Abstract
Free full text

Germline mutations in BAP1 predispose to melanocytic tumors
Abstract
Common acquired melanocytic nevi are benign neoplasms that are composed of small uniform melanocytes and typically present as flat or slightly elevated, pigmented lesions on the skin. We describe two families with a new autosomal dominant syndrome characterized by multiple skin-colored, elevated melanocytic tumors. In contrast to common acquired nevi, the melanocytic neoplasms in affected family members ranged histopathologically from epithelioid nevi to atypical melanocytic proliferations that showed overlapping features with melanoma. Some affected patients developed uveal or cutaneous melanomas. Segregating with this phenotype, we found inactivating germline mutations of the BAP1 gene. The majority of melanocytic neoplasms lost the remaining wild-type allele of BAP1 by various somatic alterations. In addition, we found BAP1 mutations in a subset of sporadic melanocytic neoplasms showing histologic similarities to the familial tumors. These findings suggest that loss of BAP1 is associated with a clinically and morphologically distinct type of melanocytic neoplasm.
We report a type of melanocytic neoplasm that was inherited in an autosomal dominant pattern in two unrelated families and was clinically, histopathologically, and genetically distinct from common acquired nevi (Fig. 1a). Beginning in the second decade of life, affected family members progressively developed skin-colored to reddish-brown, dome-shaped to pedunculated, well-circumscribed papules with an average size of 5mm (Fig. 1b; Supplementary Fig. 1–4). The number of tumors per patient varied markedly, ranging from 5 to over 50. No intellectual disabilities or dysmorphic features were identified in affected individuals.
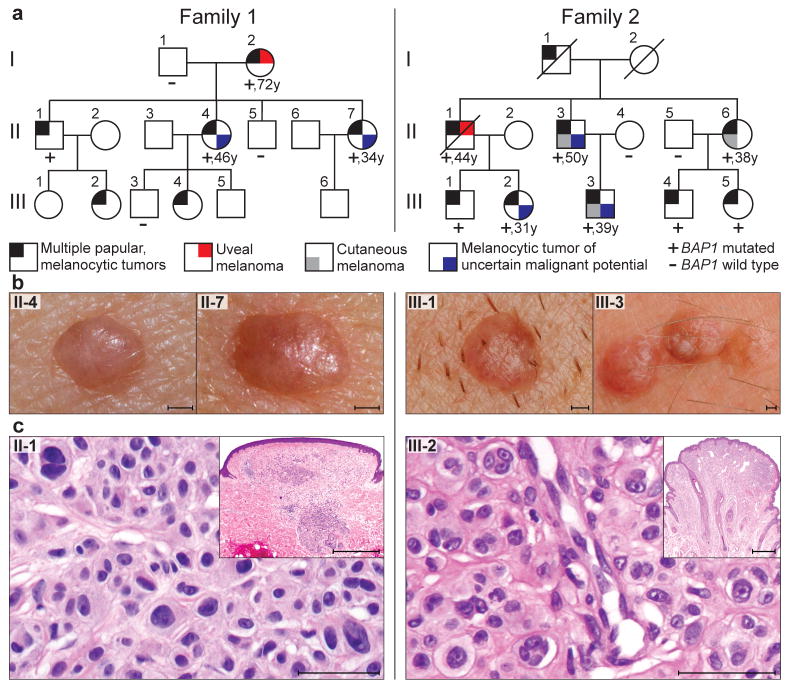
BAP1 germline mutations in two families; left panel: family 1; right panel: family 2. (a) Pedigrees. Ages of onset of the first melanoma or ‘melanocytic tumor of uncertain malignant potential’ are written below the symbols. Only subjects older than 18 years were tested for mutations. An extended pedigree of family 1 is shown in Supplementary Fig. 15. (b) Representative melanocytic neoplasms from affected individuals (II-4 and II-7 from family 1; III-1 and III-3 from family 2). Note the characteristic inconspicuous, skin-colored to reddish-brown, dome-shaped appearance. Scale bars, 1 mm. (c) Histopathology of representative melanocytic tumors from both families showing relatively symmetrical intradermal proliferations of large epithelioid melanocytes with abundant cytoplasm and enlarged, irregularly-shaped, vesicular nuclei, some with prominent nucleoli. Scale bars, 50 μm. Scale bars inset, 1 mm.
