based on an ISRE-CAT-GFP reporter system that is activated by a construct encoding hSTAT2-FLAG fused downstream of IRF9 (IRF9-hSTAT2-FLAG 1-851). This fusion is based on work utilizing a similar fusion protein encoding the hSTAT2 trans-activation domain fused downstream of IRF9, (IRF9-hSTAT2 747-851) (Kraus et al., 2003). The IRF9-hSTAT2-747-851 is constitutively active and does not require IFN treatment for its transcriptional activity (Kraus et al., 2003). We observed that, similar to IRF9-hSTAT2-747-851, transfection of IRF9-hSTAT2-FLAG 1-851 into 293T cells resulted in robust activation of the ISRE reporter (Fig. 6A) in the absence of IFN treatment. Transfection of full length NS5-HA prevents reporter activation based on its ability to bind to the STAT2 portion of the IRF9-hSTAT2 transcription factor. In contrast, the negative control protein Core-HA, failed to inhibit IRF9-hSTAT2 dependent transcriptional activity. We proceeded to make N-terminal truncations of hSTAT2 within the IRF9-hSTAT2 fusion protein to ascertain at which point NS5 would lose the ability to block transcriptional activity. As can be seen in Fig. 6A, NS5-HA is capable of preventing transcriptional activity of hSTAT2-FLAG 100-851. However, this antagonist function is lost when we transfected IRF9 fused to hSTAT2 residues 572–851 or 747–851. This indicates that NS5 requires residues somewhere between hSTAT2 region 100–572 for its antagonism. It is unlikely that the loss of antagonism is due to defective folding of hSTAT2 resulting from large truncations as the fusion protein is still capable of activating transcription and western blot controls show comparable levels of IRF9-hSTAT2 expression in all transfections (Fig. 6B).

Residues found within region 100 to 572 of hSTAT2 are required for NS5 mediated inhibition of STAT2 dependent transcription. A) 293T cells were co-transfected with plasmids encoding an ISRE-54-CAT-GFP reporter, pCAGGS-Firefly luciferase, the IRF9-hSTAT2-FLAG construct stated and either Empty, NS5-HA or Core-HA protein. 24hpt, cells were lysed and analyzed for CAT activity. Percent activation is calculated relative to samples transfected with ISRE-54-CAT-GFP, pCAGGS-Firefly luciferase and empty plasmid with no IRF9-hSTAT2-FLAG construct. Data was obtained from three independent experiments and error bars indicate standard deviation. B) Western blot of one representative experiment from the reporter assay. Lysates were analyzed by SDS-PAGE and immune-blot analysis using antibodies against FLAG, HA, GFP and Tubulin.
Next, to more finely map the area of NS5 interaction on hSTAT2, we generated chimeric STAT2 proteins using hSTAT2 and mSTAT2 sequence. Portions of the N-terminus of mSTAT2-FLAG were replaced with the homologous region of hSTAT2 (h/mSTAT2). These chimeras were then placed into an IP assay with NS5-HA. As expected, immune-precipitation against the FLAG epitope of mSTAT2 failed to pull down NS5-HA (Fig. 7A lane 3). IP of h/mSTAT2 containing the first 181 residues of hSTAT2-FLAG (h/mSTAT2-FLAG 1-181) did not pull down NS5, indicating that the first 181 residues of hSTAT2 were not sufficient for interaction (Fig. 7A lane 4). However, h/mSTAT2-FLAG expressing the first 301 residues of hSTAT2 co-precipitated NS5 (Fig. 7A lane 5). Thus, species specific sequence between residues 181–301 is required for interaction with NS5.
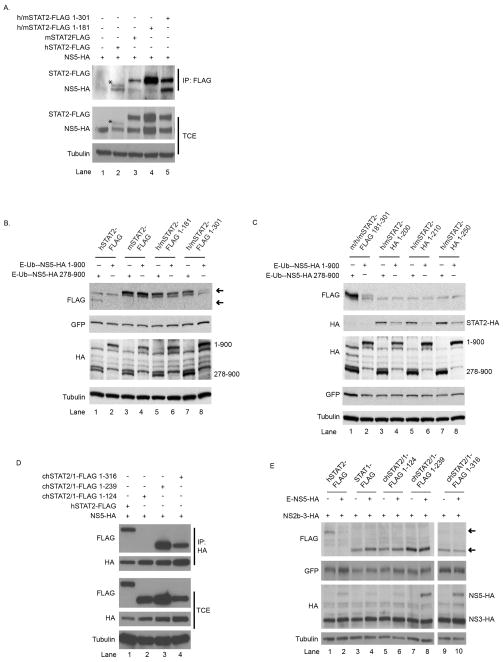
NS5 binding domain maps to the N-terminus of hSTAT2. A. U6A cells were co-transfected with NS5-HA and the FLAG tagged STAT2 constructs stated. The chimeras include N-terminal regions of hSTAT2 (1–181 and 1–301) in place of the mSTAT2 homologous region with the remainder of the downstream sequence derived from mSTAT2. 24hpt, cells were lysed and immune-precipitation performed against the FLAG epitope. Immune-precipitated samples were analyzed by SDS-PAGE and immune-blotting using antibodies against FLAG and HA in the top two panels and Tubulin in the bottom panel. Asterisks denote full length hSTAT2-FLAG which has a faster mobility in SDS-PAGE compared to the h/mSTAT2-FLAG chimeras. B. BHK21 cells were transfected with the stated FLAG-tagged STAT2, STAT1-GFP and NS5-HA-tagged constructs. 24hpt, cells were lysed and protein levels analyzed by SDS-PAGE and immune-blotting using antibodies against FLAG, GFP, HA and Tubulin. Upper arrow indicates expected mobility of mSTAT2-FLAG and chimeras and the lower arrow indicates expected mobility of hSTAT2-FLAG. C. BHK21 cells were transfected with the stated FLAG-tagged STAT2 or HA-tagged STAT2, STAT1-GFP and NS5-HA-tagged constructs. These chimeras include N-terminal regions of hSTAT2 (1–200, 1–210 and 1–250) in place of the mSTAT2 homologous region with the remainder of the downstream sequence derived from mSTAT2. In the case of m/h/mSTAT2-FLAG 181-301, the internal sequence of mSTAT2 has been replaced with the analogous sequence from hSTAT2. 24hpt, cells were lysed and protein levels analyzed by SDS-PAGE and immune-blotting using antibodies against FLAG, GFP, HA and Tubulin. E. BHK21 cells were co-transfected with NS5-HA and FLAG containing STAT constructs. The chimeras include N-terminal regions of hSTAT2 (1–124, 1–239 and 1–316) in place of the STAT1 homologous region with the remainder of the downstream sequence derived from STAT1. 24hpt, cells were lysed and immune-precipitation performed against the HA epitope. Immune-precipitated samples were analyzed by SDS-PAGE and immune-blotting using antibodies against FLAG, HA and Tubulin. F. U6A cells were transfected with the FLAG-tagged STAT2, STAT1-GFP and HA-tagged constructs. The chimeras include N-terminal regions of hSTAT2 (1–124, 1–239 and 1–316) in place of the STAT1 homologous region with the remainder of the downstream sequence derived from STAT1. 24hpt, cells were lysed and protein levels analyzed by SDS-PAGE and immune-blotting using antibodies against FLAG, GFP, HA and Tubulin. Upper arrow indicates expected mobility of hSTAT2-FLAG and lower arrows indicates expected mobility STAT1-FLAG and chimeras. All western blots are representative of experiments performed several times.
