Abstract
Free full text
Piwi and piRNAs Act Upstream of an Endogenous siRNA Pathway to suppress Tc3 Transposon Mobility in the Caenorhabditis elegans germline
Associated Data
Abstract
The Piwi proteins of the Argonaute superfamily are required for normal germline development in Drosophila, zebrafish and mice, and associate with 24-30 nucleotide RNAs termed piRNAs. We identify a class of 21 nucleotide RNAs, previously named 21U-RNAs, as the piRNAs of C. elegans. Piwi and piRNA expression is restricted to the male and female germline and independent of many proteins in other small RNA pathways, including DCR-1. We show that Piwi is specifically required to silence Tc3, but not other Tc/mariner DNA transposons. Tc3 excision rates in the germline are increased at least 100 fold in piwi mutants as compared to wild type. We find no evidence for a Ping-Pong model for piRNA amplification in C. elegans. Instead, we demonstrate that Piwi acts upstream of an endogenous siRNA pathway in Tc3 silencing. These data might suggest a link between piRNA and siRNA function.
INTRODUCTION
Piwi proteins are part of a superfamily of Argonaute proteins that is defined by the presence of PAZ and Piwi domains (Cerutti et al., 2000). The PAZ domain has been shown to bind to 3′ ends of short RNAs and the Piwi domain is similar to the catalytic domain of RNase H. The Argonaute superfamily can easily be divided into two clades according to sequence similarity (Carmell et al., 2002). The AGO clade is found in fission yeast, plants and animals, whereas the Piwi clade is found in ciliates, slime molds and animals. Members of the AGO clade directly bind to siRNAs and miRNAs, are involved in many aspects of transcriptional (TGS) and post-transcriptional gene silencing (PTGS) in many species and are generally ubiquitously expressed (Joshua-Tor, 2006).
The expression of proteins of the Piwi clade might be restricted to germ cells in vertebrates (Carmell et al., 2007; Deng and Lin, 2002; Houwing et al., 2007; Kuramochi-Miyagawa et al., 2004; Kuramochi-Miyagawa et al., 2001), but not in Drosophila, where Piwis are also found in somatic cells associated with the germline (Brennecke et al., 2007; Cox et al., 1998; Gunawardane et al., 2007; Saito et al., 2006). In flatworms, Piwis are expressed in germ cells and somatic stem cells (Reddien et al., 2005). A number of functions have been assigned to Piwi proteins: Drosophila Piwi is required for oogenesis and stem cell maintenance (Cox et al., 1998; Cox et al., 2000; Lin and Spradling, 1997); a Tetrahymena Piwi protein, Tiwi, is required for DNA elimination in the macronucleus (Mochizuki et al., 2002); in planaria the Piwi protein SMEDWI-1 is required for stem cell regulation (Reddien et al., 2005); in vertebrates, Piwi proteins are required for germline development in the zebrafish (Houwing et al., 2007) and male germline development in the mouse (Aravin et al., 2006; Deng and Lin, 2002; Girard et al., 2006; Grivna et al., 2006; Kuramochi-Miyagawa et al., 2001). Piwi proteins are also required for transposon silencing in the Drosophila germline (Aravin et al., 2001; Savitsky et al., 2006; Vagin et al., 2006) and several studies suggest that many of the phenotypes observed in piwi mutants might be due to loss of germline integrity (Chen et al., 2007; Klattenhoff et al., 2007; Pane et al., 2007).
Recently, a class of 24-30 nucleotide RNAs has been found to interact with Piwi proteins in Drosophila, zebrafish, mice and rats and has been named Piwi-associated RNAs or piRNAs (Klattenhoff and Theurkauf, 2008). piRNA populations are complex; there are hundreds of thousands of unique piRNAs in mammals. The identification of piRNAs has improved our understanding of Piwi function, in particular with regard to transposon silencing: for example, in Drosophila the piRNAs of the flamenco locus control the gypsy retrotransposon (Brennecke et al., 2007; Desset et al., 2003; Prud’homme et al., 1995).
The best studied transposable elements of C. elegans are the DNA transposons of the Tc/mariner superfamily (Moerman and Waterston, 1984), in particular Tc1 and Tc3 (van Luenen et al., 1994; Vos et al., 1996). Tc1 and Tc3 are also the most abundant transposons of the Tc family in the N2 Bristol strain of C. elegans with 31 and 22 copies, respectively (Consortium, 1998). Tc1 and Tc3 are autonomous elements encoding a transposase specific to each element (van Luenen et al., 1993; Vos et al., 1993).
Here we demonstrate a role for C. elegans Piwi in germline development and germline transposon silencing. We identify the recently named 21U-RNAs as the piRNAs of C. elegans. We demonstrate that in C. elegans piRNAs act upstream of an endogenous siRNA pathway for Tc3 silencing. These data shed light on piRNA function.