Histopathological examination revealed primarily dermal tumors composed entirely or predominantly of epithelioid melanocytes with abundant amphophilic cytoplasm and prominent nucleoli. The melanocytes often contained large, vesicular nuclei that varied significantly in size and shape (Fig. 1c; Supplementary Fig. 5–7). The cytological features of some of the cells were reminiscent of Spitz nevi; however, characteristic features (such as epidermal hyperplasia, hypergranulosis, Kamino bodies, clefting around junctional melanocytic nests, and spindle-shaped melanocytes) frequently seen in Spitz nevi were consistently absent. In addition, 37 of 42 (88%) tumors in the families showed BRAF mutations, which are typically absent in Spitz nevi.1
Some of the neoplasms exhibited one or more atypical features, such as high cellularity, considerable nuclear pleomorphism, and several chromosomal aberrations. These tumors were classified as ‘neoplasms of uncertain malignant potential’ and the patients were managed as if they had melanoma (Supplementary Fig. 8). Both families were identified because of the occurrence of multiple epithelioid melanocytic tumors, but in each family, one affected individual had uveal melanoma, and three members of family 2 had been diagnosed with cutaneous melanoma (Fig. 1a, Supplementary Table 1).
Twenty-two melanocytic neoplasms from three affected individuals (II-1, II-4, II-7) in family 1 were analyzed with array-based comparative genomic hybridization (aCGH). Losses affecting the entire chromosome 3 or portions of the short arm of chromosome 3 were found in 50% of tumors. The smallest overlap of the deleted regions encompassed 5.8 megabases, extending from base-pair position 47,976,758 to 53,848,761 (hg 18 assembly) and encoded at least 150 known genes (Fig. 2a).
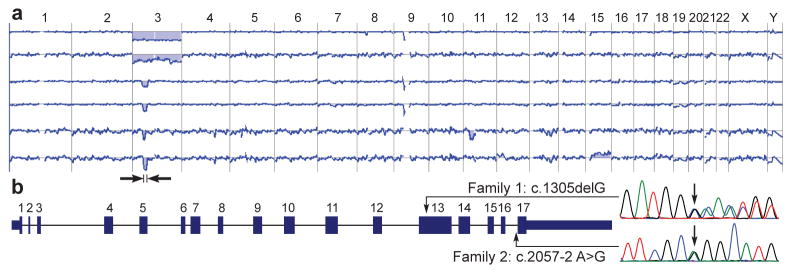
Identification of the syndrome-causing gene. (a) Recurrent deletions affecting chromosome 3 in the melanocytic neoplasms of family 1. The two uppermost array-CGH profiles illustrate deletions of the entire chromosome 3, whereas the other profiles show focal deletions in 3p21. The minimally deleted region (black arrows) encompassed approximately 6 megabases. The next to last profile shows in addition a loss of chromosome 11 and the last profile a gain of chromosome 15. (b) Gene structure of BAP1 and germline mutations observed in affected individuals from family 1 and 2.
The frequent loss of the 3p21 region suggested a second hit2, resulting in the elimination of the remaining wild-type allele of a mutated tumor suppressor gene in this region. To support this hypothesis, the haplotypes of 6 members of family 1 were reconstructed with SNP-arrays. Affected siblings in the second generation (II-1, II-4, II-7) inherited the same maternal copy of the 3p21 region, whereas the non-affected brother (II-5) received the other maternal 3p21 copy (Supplementary Fig. 9a). SNP-arrays showed that in all tumors with chromosome 3 loss, the paternal copy of chromosome 3 was lost, while the maternal copy was retained (Supplementary Fig. 9b). In conclusion, these data strongly suggested that a mutated gene in the 3p21 region was inherited from the maternal side of the family.