We wanted to determine whether the h/mSTAT2-FLAG constructs that could bind to NS5 resulted in them becoming susceptible to NS5 mediated degradation. We therefore placed these chimeras into a STAT2 degradation assay. Cells were co-transfected with the h/mSTAT2 construct and either E-Ub-NS5-1-900 or as a negative control, E-Ub-NS5-278-900. As we expected, mSTAT2-FLAG in this assay showed no detectable changes in protein level upon co-transfection with E-Ub-NS5-1-900-HA as compared to the negative control (Fig 7B lane 4 versus lane 3). Similar results were obtained with h/mSTAT2-FLAG 1-181 (Fig. 7B lane 6 versus lane 5). However, co-transfection of h/mSTAT2-FLAG 1-301 with E-Ub-NS5-HA 1-900 resulted in a significant decrease in h/mSTAT2 (Fig. 7B lanes 8 versus 7). Therefore, the ability of STAT2 to bind NS5 correlates with the susceptibility to NS5 mediated degradation. To more precisely map the hSTAT2 region required for degradation, we constructed additional h/mSTAT2 chimeras and placed them into the degradation assay. We first generated m/h/mSTAT2-FLAG in which the internal mouse STAT2 residues 181–301 were replaced with the analogous human sequence. As can be seen in figure 7C, lanes 2 versus 1, this construct was susceptible to NS5 mediated degradation. Finally, HA-tagged chimeras in which only the first 200, 210 and 250 residues of mSTAT2 were replaced with the analogous human STAT2 sequence were all susceptible to degradation (Fig. 7C lanes 3–8). This data indicates that mSTAT2 and hSTAT2 amino acid differences between region 181 and 200 play a role in NS5 mediated association and degradation of STAT2.
We next generated a separate panel of chimeras where the N-terminal portion of STAT1 was replaced using increasing N-terminal sequence of hSTAT2 (chSTAT2/1). These chSTAT2/1 chimeras were then individually co-transfected with NS5-HA. 24hpt, we performed immune-precipitation against the HA epitope. As expected, NS5-HA co-precipitated with hSTAT2-FLAG (Fig. 7D lane 1). However, chSTAT2/1-FLAG 1-124, which expresses the first 124 residues of hSTAT2 and the remaining 125–751 residues derived from STAT1, failed to be pulled down by NS5-HA (Fig. 7D lane 2). Increasing the amount of N-terminal hSTAT2 sequence so that we express the first 239 or 316 residues (chSTAT2/1-FLAG 1-239, chSTAT2/1-FLAG 1-316), resulted in the ability to co-precipitate the NS5-HA protein (Fig. 7D lanes 3 and 4 respectively). Thus, the mapped region in the h/mSTAT2 chimeras required for binding/degradation (181–200) is consistent with the binding region mapped using the STAT2/1 chimeras (124–239). This falls within the coiled-coil region of hSTAT2 which encompasses residues 138–230 (Martinez-Moczygemba et al., 1997).
We next placed these chSTAT2/1 chimeras into the STAT2 degradation assay to determine whether additional STAT2 sequence outside of this N-terminal region was required for NS5 mediated degradation. Co-transfection of E-NS5-HA, NS2b-3-HA, hSTAT2-FLAG and STAT1-GFP resulted in significant decreases in hSTAT2-FLAG protein level, as compared to the negative control lane where no E-NS5-HA was included (Fig. 7E lane 2 versus lane 1). No similar change was observed in the levels of STAT1-FLAG (Fig. 7E lane 4 versus 3). We next tested the chimeras for their susceptibility to NS5 mediated degradation. ChSTAT2/1-FLAG 1-124 levels were not decreased in the presence of NS5. STAT2/1-FLAG chimeras 1–239 and 1–316 which bind NS5 also failed to be degraded. This suggests that additional hSTAT2 sequence downstream of the first 316 residues is important for STAT2 degradation. These sequences are shared with mSTAT2, as similar h/mSTAT2 chimeras are degraded (Fig. 7B). Overall, our results indicate that the N-terminal region of hSTAT2 is needed for interaction with NS5, but that additional STAT2 specific sequences are required for degradation.
Discussion
In this manuscript, we have demonstrated that DENV NS5 mediated antagonism of the IFN pathway is species specific and is non-functional in the presence of mSTAT2. Whereas NS5 is able to associate with hSTAT2 and target it for degradation in both human and mouse cells, NS5 is incapable of performing either function with respect to mSTAT2 in both cell types. In addition, we have demonstrated that the loss of hSTAT2 expression during DENV infection does not require IRF9 or STAT1. Taken together, these results strongly suggest that species specificity occurs at the level of hSTAT2 and is independent of additional species specific host factors. While additional host factors are most certainly required, their identities remain unknown, though we believe they will most likely include components of the ubiquitin/proteasome degradation pathway. DENV strain variation has been observed with respect to manipulation of the IFN pathway (Umareddy et al., 2008). Importantly, we have shown that the species specific nature of NS5 association with STAT2 is not unique to DENV2 (strain 16681) and can be extended to DENV1. Given the relatively high percentage NS5 sequence identity when comparing DENV2 to the other three serotypes (77% for DENV1, 77% for DENV3 and 72% for DENV4) (Khan et al., 2006), we surmise that our results could extend to DENV3 and DENV4. We note that DENV encodes multiple IFN antagonists including NS2a, NS4a, NS4b and proteolitically active NS2b-3 (Leitmeyer et al., 1999; Munoz-Jordan et al., 2005; Munoz-Jordan et al., 2003; Rodriguez-Madoz JR et al., 2010) and while they were not a focus for this study, their antagonist function may influence species tropism as well.