RESULTS
Piwi is Required for Normal Germline Development
To investigate the roles of Piwi proteins in C. elegans we generated mutants lacking Piwi function. The C. elegans genome encodes two Piwi-related genes, prg-1 and prg-2. These genes are likely the result of a recent gene duplication as the genomes of the related nematode species C. briggsae and C. remanei each contain a single prg gene. PRG-1 and PRG-2 are 91% identical at the amino acid level, which suggests that they might act redundantly. We generated two deletion alleles each for prg-1 (n4357, n4503) and prg-2 (n4358, nDf57) and generated doubly mutant strains after outcrossing of the single mutants (see Supplemental Data). For brevity, we will refer to PRG-1 and PRG-2 as Piwi and we will refer to prg-1; prg-2 double mutants as piwi mutants, i.e. piwi(n4357; n4358) instead of prg-1(n4357); prg-2(n4358). prg-1 and prg-2 single mutants and piwi mutants were homozygous viable and showed neither a defect in exogenous RNAi in either the soma or germline nor defects in miRNA biogenesis or function (data not shown). However, all mutant strains showed reduced fertility (Figure S1A in Supplemental Data). Of the single mutants, prg-1 mutant animals showed the most pronounced effect with fertility reduced to 25% of that of wild-type animals (Figure S1A). The fertility defect is enhanced in piwi mutants (Figure S1A). These observations agreed with previous studies of prg-1 using RNAi and an independent prg-1 allele (Cox et al., 1998) (Yigit et al., 2006). As prg-1 RNAi had suggested a role for PRG-1 in spermatogenesis (Cox et al., 1998) we counted hermaphrodite sperm in wild-type and prg-1 and prg-2 mutant animals. Sperm counts were reduced to approximately 50% in both single and double mutants (Figure S1B). However, fertility of piwi mutants was not restored to wild-type levels by introducing wild-type sperm through mating, suggesting that Piwi function is not restricted to spermatogenesis (Figure S1C). Finally, piwi mutant germlines showed abnormal mitotic to meiotic transitions (data not shown). These data confirm a conserved role for PRG-1 and PRG-2 in C. elegans germline development (Cox et al., 1998).
The piRNAs of C. elegans Are 21 Nucleotide RNAs
In Drosophila, zebrafish, mice and rats, Piwi proteins are associated with 24-30 nt piRNAs (Aravin et al., 2006; Brennecke et al., 2007; Girard et al., 2006; Grivna et al., 2006; Gunawardane et al., 2007; Houwing et al., 2007; Lau et al., 2006; Saito et al., 2006; Vagin et al., 2006; Watanabe et al., 2006). Searching for piRNAs in C. elegans we were unable to identify an abundant class of RNAs in this size range (data not shown). We therefore searched for piRNAs among the small RNAs previously identified in C. elegans: miRNAs, tncRNAs, endogenous siRNAs and 21U-RNAs (Ambros et al., 2003; Lau et al., 2001; Lee and Ambros, 2001; Lim et al., 2003; Ruby et al., 2006). To identify candidate piRNAs we tested if any of these short RNAs were dependent on Piwi. Surprisingly, we found that a 21U-RNA, 21UR-1, which was detected in RNA from wild-type animals was absent in RNA from two independent piwi mutants by northern blotting (Figure 1A). In contrast, expression of a ubiquitous miRNA, miR-52, was unaffected. To test if 21U-RNAs were generally absent in piwi mutants we generated libraries of 5′ monophosphate small RNAs from wild-type and piwi(n4357; n4358) mutants. High-throughput sequencing identified 1398 out of 5454 previously known 21U-RNAs (Ruby et al., 2006). We also identified a large number of candidate 21U-RNAs (data not shown). 21U-RNAs were either absent or dramatically under-represented in the piwi mutant library as compared to the wild-type library (Figure 1B). The most abundant 21U-RNA in the piwi sample had 8 reads as compared to 2127 reads in wild-type. We also assessed the expression levels of other small RNAs in wild-type versus piwi mutants and found no differences in miRNA expression, tncRNA expression, or a number of siRNA species including a 26 nucleotide siRNA (Figure 1C and data not shown). These data suggest that 21U-RNAs might be the piRNAs of C. elegans. To test this hypothesis directly, we generated a rabbit polyclonal antibody against PRG-1. After immunoprecipitation of PRG-1 using αPRG-1 serum from wild-type C. elegans adult whole cell extracts we detected 21U-RNAs by RT-PCR, but were not able to detect them from piwi mutant extracts or when using pre-immune serum (Figure 1D). Overall, 21U-RNAs are 100-fold enriched in PRG-1 immunoprecipitates ((Batista et al., 2008) and Discussion). As the high-throughput sequencing data suggested that 21U-RNAs were dramatically reduced in piwi mutants, we independently quantified the expression of seven 21U-RNAs by quantitative RT-PCR. As shown in Figure 1E, while the expression of a number of 21U-RNAs is dramatically reduced, some 21U-RNAs, including 21UR-1 were still detected in piwi mutants. For 21UR-1 we verified that the signal was specific by cloning and sequencing of RT-PCR products (data not shown). Taken together these data suggest that the 21U-RNAs are the piRNAs of C. elegans and we will refer to them as piRNAs below.