To identify the mutated gene, we sequenced the minimally deleted region of chromosome 3 in 2 affected (I-2, II-4) and 2 unaffected (I-1, II-5) subjects from family 1 using an in-solution hybrid capture technique followed by massively parallel sequencing.3 This analysis revealed a frameshift mutation in the BAP1 gene (c.1305delG, p.Gln436Asnfs*135) that was subsequently found to segregate with the phenotype (Fig. 2b, Supplementary Fig. 10). To rule out BAP1 germline mutations in the general population, we reviewed sequence data from 629 individuals in the 1000genomes database (http://www.1000genomes.org/home). No truncating mutations were found indicating that BAP1 germline mutations are infrequent in the general population.
The status of the second BAP1 allele was assessed in 29 skin tumors and in the uveal melanoma (I-2) from family 1. All tumors in which a loss of 3p21 was detected previously by aCGH, also showed a loss of the wild-type BAP1 allele in the sequencing electropherogram. In five additional cases without loss of the 3p21 region, the electropherogram showed markedly suppressed residual wild-type sequences, indicating that the neoplastic cells had lost the wild-type BAP1 allele through copy number neutral mechanisms, resulting in maternal uniparental disomy. In four other neoplasms without 3p21 deletions, we found additional acquired somatic nonsense (2 cases), frameshift (1 case) and missense (1 case) mutations in BAP1. Immunohistochemistry for BAP1 showed loss of nuclear expression in all melanocytic neoplasms, including the tumors without detectable alteration of the wild-type BAP1 allele. In summary, these data suggest that the remaining wild-type allele of BAP1 is lost by various somatic alterations in the melanocytic tumors (Supplementary Table 2, Supplementary Fig. 11).
In family 2, we found a different germline mutation in BAP1 that segregated with the phenotype and removed the acceptor splice site at the last exon (c.2057-2A>G, p.Met687Glufs*28, Fig. 2b). Analysis of cDNA from 2 affected family members confirmed that the last intron was not removed by splicing (Supplementary Fig. 12). In family 2, inactivation of the remaining wild-type BAP1 allele was found in 9 of 13 skin tumors, in the uveal melanoma (II-1), and in the cutaneous melanoma from patient II-3 (Fig. 3a–d, Supplementary Table 3). The metastatic melanoma of individual II-6 did not show loss of heterozygosity of BAP1, but no additional tissue was available to investigate alternative mechanisms of BAP1 inactivation. Besides the elevated, skin-colored melanocytic neoplasms, common acquired nevi (flat, brown maculae) were also excised from 4 patients (II-3, III-3, III-4, III-5). Histopathologically these nevi were composed of small uniform melanocytes and showed strong nuclear expression of BAP1 by immunohistochemistry (Supplementary Figure 13).
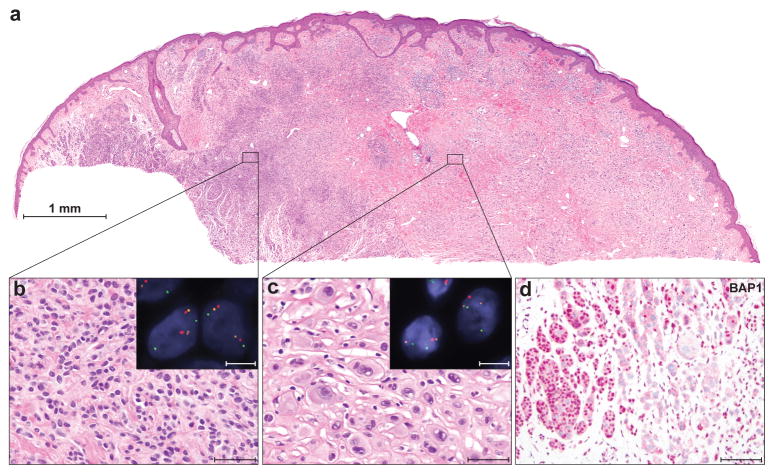
Bi-allelic BAP1 loss is associated with characteristic histological features in familial melanocytic neoplasms. (a) A melanocytic neoplasm from patient III-3, family 2, presents as a combined lesion with (b) an area of small cells (common acquired nevus) on the left, and (c) an area of large epithelioid cells on the right. Scale bars, 50 μm. Fluorescence in-situ hybridization (red signal: chr. 3p21 [BAP1]; orange: chr. 3p25 [control]; green: chr. 4p12 [control]) shows loss of BAP1 in the area with large epithelioid melanocytes (insert in c: one red BAP1 signal, but 2 orange and green control signals per nucleus. Scale bar, 10 μm), but no loss in the region of the common nevus (insert in b: two signals of each probe. Scale bar, 10 μm.). (d) BAP1 immunohistochemistry at the transition area between common nevus and epithelioid cell area shows a strong nuclear staining in the common nevus component (left) and loss of nuclear staining in the epithelioid component (right). Scale bar, 100 μm.