DENV is not the only virus whose replication is restricted by species dependent differences of STAT2 or the IFN pathway. PIV5, a member of the Paramyxoviridae family, utilizes its V protein to bind STAT2 and the E3 ligase DDB1 (Precious et al., 2005; Ulane and Horvath, 2002). This induces the recruitment of STAT1 which is subsequently targeted for degradation. This pathway functions in cells of human and non-human primate origin but not in murine cells, a result of the inability of the V protein to associate with mSTAT2 (Kraus et al., 2008). Consistent with this, hSTAT2 transgenic knock-in mice showed higher levels of PIV5 replication in vivo (Kraus et al., 2008). In related fashion, the V protein of the paramyxovirus Newcastle Disease Virus (NDV) is able to block IFN in chicken cells but not human cells, correlating with the ability of NDV to grow in these cells. However, replication in human cells is facilitated through swapping the NDV V protein with the influenza NS1 protein, an IFN antagonist that functions in human cells (Park et al., 2003a). Thus, the ability to antagonize the IFN pathway can be a limiting factor in determining species host range.
We were able to successfully exploit the observed species specificity of NS5 antagonism and measure the biological significance of this event on virus replication. While it may be possible to generate mutant virus strains expressing an NS5 protein incapable of associating with or degrading hSTAT2, we were concerned that this would also affect NS5 polymerase activity independent of the IFN antagonist function. Therefore, we generated a series of human and murine cell lines which lack endogenous STAT2 but stably over-express a tagged form of hSTAT2 or mSTAT2. This system allowed us to directly compare the impact of IFN signaling in the presence of an intact mSTAT2 versus a STAT2 capable of being degraded by NS5 (hSTAT2). The results of these experiments, performed in both human and murine cell backgrounds, demonstrated that virus replication was strongly inhibited after IFN treatment only in cells with intact mSTAT2. Thus, NS5 mediated degradation of STAT2 is a crucial event during DENV infection. The ability of DENV to replicate in BMDMs from STAT2 KO mice, but not BMDMs from wild type mice is consistent with mouse STAT2 being a factor which limits DENV replication in murine species.
While the degradation function of NS5 is crucial for efficient virus replication, NS5 association with STAT2 is sufficient for prevention of ISGF3 signal transduction. This activity is most likely due to an NS5 mediated inhibition of STAT2 phosphorylation, as has been reported by Mazzon et al. (Mazzon et al., 2009). We directly compared the IFN antagonist capability of full length NS5 protein which could associate with and degrade hSTAT2 versus a truncated form that could associate with but not degrade hSTAT2. We found that while both forms were capable of inhibiting IFN dependent signaling, the degradation function enhanced the potency of the antagonist function allowing full length NS5 to block signal transduction in the presence of higher levels of hSTAT2. In addition, the fact that mSTAT2 expression was sufficient to block NS5 mediated inhibition in these experiments suggests that STAT2 is the primary target of the IFN antagonist activity of NS5.
The results we have obtained using WT and STAT2−/− mice demonstrate, for the first time, that mSTAT2 is important for efficient control of virus replication in vivo and in primary macrophages. While this conclusion is not surprising, as STAT2 is required for IFN signaling, it is clear from the in vivo experiments that given a cellular environment marked by the absence of mSTAT2, DENV is capable of establishing an infection with detectable viral load up to 72 hours post-infection. Human STAT2 is degraded rapidly upon DENV infection regardless of the species origin of the host cell as demonstrated in this manuscript (Fig. 1B, 1D and 1F) and in previously published data (Ashour et al., 2009). It is likely that a similar STAT2 deficient cellular environment in vivo can be achieved, permitting detectable early levels of DENV replication, after DENV infection in an mSTAT2−/− mouse expressing endogenous levels of hSTAT2. Thus, although the substitution of mSTAT2 by hSTAT2 certainly will not overcome all murine restrictions of DENV replication, especially at late times of infection, it may be a first step towards the development of a mouse model, which would be advantageous as it would facilitate study of in vivo DENV replication in the context of a functioning immune system. It should be noted that the remaining DENV restriction at late times in STAT2−/− mice might be mediated by type-II IFN, as this antiviral factor does not require STAT2 for signaling, and it is known to contribute to decreased disease in DENV-infected mice (Shresta et al., 2004a; Shresta et al., 2004b; Shresta et al., 2005).
Finally, through the generation of STAT chimeras, we identified two separate regions within STAT2 that contain sequences required for NS5 association and NS5 mediated degradation. When sequences derived from the N-terminal portion of hSTAT2 were placed into mSTAT2, it resulted in a chimera that was susceptible to both NS5 association and NS5 mediated degradation. However, when this same region was placed into STAT1, it resulted in NS5 association but the chimera remained resistant to NS5 mediated degradation, therefore indicating additional STAT2 specific sequences towards the C-terminus were required for the degradation step. Thus, two regions within STAT2 are involved in the NS5 dependent degradation pathway. Additional experiments utilizing this chimera approach will refine the sequence required for both activities. It is interesting to note that the coiled-coil domain of STAT2, involved in cellular protein-protein interactions, lies within the area we have mapped for NS5 association. This region, though highly similar between mouse and human, contains several patches of sequence which diverge and remains an attractive target for future mutational analysis. Regardless, based on our results, and on the fact that the first 10 amino acids of NS5 are required for degradation but not association of STAT2 (Ashour et al., 2009), we hypothesize that NS5 binds to the N-terminal region of hSTAT2 and that an as yet unidentified third protein containing E3 ligase activity is recruited to the complex through a motif located at the N-terminus of NS5. This protein would then interact with the C-terminal half of STAT2 resulting in STAT2 ubiquitin conjugation and degradation. Further investigation of the cellular proteins required for STAT2 degradation in the presence of DENV is necessary to test this hypothesis.