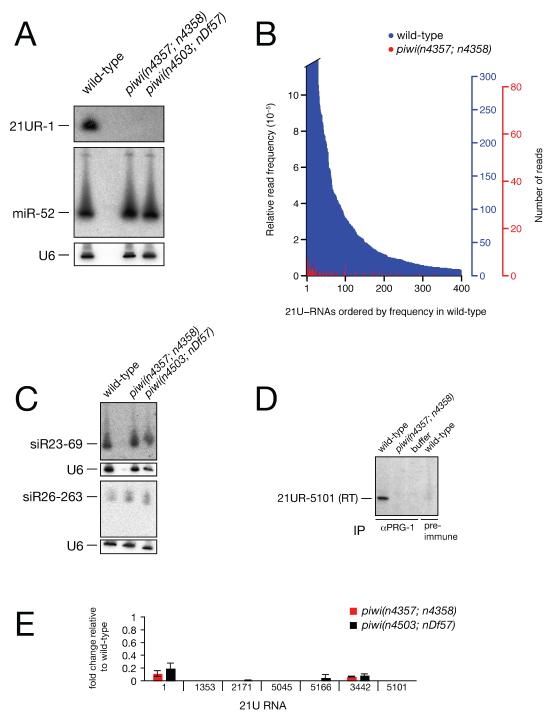
(A) Northern blot showing that 21UR-1 is not detected in RNA (40 μg) isolated from two independent piwi double-mutants: piwi(n4357; n4358) and piwi(n4503; nDf57), whereas miR-52 is expressed in both mutants (same blot re-probed). Antisense DNA probes were used for 21UR-1 and miR-52. A U6 northern blot is shown as loading control. piwi(n4357; n4358) is an abbreviation for prg-1(n4357); prg-2( n4358). piwi(n4503; nDf57) is an abbreviation for prg-1(n4503); prg-2( nDf57).
(B) High-throughput sequencing reveals that the expression of many 21U-RNAs is dramatically reduced in piwi mutants. 21U-RNAs cloned from 5′ dependent wild-type and piwi(n4357; n4358) mutant libraries. Frequencies are shown for wild-type (blue) and piwi mutant (red) for the 400 most abundant 21U-RNAs in wild-type, plotted in the order of their wild-type frequency. Read frequencies were obtained by dividing the number of reads for a given 21U-RNA by the total number of reads from the same library (left-hand y-axis). The corresponding absolute number of reads are indicated in the right-hand y-axes. 21U-RNAs for which frequencies are shown include the 21U-RNA with most reads in the piwi mutant library (21UR-3224), which was sequenced 8 times in the piwi mutant and 2,127 times in wild-type.
(C) Expression of a 23 nucleotide antisense RNA (siR23-69) and a 26 nucleotide antisense RNA (siR26-263) is not affected in piwi mutants (northern blotting, 40 μg total RNA, antisense DNA probes).
(D) Immunoprecipitation followed by RT-PCR for 21UR-5101 reveals that 21U-RNAs are associated with PRG-1 in C. elegans extracts.
(E) Quantitative RT-PCR of seven 21U-RNAs demonstrates that Piwi is not essential for 21U-RNA biogenesis. Total RNA was extracted from 12 hour adult C. elegans. Expression levels shown are relative to levels in wild-type RNA. miR-52 expression was used as an internal control. Data are from three independent biological replicates. Error bars represent standard error of the mean.
piRNA Biogenesis is Independent of Many Genes in Other Small RNA Pathways
To identify additional genes involved in piRNA pathways in C. elegans we tested a panel of genes using mutants and RNAi for their effect on piRNA expression by northern blotting or quantitative RT-PCR (Figure 2 and Table S1). First, we checked piRNA expression in prg-1 and prg-2 single mutants. Using three independent alleles and RNAi experiments we found that only PRG-1 but not PRG-2 is required for piRNA expression (Figure 2 and Table S1). Next, we tested other Argonaute proteins; these included RDE-1, which is required for exogenous RNAi (Tabara et al., 1999), ALG-1 and ALG-2, which are redundantly required for miRNA function (Grishok et al., 2001), ERGO-1, SAGO-1, SAGO-2 and a group of five “MAGO” Argonaute proteins associated with endogenous siRNA pathways (Yigit et al., 2006). None of these Argonaute proteins were required for piRNA expression emphasizing the specificity of the requirement for PRG-1. In addition, we tested a number of other proteins involved in small RNA biology in C. elegans including all four known RNA-dependent RNA polymerases (RdRPs), RRF-1, RRF-2, RRF-3 and EGO-1 (Simmer et al., 2002; Smardon et al., 2000), none of which were required for piRNA expression. We also tested if the RNase III enzyme DCR-1, which is essential for the generation of siRNAs and germline development (Grishok et al., 2001; Ketting et al., 2001; Knight and Bass, 2001), is required for piRNA expression. As dcr-1 mutants are sterile, these experiments were carried out using homozygous mutant animals derived from heterozygous mothers or using RNAi. We found that DCR-1 is not required for piRNA expression (Figure 2B, Table S1). These data were also confirmed by high-throughput sequencing in dcr-1 mutants (Figure 2C). These data suggest that piRNAs are independent of many proteins involved in other small RNA pathways, including DCR-1.