To address the role of BAP1 mutations in sporadic melanocytic neoplasms, we sequenced BAP1 in 156 randomly selected tumors without family history: common nevi with uniform small melanocytes (n=28); Spitz nevi (n=17); neoplasms with overlapping features between Spitz nevus and melanoma (so-called “atypical Spitz tumors”, n=18); primary melanomas originating from acral skin (n=15), mucosa (n=15), or skin with (n=15) or without (n=15) chronic sun-induced damage, and uvea (n=33). Thirteen (40%) uveal melanomas, two (11%) atypical Spitz tumors and three (5%) of the melanomas (2 melanoma on skin without chronic sun-induced damage and 1 acral melanoma) had somatic BAP1 mutations. No mutations were found in the other categories (Supplementary Table 4–8). The mutation frequency seen in uveal melanoma is similar to a recent report.4 Two of five sporadic cutaneous melanomas that arose in nevi harbored BAP1 mutations. In both cases the BAP1 mutations were absent in the nevus portion, suggesting that loss of function of BAP1 may play a role in progression from nevus to melanoma in some cases (Fig. 4a–g).
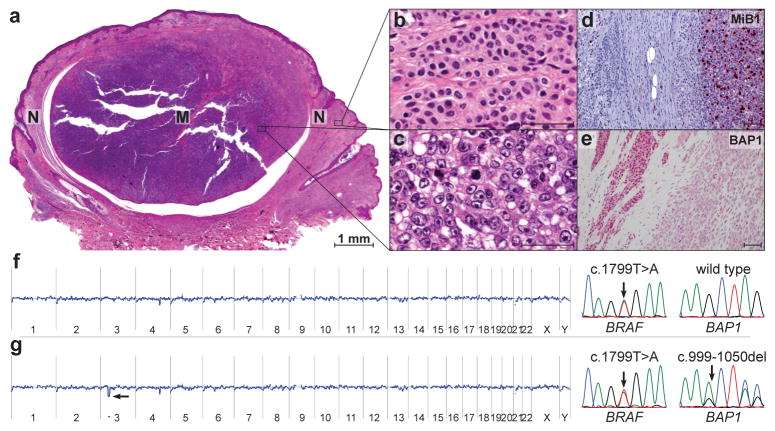
Progression of a nevus to melanoma is associated with loss of BAP1. (a) Scanning magnification of a sporadic melanoma (M) arising within a nevus (N) (hematoxylin-eosin stain). (b) Nevus component with bland melanocytes containing monomorphous round and oval-shaped nuclei. Scale bar, 50 μm. (c) Melanoma component showing melanocytes with large vesicular nuclei and prominent nucleoli. Scale bar, 50 μm. (d) MIB-1 immunohistochemistry shows a high proliferation rate in the melanoma compared to the nevus component. Scale bar, 100 μm. (e) BAP1 immunohistochemistry displays a conspicuous nuclear staining in the nevus contrasted with absent nuclear staining in the melanoma. Scale bar 100 μm. (f) The nevus component shows a BRAFV600E mutation, but no BAP1 mutation or chromosomal aberrations in aCGH. (g) The melanoma component shows the same BRAF mutation and in addition a focal loss of chromosome region 3p21 spanning the BAP1-locus in aCGH. The electropherogram of BAP1 demonstrates a frameshift mutation of the second BAP1 allele.