Our understanding of DENV pathogenesis has benefited greatly through the use of animal models. Specifically, through utilization of several genetically engineered mouse strains, the critical roles of the type-I and type-II IFN pathways in murine resistance to DENV infection have been elucidated (Shresta et al., 2004b); we note that while both STAT1 and STAT2 activity have been implicated in type-III IFN signaling, it remains to be seen whether the activation of this pathway influences DENV replication (Dumoutier et al., 2003; Kotenko et al., 2003). Nevertheless, while the type-I and type-II IFN deficient mice have proven invaluable with respect to our understanding of some aspects of dengue pathogenesis, their immune-deficiencies limit the scope of questions that can be addressed, particularly those relevant to the development of vaccines and therapeutics. The use of humanized SCID or NOD/SCID mouse strains has overcome this barrier to some degree though this approach requires a degree of skill and time (An et al., 1999; Bente et al., 2005; Blaney et al., 2002; Kuruvilla et al., 2007; Lin et al., 1998; Wu et al., 1995). Therefore, the development of an immune-competent mouse strain that is susceptible to DENV infection is desirable. Given the results obtained in our study, we speculate that initial resistance to DENV in mice results from an inability of NS5 to associate with and target STAT2 for degradation. Therefore, development of a transgenic mouse expressing a functional chimeric STAT2 capable of being degraded by an NS5 dependent mechanism could result in an immune-competent mouse susceptible to early DENV infection. While one would most certainly need to address additional mouse restrictions, especially at late times, such a mouse may represent the first step towards the generation of an immune-competent DENV-susceptible mouse strain that in the future will prove a valuable resource to the DENV field.
Materials and Methods
Cells and viruses
All cell lines used in this work; 293T cells, BHK21 cells, mouse embryonic fibroblasts (MEFs) including wtMEFs, STAT2−/− and stable cell line derivatives, 2FTGH cells and 2FTGH derivatives: U2A (IRF9 deficient), U3A (STAT1 deficient) U6A cells (STAT2 deficent) and U6A stable cell line derivatives were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum. All clonal cell lines derived from the U6A and MEF STAT2−/− cell lines (U6A-STAT2-GFP, U6A-hSTAT2-FLAG, U6A-mSTAT2-FLAG, MEF-STAT2−/− hSTAT2-FLAG, MEF-STAT2−/− mSTAT2-FLAG) were generated by co-transfection of the STAT2 encoding pCAGGS plasmid and pCDNA-zeo and subsequently selecting for zeocin resistance. Recombinant Newcastle Disease Virus (NDV) expressing GFP was grown in 10 day old embryonated chicken eggs. High titer stocks of dengue virus (DENV2 16681) were obtained by passage in C6/36 cells. DENV1 D2S10 was kindly provided by Eva Harris. All transfections were performed using Lipofectamine 2000 (Invitrogen).
Primary monocyte isolation and differentiation
For derivation of BMDMs from wild type and STAT2 knockout mice, cells were isolated from the bone marrow and cultured for 5 days in RPMI 1640, 10%FCS, Pencillin/Streptomyocin and 20% L929 conditioned media.
In vivo DENV infection and quantification from organs
Intravenous delivery of DENV: 24 hours prior to infection, 12 WT C57BL/6 and 12 STAT2−/− C57BL/6 mice were injected intraperitoneally with 200ul anti-DENV1 serum that has previously been shown to enhance DENV2 infection (Balsitis et al., 2010). The mice were then infected with 106 pfu of a mouse-adapted DENV2 strain, D2S10, via tail-vein injection. Organs were harvested and weighed at 8, 18, 32 and 60 hours. The organs were homogenized in RLT buffer (Qiagen, Valencia, CA) and RNA was extracted using the RNeasy® kit (Qiagen) according to the manufacturer’s protocol. DENV2 copy number was measured by the PCR Core Facility of Mount Sinai School of Medicine using a SYBR green qPCR assay and primers against DENV NS5 and three house-keeping genes (rps11, β-actin and α-tubulin). Intracranial delivery of DENV: Sixteen WT C57BL/6 and 16 STAT2−/− C57BL/6 mice were injected intracranially with 105 pfu of D2S10. Their brains were harvested at 24, 48 and 72 hours post infection and then weighed. Tissues were homogenized in DMEM with 10% FBS and 1% penicillin-streptomycin to release virus, and the homogenates were cleared by centrifugation. The viral loads in the cleared homogenates were measured by plaque assay on BHK21 cells. Statistical analysis was performed to analyze the effect of genotype using 2-way ANOVA (Bonferroni post-test). Values were log10 transformed to approximate Gaussian distribution. Values below the limit of detection were substituted for the limit of detection value (for example, a value of 0.025 in Figure 5A would be replaced with a value of 0.03), thus giving a more conservative calculation of p-value.
Dengue virus infection of cell cultures
Unless otherwise stated, all DENV infections were performed with DENV at an MOI of 10 using the DENV2 strain 16681 virus for 1 hour. Cells were then washed and subsequently maintained in DMEM 10% FCS at 37 degrees. 24hpi cells were lysed and analyzed via western blot.
Virus quantification from cell supernatant
DENV titer present in infected cell supernatant was calculated by overlaying 250 μl of 1ml total volume serial dilutions of the supernatant onto Vero cells and incubating for 2 hours at room temperature. Supernatant was then removed and replaced with fresh media containing 1% agar. The infection was then continued for 72 hours at 37° C. Infected cells were then fixed and stained for DNA (using DAPI) and for NS5 protein using anti-NS5 antibody and a secondary anti-rabbit FITC fluorophore. The number of positive foci was counted by eye under a fluorescent microscope. Infectious particles are expressed as fluorescing foci forming units/mL (FFU/mL). The limit of detection value reflects the sensitivity obtained when titrating virus stocks of known quanitity alongside the infected cell supernatants (i.e. 1 FFU is observed with 250μl of virus stock diluted to 4 FFU/mL). To calculate percentage of inhibition of dengue virus replication in IFN treated cells, the FFU/ml obtained in IFN treated samples was divided by the FFU/ml obtained in IFN untreated samples, and this number was subtracted from 1 and expressed as %.