(A) 21UR-1 northern blotting of total RNA of wild-type and mutant young adult C. elegans. In the case of alg-1; alg-2(RNAi), alg-1 mutant L1 larvae were transferred to alg-2 RNAi feeding plates and young adult animals were harvested. A U6 northern blot is shown as loading control. See Table S1 in Supplemental Data for quantification of these results.
(B) 21UR-1 northern blotting of total RNA of wild-type and mutant young adult C. elegans. dcr-1 mutant animals used were homozygous animals derived from heterozygous mothers. To test for loss of DCR-1 activity in dcr-1 mutants, let-7 miRNA and pre-miRNA is shown. A U6 northern blot is shown as loading control.
(C) Distribution of 21U-RNAs on chromosome IV as detected by high-throughput sequencing of 5′ dependent wild-type, piwi(n4357; n4358), dcr-1 and mut-7 mutant libraries. Frequencies for a given 21U-RNA and locus were obtained by correcting the number of reads for multiple alignments and dividing by the total number of reads from the same library. Cumulative frequencies were plotted for non-overlapping 100 kb windows along chromosome IV.
Piwi and piRNAs are Restricted to the Male and Female Germline
We found that 21UR-1 expression is developmentally regulated with highest expression detected in young adults and adults by northern blot (Figure 3A). Additional piRNAs showed a similar expression pattern, as determined by quantitative RT-PCR (e.g. 21UR-5101, Figure 3B). Interestingly, prg-1 and prg-2 mRNAs show a similar pattern (Figure 3C, Figure S2). The observed expression pattern for piRNAs and piwi mRNA is consistent with expression in the germline. As Piwi proteins and piRNAs are thought to be exclusively expressed in germ cells in vertebrates (Carmell et al., 2007; Deng and Lin, 2002; Kuramochi-Miyagawa et al., 2004; Kuramochi-Miyagawa et al., 2001), but not in Drosophila (Brennecke et al., 2007; Cox et al., 1998; Gunawardane et al., 2007; Saito et al., 2006), we decided to test their germline restriction in C. elegans using a set of temperature-sensitive mutants (Figure 3D). 21UR-1 was absent from glp-4(bn2ts) and glp-1(e2144lf) mutant animals at the restrictive temperature, which are devoid of germ cells (Beanan and Strome, 1992). However, 21UR-1 levels were unchanged in glp-1(ar202gf,ts) mutants at the restrictive temperature, which are highly enriched in germ cells that have not yet entered meiosis (Pepper et al., 2003). 21UR-1 RNA was also detected in RNA from fem-1(hc17ts) (Kimble et al., 1984; Nelson et al., 1978) and fem-3(q22sd,ts) (Barton et al., 1987) mutants at the restrictive temperature that are devoid of sperm and oocytes respectively. The same restricted pattern was also observed for piwi mRNA (Figure 3E and Figure S2). Together these data suggested that C. elegans Piwi and piRNAs are restricted to the male and female germline.
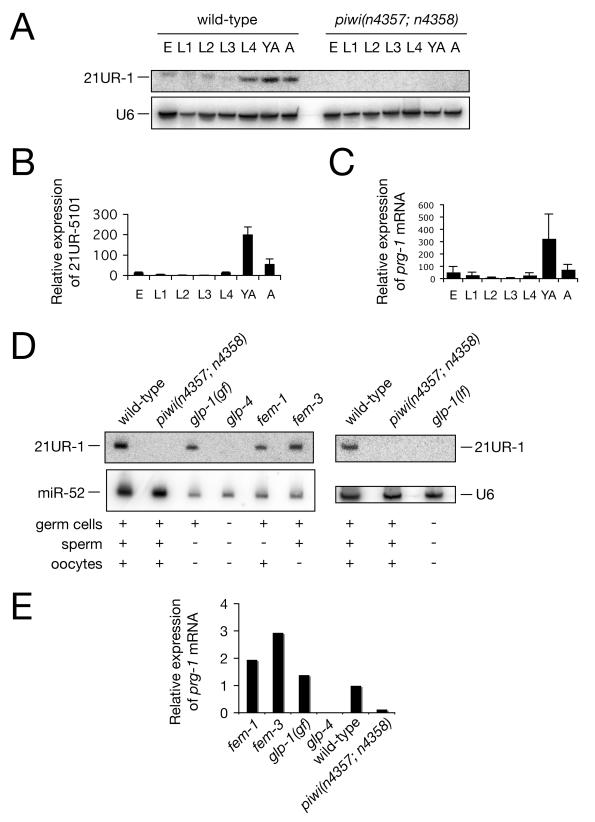
(A) Profile of 21U-R1 expression during development. E, embryo. L1-L4, larval stages 1-4. YA, 12-hour adult. A, 24-48-hour adult. A U6 northern blot is shown as loading control.