The two atypical Spitz tumors with somatic BAP1 mutations had similar morphologic features to the melanocytic neoplasms seen in both families, lacked immunohistochemical expression of BAP1, and harbored BRAF mutations (Supplementary Figure 14). This finding suggests that bi-allelic inactivation of BAP1 is associated with a clinically and morphologically distinct type of melanocytic neoplasm. Histopathologically, the tumors range from intradermal nevi composed of bland epithelioid melanocytes to atypical proliferations of epithelioid melanocytes with morphological and cytogenetic features overlapping with melanoma. These tumors have been previously subsumed under the category of “spitzoid” melanocytic neoplasms because they share cytologic features with Spitz nevi.
As illustrated by the families, inheriting one mutant copy of BAP1 results in a markedly increased number of these neoplasms. In aggregate, several hundred papular melanocytic tumors were present in affected family members, while the number of melanomas was substantially lower. This indicates that the risk of malignant progression in individual tumors is low, and that bi-allelic loss of BAP1 in conjunction with mutations in BRAF is not sufficient for melanoma formation in the skin. Harbour et al.4 proposed that bi-allelic loss of BAP1 in uveal melanoma, which carry mutations in GNAQ5 or GNA116 but not in BRAF, marks the transition to metastatic disease. Our findings may indicate that the role of BAP1 in melanocytic neoplasia depends on the associated oncogene and/or the cell of origin.
BAP1 was originally discovered as a binding partner of BRCA1 and has been functionally implicated in DNA damage response7,8, as well as in regulation of apoptosis, senescence, and the cell cycle.9 Its Drosophila counterpart Calypso is involved in chromatin remodeling, which was shown to oppose the mono-ubiquitination activity of the polycomb repressive complex 1, a critical component of transcriptional silencing.10 In cancer, mutations and deletions in BAP1 have been reported in breast and lung cancers11–13, but none of the individuals in our study developed breast or lung cancers.
In summary, we describe a novel autosomal dominant syndrome that is caused by germline mutations of BAP1, characterized by a high penetrance of melanocytic neoplasms with distinctive clinical and histopathological features, and possibly associated with an increased risk for uveal and cutaneous melanoma.
ONLINE METHODS
Patients
The study was approved by the Ethics Committees of the Medical University of Graz, Austria, and Memorial Sloan-Kettering Cancer Center, New York, and was conducted according to the Declaration of Helsinki. Written consent was obtained from all participating family members and only subjects older than 18 years were included. Archival, paraffin-embedded tissue from both families, sporadic cutaneous and uveal melanomas, sporadic common melanocytic nevi, Spitz nevi, and atypical Spitz tumors were retrieved from the Department of Dermatology, Medical University of Graz, Austria.
Family 1 was from southern Germany. We collected blood from 7 individuals (I-1, I-2, II-1, II-4, II-5, II-7, III-3), 29 melanocytic tumors from 3 affected subjects (II-1, II-4, II-7), and one uveal melanoma from patient I-2 (Fig. 1a, Supplementary Table 1 and 2). The melanocytic tumor of uncertain malignant potential of subject II-4 and II-7 were not available for molecular genetic analysis.
Family 2 was from southern Austria. We collected blood from 9 individuals (II-3, II-4, II-5, II-6, III-1, III-2, III-3, III-4, III-5), 11 melanocytic tumors from 3 affected subjects (II-3, III-2, III-3), 2 melanocytic tumors of uncertain malignant potential from patient II-3 and III-2, 2 cutaneous melanomas from patient II-3 (non-metastasizing melanoma) and II-6 (primary tumor and lymph node metastasis), and one uveal melanoma from patient II-1 (Fig. 1a, Supplementary Table 1 and 3). The melanoma and the melanocytic tumor of uncertain malignant potential of subject III-3 were not available for molecular genetic analysis.