Plasmids and antibodies
All DENV protein encoding plasmids were generated in the pCAGGS (chicken β-actin promoter) background. Primer sequences used in the generation of these constructs are available upon request. Plasmids encoding human STAT1 and STAT2 were a kind gift from Dr. Megan Shaw. Plasmid encoding for human IRF9 was kindly provided by Estanislao Nistal-Villan. ISRE-54-CAT-GFP reporter, pCAGGS-Firefly and luciferase plasmids were kind gifts from Dr. Luis Martinez-Sobrido. pCDNA-zeo was a kind gift from Dr. Ben tenOever. Mouse STAT2 cDNA was kindly provided by Dr. David Levy. Antibodies utilized for this manuscript include those raised against HA (Sigma), FLAG (Sigma), STAT1 (BD), STAT2 (Santa Cruz SC476), mSTAT2 (Santa Cruz SC839), GFP (Sigma), GAPDH (Research Diagnostics Incorporated), and tubulin (Sigma). Rabbit antibody raised against DENV2 NS5 was generated by serial injection of bacterially expressed and purified DEN2 NS5 protein.
IFN reporter assay
293T cells were co-transfected with an HA-tagged plasmid encoding for various viral proteins, the IFN inducible CAT reporter (ISG54-CAT-GFP), and a plasmid constitutively expressing the firefly luciferase protein (pCAGGS-Firefly luciferase). 24 hours post-transfection cells were treated with 100u/mL of u-IFN (universal interferon) (PBL). 24 hours post-treatment cells were visualized for GFP expression or were lysed and measured for CAT activity. Fold induction of the sample is calculated as the CAT activity of the treated sample normalized to the firefly luciferase value of that sample; which is then divided by the normalized value of untreated empty-HA transfected cells. Students T-test (unpaired, one-tail) was utilized for the statistical analysis.
NDV based bioassay
2FTGH, U6A, U6A(hSTAT2-FLAG), U6A(mSTAT2-FLAG), wtMEF’s, STAT2−/−MEF, STAT2−/−(hSTAT2-FLAG) and STAT2−/−(mSTAT2-FLAG) cells were treated with the stated amounts of u-IFN (universal interferon) (PBL). 24 hours post-treatment, cells were challenged with NDV-GFP and subsequent fluorescence images obtained 12 hours post-infection.
Immune-precipitation assays
293T or BHK21 cells were transfected with HA-tagged plasmid encoding for the viral protein and FLAG tagged construct(s). Lysis, immune-precipitation and washes were performed using buffer containing 50mM Tris (pH 7.5), 280mM NaCl, 0.2mM EDTA, 2mM EGTA, 0.5% NP-40, 10% glycerol, 1mM DTT, and 1mM sodium orthovanadate.
STAT2 degradation assays
Analysis of degradation of over-expressed STAT2 was performed in MEF STAT2−/−, BHK21 or U6A cells using viral protein encoding HA-tagged plasmids co-transfected with STAT2-FLAG. STAT1-GFP is also transfected in these experiments and used as a negative control.
Acknowledgments
This work was supported by grant U54 AI57158 (to A.G.-S and Pei-Yong Shi.) and grant 5R01 AI073450 (A F-S) from the National Institutes of Health and by a National Institutes of Health fellowship (to M.L-R.). We thank Megan Shaw, Estanislao Nistal-Villan, Benjamin tenOever, David Levy and Luis Martinez-Sobrido for kindly providing plasmids. We thank Richard Cádagan for excellent technical assistance and Valmas Charalampos and Inma Barasa for helping with the statistics. We thank Domenico Tortorella for his sage advice.
References
- Aebi M, Fah J, Hurt N, Samuel CE, Thomis D, Bazzigher L, Pavlovic J, Haller O, Staeheli P. cDNA structures and regulation of two interferon-induced human Mx proteins. Mol Cell Biol. 1989;9:5062–5072. [Europe PMC free article] [Abstract] [Google Scholar]
- An J, Kimura-Kuroda J, Hirabayashi Y, Yasui K. Development of a novel mouse model for dengue virus infection. Virology. 1999;263:70–77. [Abstract] [Google Scholar]
- Andrejeva J, Young DF, Goodbourn S, Randall RE. Degradation of STAT1 and STAT2 by the V proteins of simian virus 5 and human parainfluenza virus type 2, respectively: consequences for virus replication in the presence of alpha/beta and gamma interferons. J Virol. 2002;76:2159–2167. [Europe PMC free article] [Abstract] [Google Scholar]
- Ashburn PM, Craig CF. Experimental investigations regarding the etiology of dengue fever. 1907. J Infect Dis. 2004;189:1747–1783. discussion 1744–1746. [Abstract] [Google Scholar]
- Ashour J, Laurent-Rolle M, Shi PY, Garcia-Sastre A. NS5 of dengue virus mediates STAT2 binding and degradation. J Virol. 2009;83:5408–5418. [Europe PMC free article] [Abstract] [Google Scholar]
- Balsitis SJ, Williams KL, Lachica R, Flores D, Kyle JL, Mehlhop E, Johnson S, Diamond MS, Beatty PR, Harris E. Lethal antibody enhancement of dengue disease in mice is prevented by Fc modification. PLoS Pathog. 2010;6:e1000790. [Europe PMC free article] [Abstract] [Google Scholar]
- Bente DA, Melkus MW, Garcia JV, Rico-Hesse R. Dengue fever in humanized NOD/SCID mice. J Virol. 2005;79:13797–13799. [Europe PMC free article] [Abstract] [Google Scholar]
- Blaney JE, Jr, Johnson DH, Manipon GG, Firestone CY, Hanson CT, Murphy BR, Whitehead SS. Genetic basis of attenuation of dengue virus type 4 small plaque mutants with restricted replication in suckling mice and in SCID mice transplanted with human liver cells. Virology. 2002;300:125–139. [Abstract] [Google Scholar]
- Brooks AJ, Johansson M, John AV, Xu Y, Jans DA, Vasudevan SG. The interdomain region of dengue NS5 protein that binds to the viral helicase NS3 contains independently functional importin beta 1 and importin alpha/beta-recognized nuclear localization signals. J Biol Chem. 2002;277:36399–36407. [Abstract] [Google Scholar]