(B) Quantitative RT-PCR of 21UR-5101. miR-52 expression was used as an internal control. Data are from three independent biological replicates. Error bars represent standard error of the mean.
(C) Quantitative RT-PCR of prg-1 mRNA. Actin mRNA was used as an internal control. Data are from three independent biological replicates. Error bars represent standard error of the mean.
(D) Northern blot showing that piRNA expression is restricted to the male and female germline. glp-1(gf), glp-1(lf), glp-4, fem-1 and fem-3 L1 larvae were grown to 12-hour adult stage at 25°C. 20 μg of total RNA was loaded in each lane. U6 and miR-52 northern blots are shown as loading controls.
(E) Quantitative RT-PCR of prg-1 mRNA. Actin mRNA was used as an internal control.
Piwi is Specifically Required for Silencing of Tc3 DNA Transposons
As Piwi proteins are required for transposon silencing in Drosophila (Aravin et al., 2001; Savitsky et al., 2006; Vagin et al., 2006) we examined transposon silencing in piwi mutants in C. elegans. We focused on the two most abundant DNA transposons in C. elegans, Tc1 and Tc3 (Figure 4A, B) (Consortium, 1998). First, we examined Tc1 transposase expression by quantitative RT-PCR using primers specific for 15 Tc1 loci (Figure 4C). We observed an approximately 50 fold increase in Tc1 transposase mRNA levels in mut-7 mutants, which show an elevated rate of transposition of Tc/mariner elements in the germline (Ketting et al., 1999). We did not, however, observe an increase in Tc1 transposase mRNA in piwi mutants. Surprisingly then, we found increased Tc3 transposase mRNA in two independent piwi mutants and three independent prg-1 mutants using primer pairs specific for 20 Tc3 loci (Figure 4D). Next, we assayed the germline excision rate of Tc1 and Tc3 transposons directly by the phenotypic reversion of unc-22 transposon insertion alleles (Ketting et al., 1999). As shown in Table 1, unc-22 reversion rates of Tc1, Tc3 or Tc4 insertion alleles were less than 10−6 in an otherwise wild-type background. In mut-7 mutant animals the reversion rate was increased 100 fold, as reported previously (Ketting et al., 1999). Strikingly, in prg-1 and piwi mutants Tc1 and Tc4 excision rates were not affected, but reversion of the Tc3 allele was increased 100 and 1000 fold, respectively (Table 1). These data suggest that PRG-1 and Piwi are powerful and specific suppressors of Tc3 transposition in the germline of C. elegans. As we found that Piwi and MUT-7 were both involved in germline transposon silencing we wondered if these proteins shared additional functions. We therefore tested if Piwi, like MUT-7, is required for germline transgene silencing (Kim et al., 2005). However, we found that germline transgene silencing is intact in piwi mutants (Figure S3). These data suggest that Piwi and MUT-7 have overlapping and distinct roles in the C. elegans germline.
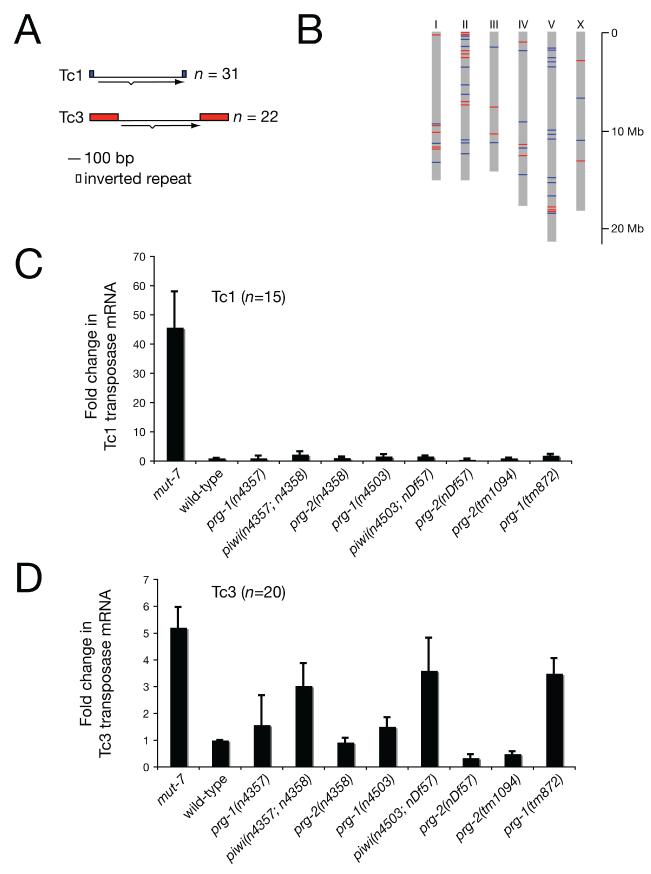
(A) Diagram of the genomic structure of the two most common DNA transposons in C. elegans, Tc1 and Tc3. Tc1 and Tc3 are flanked by inverted repeats and encode a single spliced transcript for transposase. bp, base pairs. n, number of copies in the wild-type strain N2.