Histopathology and immunohistochemistry
Excised skin lesions were fixed in 4% neutral buffered formalin. The fixed tissues were processed using routine histologic methods and embedded in paraffin. 5μm-thick sections were cut from the paraffin blocks and stained with hematoxylin-eosin. Immunohistochemistry using an antibody against BAP1 (clone C4, Santa Cruz Biotechnology, Santa Cruz, CA) was performed on tissue sections using standard methods.
DNA extraction
Tumor-bearing tissue was manually microdissected from sections of archival paraffin-embedded samples of melanocytic tumors and melanomas. In the case of small tumors, tumor-bearing tissue was microdissected using a ‘PALM Laser Microdissection and Pressure Catapulting system’ (P.A.L.M., Zeiss, Germany). DNA was extracted and purified from the microdissected tissue using the QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany) or the Chemagic DNA Tissue Kit (Chemagen, Baesweiler, Germany) according to the manufacturer’s instructions. DNA was also isolated from adjacent normal tissue to distinguish somatic from germline mutations.
Array-based comparative genomic hybridization (aCGH)
DNA samples were labeled using a Bioprime Array CGH Genomic Labeling Kit according to the manufacturer’s instructions (Invitrogen, Carlsberg, CA). Briefly, 250 to 500ng test and reference DNA (Promega, Madison, WI) were differentially labeled with dCTP-Cy5 and dCTP-Cy3, respectively (GE Healthcare, Piscataway, NJ). Genome-wide analysis of DNA copy number changes was conducted using oligonucleotide arrays containing 60,000 or 180,000 probes according to the manufacturer’s protocol version 6.0 (Agilent, Santa Clara, CA). Slides were scanned using Agilent’s microarray scanner G2505B and analyzed using Agilent Feature Extraction and DNA Workbench software 5.0.14.
Sanger sequencing
The exonic regions of BAP1 were divided into amplicons of 300 bp or less. Specific primers were designed using primer 3 and tagged with an M13 tail to facilitate Sanger sequencing (Supplementary Table 9). Primers for GNAQ, GNA11, BRAF were described previously.5,6,14 The PCR reaction conditions were 0.25 mM dNTPs, 0.4x BSA (New England Biolabs, Ipswich, MA), 1 U Hotstar Taq (Qiagen), 1X Hotstar Taq buffer (Qiagen), and 0.4 μM primer. PCR consisted of 35 cycles of 95°C (45 seconds), 57°C (45 seconds), and 72°C (45 seconds) after initial denaturation at 95°C for 5 minutes. PCR reaction products were purified using QIAquick PCR Purification kit (Qiagen) and then used as templates for sequencing in both directions using Big Dye v3.1 (Applied Biosystems, Foster City, CA). Dye terminators were removed using the CleanSEQ kit (Agencourt Biosciences), and subsequent products were run on the ABI PRISM 3730xl (Applied Biosystems). Mutations were identified by using Sequencher Software (http://www.genecodes.com/) and only considered when variants were called in reads in both directions. All identified mutations were replicated at least twice. In sporadic tumors, germline DNA was sequenced from the adjacent normal tissue or blood to determine whether the mutations were somatically acquired.
RNA analysis
To evaluate the consequences of the splice-site germline mutation in family 2, RNA was isolated from whole blood of affected subjects and normal controls. The isolated RNA was reverse transcribed using Qiagen’s Omniscript transcriptase and subsequently amplified using cDNA specific primers for BAP1 (Supplementary Table 9) employing the same PCR reaction described in the previous paragraph. PCR products were electrophoretically separated on a 1% agarose gel. Bands were cut out, purified using gel purification (Promega Wizard SV-Gel and PCR cleanup system), and sequenced on a 3730xl capillary genetic analyzer (Applied Biosystems).