- Chaturvedi UC, Shrivastava R, Nagar R. Dengue vaccines: problems and prospects. Indian J Med Res. 2005;121:639–652. [Abstract] [Google Scholar]
- Cleary CM, Donnelly RJ, Soh J, Mariano TM, Pestka S. Knockout and reconstitution of a functional human type I interferon receptor complex. J Biol Chem. 1994;269:18747–18749. [Abstract] [Google Scholar]
- Cleaves GR, Dubin DT. Methylation status of intracellular dengue type 2 40 S RNA. Virology. 1979;96:159–165. [Abstract] [Google Scholar]
- Clemens MJ, Williams BR. Inhibition of cell-free protein synthesis by pppA2′p5′A2′p5′A: a novel oligonucleotide synthesized by interferon-treated L cell extracts. Cell. 1978;13:565–572. [Abstract] [Google Scholar]
- Coia G, Parker MD, Speight G, Byrne ME, Westaway EG. Nucleotide and complete amino acid sequences of Kunjin virus: definitive gene order and characteristics of the virus-specified proteins. J Gen Virol. 1988;69(Pt 1):1–21. [Abstract] [Google Scholar]
- Colamonici OR, Platanias LC, Domanski P, Handa R, Gilmour KC, Diaz MO, Reich N, Pitha-Rowe P. Transmembrane signaling by the alpha subunit of the type I interferon receptor is essential for activation of the JAK kinases and the transcriptional factor ISGF3. J Biol Chem. 1995;270:8188–8193. [Abstract] [Google Scholar]
- Domanski P, Fish E, Nadeau OW, Witte M, Platanias LC, Yan H, Krolewski J, Pitha P, Colamonici OR. A region of the beta subunit of the interferon alpha receptor different from box 1 interacts with Jak1 and is sufficient to activate the Jak-Stat pathway and induce an antiviral state. J Biol Chem. 1997;272:26388–26393. [Abstract] [Google Scholar]
- Dumoutier L, Lejeune D, Hor S, Fickenscher H, Renauld JC. Cloning of a new type II cytokine receptor activating signal transducer and activator of transcription (STAT)1, STAT2 and STAT3. Biochem J. 2003;370:391–396. [Europe PMC free article] [Abstract] [Google Scholar]
- Forwood JK, Brooks A, Briggs LJ, Xiao CY, Jans DA, Vasudevan SG. The 37-aminoacid interdomain of dengue virus NS5 protein contains a functional NLS and inhibitory CK2 site. Biochem Biophys Res Commun. 1999;257:731–737. [Abstract] [Google Scholar]
- Fu XY, Kessler DS, Veals SA, Levy DE, Darnell JE., Jr ISGF3, the transcriptional activator induced by interferon alpha, consists of multiple interacting polypeptide chains. Proc Natl Acad Sci U S A. 1990;87:8555–8559. [Europe PMC free article] [Abstract] [Google Scholar]
- Graham H. The dengue: a study of its pathology and mode of propagation. Journal of Tropical Medicine. 1903:209–214. [Google Scholar]
- Greenlund AC, Morales MO, Viviano BL, Yan H, Krolewski J, Schreiber RD. Stat recruitment by tyrosine-phosphorylated cytokine receptors: an ordered reversible affinity-driven process. Immunity. 1995;2:677–687. [Abstract] [Google Scholar]
- Gubler DJ. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev. 1998;11:480–496. [Europe PMC free article] [Abstract] [Google Scholar]
- Gupta S, Yan H, Wong LH, Ralph S, Krolewski J, Schindler C. The SH2 domains of Stat1 and Stat2 mediate multiple interactions in the transduction of IFN-alpha signals. Embo J. 1996;15:1075–1084. [Europe PMC free article] [Abstract] [Google Scholar]
- Halfmann P, Ebihara H, Marzi A, Hatta Y, Watanabe S, Suresh M, Neumann G, Feldmann H, Kawaoka Y. Replication-deficient ebolavirus as a vaccine candidate. J Virol. 2009;83:3810–3815. [Europe PMC free article] [Abstract] [Google Scholar]
- Halstead SB. Dengue viruses. Philadelphia: WB Saunders; 1998. [Google Scholar]
- Horvath CM. Weapons of STAT destruction. Interferon evasion by paramyxovirus V protein. Eur J Biochem. 2004;271:4621–4628. [Abstract] [Google Scholar]
- Hu H, Huang X, Tao L, Huang Y, Cui BA, Wang H. Comparative analysis of the immunogenicity of SARS-CoV nucleocapsid DNA vaccine administrated with different routes in mouse model. Vaccine. 2009;27:1758–1763. [Europe PMC free article] [Abstract] [Google Scholar]
- Jessie K, Fong MY, Devi S, Lam SK, Wong KT. Localization of dengue virus in naturally infected human tissues, by immunohistochemistry and in situ hybridization. J Infect Dis. 2004;189:1411–1418. [Abstract] [Google Scholar]
- Jones M, Davidson A, Hibbert L, Gruenwald P, Schlaak J, Ball S, Foster GR, Jacobs M. Dengue virus inhibits alpha interferon signaling by reducing STAT2 expression. J Virol. 2005;79:5414–5420. [Europe PMC free article] [Abstract] [Google Scholar]
- Kabra SK, Jain Y, Singhal T, Ratageri VH. Dengue hemorrhagic fever: clinical manifestations and management. Indian J Pediatr. 1999;66:93–101. [Abstract] [Google Scholar]
- Kapoor M, Zhang L, Ramachandra M, Kusukawa J, Ebner KE, Padmanabhan R. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J Biol Chem. 1995;270:19100–19106. [Abstract] [Google Scholar]
- Kawai T, Akira S. Antiviral signaling through pattern recognition receptors. J Biochem. 2007;141:137–145. [Abstract] [Google Scholar]
- Kessler DS, Levy DE, Darnell JE., Jr Two interferon-induced nuclear factors bind a single promoter element in interferon-stimulated genes. Proc Natl Acad Sci U S A. 1988;85:8521–8525. [Europe PMC free article] [Abstract] [Google Scholar]