(B) Distribution of Tc1 and Tc3 transposons in the C. elegans genome.
(C), (D) Quantitative RT-PCR of Tc1 or Tc3 transposase mRNA. As the genomic copies of Tc1 and Tc3 have minor sequence variations, the number of transposon loci amplified by each qRT-PCR primer pair are shown (n). Actin mRNA was used as an internal control. Expression levels shown are relative to levels from wild-type RNA. Data are from three independent biological replicates. Error bars represent standard error of the mean.
Table 1
PIWI is required to inhibit Tc3 transposition in the germline
|
unc-
22::Tc1 |
unc-
22::Tc3 |
unc-
22::Tc4 | |
|---|---|---|---|
| wild-type | <10−6 a | <10−6 a | <10−6 a |
| prg-1(n4357) | 10−6 | 10−4 | 10−6 |
| prg-1(tm872) | n.d. | 10−3 | n.d. |
| piwi(n4357; n4358) | 10−6 | 10−3 | 10−6 |
| piwi(n4503; nDf57) | n.d. | 10−3 | n.d. |
| mut-7 | 10−4 | 10−4 | 10−4 |
Transposition rates were estimated by scoring unc-22 reversion rates. Germline excision was verified by scoring the progeny of revertants. Animals were grown at 20°C.
Piwi Acts Upstream of an Endogenous siRNA Pathway
Previous work demonstrated a role for MUT-7 (Ketting et al., 1999) and endogenous siRNAs (Sijen and Plasterk, 2003) in germline transposon silencing. Our findings suggested a role for Piwi and piRNAs in the same process. We therefore wanted to determine how these two pathways related to each other. First, we tested if MUT-7 and endogenous siRNAs might act upstream of piRNA expression. However, we found that piRNA levels were not affected in mut-7 mutants by northern blotting and high-throughput sequencing (Figure 2C, Figure 5A). Next, we tested if Piwi and piRNAs might act upstream of MUT-7 and endogenous siRNAs. We searched for piRNAs mapping to Tc/mariner transposons and found that no known piRNAs mapped directly to Tc1 or Tc4. However, a single piRNA, 21UR-139, mapped to the sense strand of the transposase gene of 20 out of the 22 Tc3 insertions in the N2 genome (Table S2, Figure 5B). A second 21U-RNA in the Tc3 locus has also been identified ((Batista et al., 2008) and Discussion). To identify endogenous siRNAs mapping to Tc transposons we generated 5′ independent small RNA libraries from wild type and piwi mutants, as we expected these RNAs to carry 5′ di- or triphosphates (Pak and Fire, 2007; Ruby et al., 2006; Sijen et al., 2007). High-throughput sequencing identified a large number of endogenous small RNAs that map to Tc1 and Tc3 loci (Figure 5B, Supplemental Data). Interestingly, small RNAs mapping to the terminal inverted repeat (TIR) and the transposase open reading frame (ORF) of Tc3 were nearly absent in piwi mutants, but those mapping to Tc1 were unaffected. We confirmed these observations for small RNAs antisense to the transposase transcripts using an RNase protection assay (Figure 5C). While Tc1 and Tc3 antisense siRNAs were dependent on MUT-7, only Tc3 siRNAs were also dependent on Piwi. Unfortunately, we were unable to detect endogenous small RNAs from the TIRs using the same assay (data not shown). These data suggest that Piwi and piRNAs act upstream of a MUT-7 dependent endogenous siRNA pathway for Tc3 silencing. This finding raised the possibility that additional piRNAs are functionally linked to endogenous siRNAs. To test this hypothesis we examined piRNA loci for nearby endogenous siRNAs. Interestingly, we observed a local positional bias of antisense siRNAs downstream of piRNA loci, and their accumulation appears to be dependent on Piwi (Figure 6A and Discussion).