Single nucleotide polymorphism (SNP) arrays
DNA from 4 affected (I-2, II-1, II-4, II-7) and 2 unaffected (I-1, II-5) individuals from family 1 were analyzed with the Affymetrix GeneChip Mapping 500K array NspI chip. In accordance to the manufacturer’s instructions, the arrays were scanned with a GeneChip Scanner and the data were analyzed with GeneChip Operating Software (GCOS) and GeneChip Genotyping Analysis Software (GTYPE 3.0.2) to generate SNP allele calls. Haplotyping analysis was performed with the DNA-Chip Analyzer (www.dchip.org) software under the assumption of an autosomal dominant trait.
Target enrichment and SOLiD sequencing
DNA capture was performed on 3μg of high quality genomic DNA using a custom SureSelect Target Enrichment kit according to the protocol provided by Agilent (Agilent, Santa Clara, CA). The baits library for SureSelect Target Enrichment was designed in Agilent’s eArray (https://earray.chem.agilent.com/earray), contained 49,920 unique baits covering 3,291,328 bases, extended from base-pair position 47,501,754 to 54,373,721 according to hg 18 assembly, and will be provided on request. 3p21 enriched DNA libraries of 2 affected (I-2, II-4) and 2 unaffected subjects (I-1, II-5) of family 1 were sequenced with SOLiD 4 according to the protocol provided by Applied Biosystems (Applied Biosystems, Carlsbad, California), generating an average of 73,096,487 reads per samples with a read length of 50 bp. On average, 78% of the targeted region was covered at 400x.
To detect small insertion/deletion (in/dels) events, the SOLiD reads were mapped using the BWA mapping program (v0.5.8c) with default options except for the colorspace switch. The mapped bam file was then processed to remove duplicated reads and realigned with the IndelRealigner from the Genome Analysis Toolkit (GATK) which does a more careful alignment of reads around putative in/dels. The realigned bam file was converted to a pileup with samtools and in/del variants were called with the VarScan (v2.2) program.
Interphase fluorescence in situ hybridization (FISH)
FISH probes were synthesized from BAC clones and labeled by nick translation with SpectrumGreen-dUTP, SpectrumRed-dUTP and SpectrumOrange-dUTP (Abbott, Des Plaines, IL) with standard procedures. The SpectrumRed probe mapped to 3p21 (RP11-447A21 and RP5-966M1), SpectrumOrange to 3p25 (RP11-614E19 and RP11-963B11RP), and SpectrumGreen to 4q12 (RP11-117E8 and RP11-231C18). The probes were hybridized on 5μm thick sections. The number of hybridization signals for these probes was assessed in a minimum of 200 interphase nuclei with strong and well-delineated contours.
Computational predictors of functional significance
The functional consequences of missense mutations in BAP1 were assessed with three computational functional significance predictors (Supplementary Table 7 and 8).15–17
Acknowledgments
We are indebted to all family members for their enthusiastic participation in the study. We thank Werner Stieber for clinical photography, Ulrike Schmidbauer for providing histotechnical services, Dr. Margaret Leversha for performing FISH, Dr. Adriana Heguy for supporting sequencing, Marina Asher for immunohistochemistry, Dr. Hikmat Al-Ahmadie for assistance with the histological images, and Dr. Peter Dillinger for helping to collect the tissue samples. T.W. was supported by a Max-Kade Fellowship and expresses special thanks to Professor emeritus Dr. Helmut Kerl for supporting, encouraging, and inspiring his academic interests. A.C.O is supported by the Austrian Science Fund (J3013) and K.G.G. by the Deutsche Forschungsgemeinschaft (GR 3671/1-1a). This work was funded by grants from the National Institutes of Health (R01 CA131524), Geoffrey Beene Cancer Research Center (CC 66270), the American Skin Association (all to B.C.B.), the Andrew Sabin Family Foundation (to K.O.), the European Commission (GENINCA, contract no. HEALTH-F2-2008-202230), the Jubilaeumsfonds of the Oesterreichische Nationalbank (13837).