- Khan AM, Heiny AT, Lee KX, Srinivasan KN, Tan TW, August JT, Brusic V. Large-scale analysis of antigenic diversity of T-cell epitopes in dengue virus. BMC Bioinformatics. 2006;7(Suppl 5):S4. [Europe PMC free article] [Abstract] [Google Scholar]
- Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, Donnelly RP. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4:69–77. [Abstract] [Google Scholar]
- Kraus TA, Garza L, Horvath CM. Enabled interferon signaling evasion in an immune-competent transgenic mouse model of parainfluenza virus 5 infection. Virology. 2008;371:196–205. [Abstract] [Google Scholar]
- Kraus TA, Lau JF, Parisien JP, Horvath CM. A hybrid IRF9-STAT2 protein recapitulates interferon-stimulated gene expression and antiviral response. J Biol Chem. 2003;278:13033–13038. [Abstract] [Google Scholar]
- Kuno G, Chang GJ, Tsuchiya KR, Karabatsos N, Cropp CB. Phylogeny of the genus Flavivirus. J Virol. 1998;72:73–83. [Europe PMC free article] [Abstract] [Google Scholar]
- Kuruvilla JG, Troyer RM, Devi S, Akkina R. Dengue virus infection and immune response in humanized RAG2(−/−)gamma(c)(−/−) (RAG-hu) mice. Virology. 2007;369:143–152. [Abstract] [Google Scholar]
- Leitmeyer KC, Vaughn DW, Watts DM, Salas R, Villalobos I, de C, Ramos C, Rico-Hesse R. Dengue virus structural differences that correlate with pathogenesis. J Virol. 1999;73:4738–4747. [Europe PMC free article] [Abstract] [Google Scholar]
- Lin YL, Liao CL, Chen LK, Yeh CT, Liu CI, Ma SH, Huang YY, Huang YL, Kao CL, King CC. Study of Dengue virus infection in SCID mice engrafted with human K562 cells. J Virol. 1998;72:9729–9737. [Europe PMC free article] [Abstract] [Google Scholar]
- Lo MS, Brazas RM, Holtzman MJ. Respiratory syncytial virus nonstructural proteins NS1 and NS2 mediate inhibition of Stat2 expression and alpha/beta interferon responsiveness. J Virol. 2005;79:9315–9319. [Europe PMC free article] [Abstract] [Google Scholar]
- Martinez-Moczygemba M, Gutch MJ, French DL, Reich NC. Distinct STAT structure promotes interaction of STAT2 with the p48 subunit of the interferon-alpha-stimulated transcription factor ISGF3. J Biol Chem. 1997;272:20070–20076. [Abstract] [Google Scholar]
- Mazzon M, Jones M, Davidson A, Chain B, Jacobs M. Dengue virus NS5 inhibits interferon-alpha signaling by blocking signal transducer and activator of transcription 2 phosphorylation. J Infect Dis. 2009;200:1261–1270. [Abstract] [Google Scholar]
- Munoz-Jordan JL, Laurent-Rolle M, Ashour J, Martinez-Sobrido L, Ashok M, Lipkin WI, Garcia-Sastre A. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J Virol. 2005;79:8004–8013. [Europe PMC free article] [Abstract] [Google Scholar]
- Munoz-Jordan JL, Sanchez-Burgos GG, Laurent-Rolle M, Garcia-Sastre A. Inhibition of interferon signaling by dengue virus. Proc Natl Acad Sci U S A. 2003;100:14333–14338. [Europe PMC free article] [Abstract] [Google Scholar]
- Novick D, Cohen B, Rubinstein M. The human interferon alpha/beta receptor: characterization and molecular cloning. Cell. 1994;77:391–400. [Abstract] [Google Scholar]
- Parisien JP, Lau JF, Rodriguez JJ, Sullivan BM, Moscona A, Parks GD, Lamb RA, Horvath CM. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology. 2001;283:230–239. [Abstract] [Google Scholar]
- Park C, Lecomte MJ, Schindler C. Murine Stat2 is uncharacteristically divergent. Nucleic Acids Res. 1999;27:4191–4199. [Europe PMC free article] [Abstract] [Google Scholar]
- Park MS, Garcia-Sastre A, Cros JF, Basler CF, Palese P. Newcastle disease virus V protein is a determinant of host range restriction. J Virol. 2003a;77:9522–9532. [Europe PMC free article] [Abstract] [Google Scholar]
- Park MS, Shaw ML, Munoz-Jordan J, Cros JF, Nakaya T, Bouvier N, Palese P, Garcia-Sastre A, Basler CF. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J Virol. 2003b;77:1501–1511. [Europe PMC free article] [Abstract] [Google Scholar]
- Paulson M, Pisharody S, Pan L, Guadagno S, Mui AL, Levy DE. Stat protein transactivation domains recruit p300/CBP through widely divergent sequences. J Biol Chem. 1999;274:25343–25349. [Abstract] [Google Scholar]
- Pavlovic J, Zurcher T, Haller O, Staeheli P. Resistance to influenza virus and vesicular stomatitis virus conferred by expression of human MxA protein. J Virol. 1990;64:3370–3375. [Europe PMC free article] [Abstract] [Google Scholar]
- Precious B, Childs K, Fitzpatrick-Swallow V, Goodbourn S, Randall RE. Simian virus 5 V protein acts as an adaptor, linking DDB1 to STAT2, to facilitate the ubiquitination of STAT1. J Virol. 2005;79:13434–13441. [Europe PMC free article] [Abstract] [Google Scholar]
- Qureshi SA, Salditt-Georgieff M, Darnell JE., Jr Tyrosine-phosphorylated Stat1 and Stat2 plus a 48-kDa protein all contact DNA in forming interferon-stimulated-gene factor 3. Proc Natl Acad Sci U S A. 1995;92:3829–3833. [Europe PMC free article] [Abstract] [Google Scholar]
- Rice CM, Lenches EM, Eddy SR, Shin SJ, Sheets RL, Strauss JH. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science. 1985;229:726–733. [Abstract] [Google Scholar]
- Rodriguez-Madoz JR, Belicha-Villanueva A, Bernal-Rubio D, Ashour J, Ayllon J, AF-S Inhibition of Type I IFN Response in Human Dendritic Cells by Dengue Virus Infection Requires a Catalytically Active NS2B3 Complex. Journal of Virology. 2010 10.1128/JVI.01051-10. Published ahead of print on 21 July 2010. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Sampath A, Padmanabhan R. Molecular targets for flavivirus drug discovery. Antiviral Res. 2009;81:6–15. [Europe PMC free article] [Abstract] [Google Scholar]