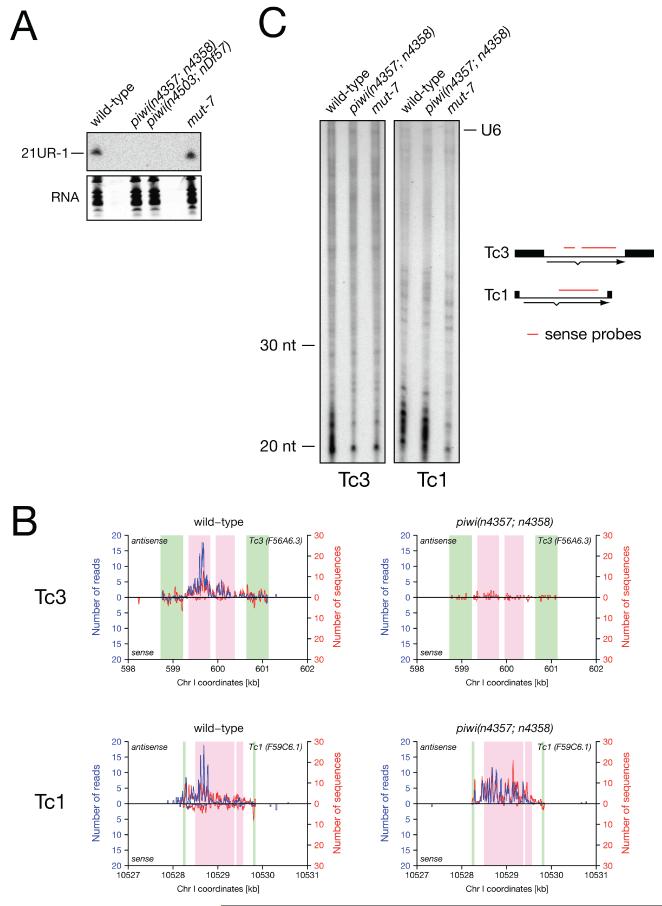
(A) piRNA expression is independent of MUT-7. 21UR-1 northern blotting of total RNA of wild-type and mutant young adult C. elegans. Total RNA is shown as loading control (GelStar).
(B) Tc3 associated small RNAs are absent in piwi mutants. Small RNAs mapping to the loci of Tc3 (top) and Tc1 (bottom) on chromosome I as identified by high-throughput sequencing of 5′ independent wild-type (left) and piwi mutant (right) libraries. Inverted repeat and exon sequences are indicated in green and pink respectively. The number of aligned sequence reads (blue) and number of aligned unique sequences (red) were plotted for each base pair position, with the top and bottom graph in each panel corresponding to the antisense and sense strand relative to the transposase transcript. Read and sequence counts were corrected for multiple alignments to the genome. The total number of reads from wild-type and piwi mutant libraries were comparable (2,963,895 and 3,017,027 of reads with perfect matches to the reference genome respectively).
(C) Tc3 transposase antisense siRNAs are dramatically reduced in piwi mutants. RNase protection assay using sense fragments of Tc1 and Tc3 transposase. Sense siRNAs were not detected above background levels using this assay (see Figure S4 in Supplemental Data). U6 was used as an internal control, but its concentration had to be titrated down due to some interference with small RNA detection (Figure S5).
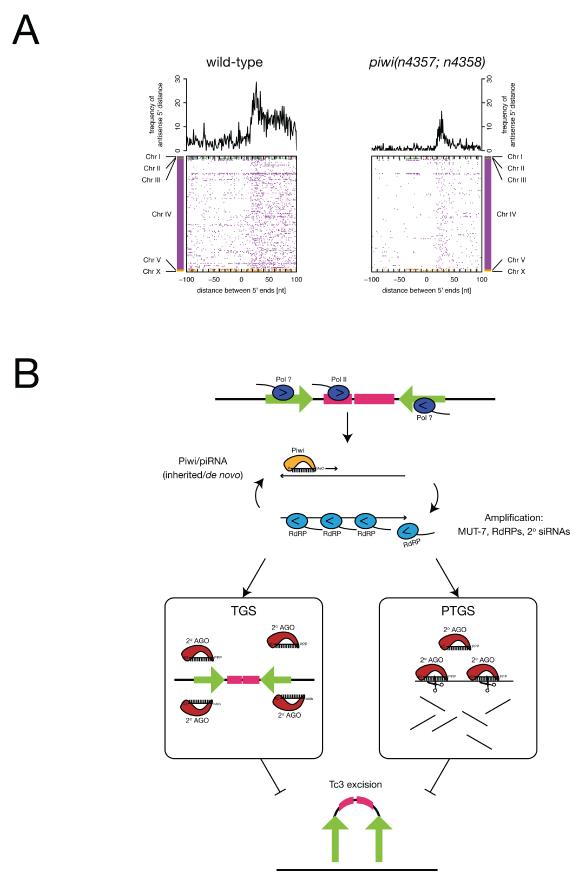
(A) Small RNAs mapping to the opposite strand of nearby 21U-RNA loci show a preference for locations downstream of the 21U-RNA locus and were reduced in the piwi mutant (right) as compared to wild-type (left). Proximate small RNAs on the same strand as the 21U-RNA were also reduced in the piwi mutant (Figure S7). (Bottom) Rows correspond to 6,021 21U-RNA loci, ordered by genomic position with colors representing different chromosomes. For a given row (21U-RNA locus) dots correspond to the relative position of nearby antisense small RNAs as defined by the distance of the 5′ end of the cloned small RNA relative to the 21U-RNA 5′ end. (Top) Shown is the frequency of distances between 5′ ends of 21U-RNAs and antisense small RNAs. Frequencies were based on the normalized number of loci for 5′ unique sequences.