Footnotes
ACCESSION CODES
The mutation nomenclature for BAP1 is based on RefSeqs NM_004656.2 and NP_004647.1, for BRAF on NM_004333.4 and NP_004324.2, for GNAQ on NM_002072.3 and NP_002063.2, and for GNA11 on NM_002067.2 and NP_002058.2.
AUTHOR CONTRIBUTION
Project planning and experimental design: T.W., B.C.B., M.R.S. Review of clinical phenotypes: T.W., I.F., I.W., J.C.B. Review of histology and immunohistology: T.W., B.C.B., H.K., R.M., L.C., I.F., A.R, A.O. FISH analysis: T.W., G.P. Sample collection: T.W., H.K., W.W., K.O., D.A., aCGH: T.W., A.C.O. Linkage analysis: A.C.O, C.W. Mutation analysis: T.W., P.U., S.L. Next generation sequencing and data analysis: T.W., A.V., A.L., N.S., M.P. Manuscript writing: T.W., B.C.B, R.M., M.R.S., K.G.G, Critical revision of the manuscript: all authors.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interest.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/ng.910
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3328403
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/ng.910
Article citations
[Dermoscopy of melanocytoma-groundbreaking or established knowledge].
Dermatologie (Heidelb), 21 Oct 2024
Cited by: 0 articles | PMID: 39433580
Multiple Onychopapillomas and BAP1 Tumor Predisposition Syndrome.
JAMA Dermatol, 160(8):838-845, 01 Aug 2024
Cited by: 0 articles | PMID: 38759225
Saturation genome editing of BAP1 functionally classifies somatic and germline variants.
Nat Genet, 56(7):1434-1445, 05 Jul 2024
Cited by: 1 article | PMID: 38969833 | PMCID: PMC11250367
Low Exposures to Amphibole or Serpentine Asbestos in Germline Bap1-mutant Mice Induce Mesothelioma Characterized by an Immunosuppressive Tumor Microenvironment.
Cancer Res Commun, 4(4):1004-1015, 01 Apr 2024
Cited by: 2 articles | PMID: 38592450 | PMCID: PMC11000687
Harnessing ferroptosis for enhanced sarcoma treatment: mechanisms, progress and prospects.
Exp Hematol Oncol, 13(1):31, 12 Mar 2024
Cited by: 1 article | PMID: 38475936 | PMCID: PMC10935897
Review Free full text in Europe PMC
Go to all (389) article citations
Other citations
Wikipedia
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
RefSeq - NCBI Reference Sequence Database (3)
- (1 citation) RefSeq - NM_002072.3
- (1 citation) RefSeq - NM_004656.2
- (1 citation) RefSeq - NM_002067.2
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Histomorphologic spectrum of germline-related and sporadic BAP1-inactivated melanocytic tumors.
J Am Acad Dermatol, 79(3):525-534, 10 May 2018
Cited by: 8 articles | PMID: 29753057
Germline BRCA1-associated protein 1 mutation presenting as BAP1 inactivated melanocytic nevi in a child of a father with fatal paraganglioma.
Pediatr Dermatol, 35(5):e316-e318, 04 Jul 2018
Cited by: 3 articles | PMID: 29974497
Histomorphologic spectrum of BAP1 negative melanocytic neoplasms in a family with BAP1-associated cancer susceptibility syndrome.
J Cutan Pathol, 42(6):406-412, 12 May 2015
Cited by: 9 articles | PMID: 25902915
The morphologic spectrum of germline-mutated BAP1-inactivated melanocytic tumors includes lesions with conventional nevic melanocytes: A case report and review of literature.
J Cutan Pathol, 46(11):852-857, 04 Jul 2019
Cited by: 3 articles | PMID: 31206729
Review
Funding
Funders who supported this work.
Austrian Science Fund FWF (1)
The impact of stochastic (epi)mutations on the aging genome
Dr Anna Obenauf, Medical University of Graz
Grant ID: J 3013
European Commission FP7 (1)
Grant ID: FP7_202230
NCI NIH HHS (3)
Grant ID: R01 CA131524-05
Grant ID: R01 CA131524-04
Grant ID: R01 CA131524