- Samuel CE. Molecular mechanisms of interferon action: interferon-mediated phosphorylation of ribosome-associated protein P1 and protein synthesis initiation factor eIF-2. Tex Rep Biol Med. 1981;41:463–470. [Abstract] [Google Scholar]
- Shresta S, Kyle JL, Robert Beatty P, Harris E. Early activation of natural killer and B cells in response to primary dengue virus infection in A/J mice. Virology. 2004a;319:262–273. [Abstract] [Google Scholar]
- Shresta S, Kyle JL, Snider HM, Basavapatna M, Beatty PR, Harris E. Interferon-dependent immunity is essential for resistance to primary dengue virus infection in mice, whereas T- and B-cell-dependent immunity are less critical. J Virol. 2004b;78:2701–2710. [Europe PMC free article] [Abstract] [Google Scholar]
- Shresta S, Sharar KL, Prigozhin DM, Beatty PR, Harris E. Murine model for dengue virus-induced lethal disease with increased vascular permeability. J Virol. 2006;80:10208–10217. [Europe PMC free article] [Abstract] [Google Scholar]
- Shresta S, Sharar KL, Prigozhin DM, Snider HM, Beatty PR, Harris E. Critical roles for both STAT1-dependent and STAT1-independent pathways in the control of primary dengue virus infection in mice. J Immunol. 2005;175:3946–3954. [Abstract] [Google Scholar]
- Shuai K, Horvath CM, Huang LH, Qureshi SA, Cowburn D, Darnell JE., Jr Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell. 1994;76:821–828. [Abstract] [Google Scholar]
- Shuai K, Stark GR, Kerr IM, Darnell JE., Jr A single phosphotyrosine residue of Stat91 required for gene activation by interferon-gamma. Science. 1993;261:1744–1746. [Abstract] [Google Scholar]
- Ulane CM, Horvath CM. Paramyxoviruses SV5 and HPIV2 assemble STAT protein ubiquitin ligase complexes from cellular components. Virology. 2002;304:160–166. [Abstract] [Google Scholar]
- Umareddy I, Tang KF, Vasudevan SG, Devi S, Hibberd ML, Gu F. Dengue virus regulates type I interferon signalling in a strain-dependent manner in human cell lines. J Gen Virol. 2008;89:3052–3062. [Abstract] [Google Scholar]
- van der Laan JW, Herberts C, Lambkin-Williams R, Boyers A, Mann AJ, Oxford J. Animal models in influenza vaccine testing. Expert Rev Vaccines. 2008;7:783–793. [Abstract] [Google Scholar]
- Wengler G, Wengler G, Gross HJ. Studies on virus-specific nucleic acids synthesized in vertebrate and mosquito cells infected with flaviviruses. Virology. 1978;89:423–437. [Abstract] [Google Scholar]
- Whitehead SS, Blaney JE, Durbin AP, Murphy BR. Prospects for a dengue virus vaccine. Nat Rev Microbiol. 2007;5:518–528. [Abstract] [Google Scholar]
- Wu SJ, Hayes CG, Dubois DR, Windheuser MG, Kang YH, Watts DM, Sieckmann DG. Evaluation of the severe combined immunodeficient (SCID) mouse as an animal model for dengue viral infection. Am J Trop Med Hyg. 1995;52:468–476. [Abstract] [Google Scholar]
- Yauch LE, Shresta S. Mouse models of dengue virus infection and disease. Antiviral Res. 2008;80:87–93. [Europe PMC free article] [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.chom.2010.10.007
Read article for free, from open access legal sources, via Unpaywall:
http://www.cell.com/article/S1931312810003446/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/106432550
Article citations
RT-qPCR Analysis of Inflammatory & Apoptotic Factors-Related Gene Expression in ZIKV-Infected IFNAR1<sup>-/-</sup> Mice.
Curr Microbiol, 81(12):418, 21 Oct 2024
Cited by: 0 articles | PMID: 39432111
Human Stimulator of Interferon Genes Promotes Rhinovirus C Replication in Mouse Cells In Vitro and In Vivo.
Viruses, 16(8):1282, 10 Aug 2024
Cited by: 0 articles | PMID: 39205256 | PMCID: PMC11358906
Evolution of STAT2 resistance to flavivirus NS5 occurred multiple times despite genetic constraints.
Nat Commun, 15(1):5426, 26 Jun 2024
Cited by: 0 articles | PMID: 38926343 | PMCID: PMC11208600
The Dynamic Relationship between Dengue Virus and the Human Cutaneous Innate Immune Response.
Viruses, 16(5):727, 04 May 2024
Cited by: 0 articles | PMID: 38793609 | PMCID: PMC11125669
Review Free full text in Europe PMC
ISG15/USP18/STAT2 is a molecular hub regulating IFN I-mediated control of Dengue and Zika virus replication.
Front Immunol, 15:1331731, 07 Feb 2024
Cited by: 2 articles | PMID: 38384473 | PMCID: PMC10879325
Go to all (136) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Host-Specific NS5 Ubiquitination Determines Yellow Fever Virus Tropism.
J Virol, 93(14):e00151-19, 28 Jun 2019
Cited by: 18 articles | PMID: 31043530 | PMCID: PMC6600188
Dengue virus co-opts UBR4 to degrade STAT2 and antagonize type I interferon signaling.
PLoS Pathog, 9(3):e1003265, 28 Mar 2013
Cited by: 148 articles | PMID: 23555265 | PMCID: PMC3610674
NS5 of dengue virus mediates STAT2 binding and degradation.
J Virol, 83(11):5408-5418, 11 Mar 2009
Cited by: 288 articles | PMID: 19279106 | PMCID: PMC2681973
Innate immunity evasion by Dengue virus.
Viruses, 4(3):397-413, 15 Mar 2012
Cited by: 93 articles | PMID: 22590678 | PMCID: PMC3347034
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NIAID NIH HHS (7)
Grant ID: R01 AI073450
Grant ID: R01 AI085607
Grant ID: U54 AI057158
Grant ID: U54 AI57158
Grant ID: AI058211
Grant ID: U54 AI057158-09
Grant ID: 5R01 AI073450