(B) A speculative model of the role of Piwi in Tc3 silencing. TGS, transcriptional gene silencing. PTGS, post-transcriptional gene silencing. See Discussion for an explanation.
DISCUSSION
21U-RNAs are piRNAs With Surprising Features
We identify the piRNAs of C. elegans as 21 nucleotide RNAs (Figure 1). Our observations are strongly supported by the finding that PRG-1 immunoprecipitates are 100-fold enriched for 21U-RNAs (Batista et al., 2008). The C. elegans 21U-RNAs/piRNAs share several features with piRNAs of other species. First, C. elegans piRNAs have 5′ uridines. This bias is common to piRNAs from Drosophila and vertebrates (Klattenhoff and Theurkauf, 2008). Second, several C. elegans piRNAs have a 5′ monophosphate and a 3′ modification blocking the 2′ or 3′ oxygen, likely a 2′-O-methyl group (data not shown, (Ruby et al., 2006)). This modification is shared by piRNAs from Drosophila (Horwich et al., 2007; Saito et al., 2007; Vagin et al., 2006), zebrafish (Houwing et al., 2007) and mice (Kirino and Mourelatos, 2007; Ohara et al., 2007). However, 3′ ends with 2′-O-methyl groups are not a unique feature of piRNAs as plant miRNAs and siRNAs and Drosophila siRNAs also carry this modification (Ghildiyal et al., 2008; Li et al., 2005).
The piRNAs of Drosophila, zebrafish, mice and rats are 24-30 nucleotides in length, and as such are distinct from miRNAs or endogenous siRNAs (Klattenhoff and Theurkauf, 2008). The reason for the size difference between 21U-RNAs and piRNAs of other organisms is unclear. Another distinction of C. elegans piRNAs is their genomic location and organization: while Drosophila and vertebrate piRNAs map to regions devoid of protein-coding genes (Klattenhoff and Theurkauf, 2008), some C. elegans piRNAs are interspersed with protein-coding genes (Ruby et al., 2006).
Biogenesis of piRNAs
The striking difference in genomic organization of C. elegans piRNA clusters compared to vertebrates and Drosophila might suggest a divergent mechanism of biogenesis. We found that C. elegans piRNAs are Dicer independent (Figure 2B), as are Drosophila and zebrafish piRNAs (Houwing et al., 2007; Vagin et al., 2006). Taken together with the strong local piRNA strand bias in all three species (Brennecke et al., 2007; Houwing et al., 2007; Ruby et al., 2006; Vagin et al., 2006) it is unlikely that piRNAs are generated through a double-stranded RNA intermediate. Instead, piRNAs might be generated directly as primary transcripts. In C. elegans, this could be achieved by an RdRP. However, two observations argue against this hypothesis. First, we found no evidence for the dependence of piRNA expression on RdRPs (Table 1), although we cannot exclude redundancy. Second, piRNAs lack 5′ triphosphates, a hallmark of other RdRP derived small RNAs. We therefore favor a model in which piRNAs are derived from longer primary transcripts as is the case for miRNAs (Bartel, 2004). Interestingly, we show that Piwi is not essential for piRNA biogenesis, but that piRNA levels are dramatically reduced in piwi mutants (Figure 1E). This is analogous to C. elegans miRNAs, whose levels are reduced but are not absent in alg-1; alg-2 Argonaute mutants (Grishok et al., 2001). We therefore postulate that Piwi is not an essential component of the piRNA biogenesis pathway, but is required for piRNA accumulation or stability.
The piRNA genomic organization in Drosophila suggests that whole piRNA clusters could be transcribed as single primary transcripts and then processed into many mature piRNAs (Brennecke et al., 2007). Our finding that 21U-RNAs are piRNAs now suggests an alternative model. Many 21U-RNA loci are associated with a conserved upstream motif containing an eight nucleotide core consensus sequence CTGTTTCA ((Ruby et al., 2006) and Supplemental Data). This motif is located approximately 38 bases upstream of the base corresponding to the 5′ uridine of the 21U-RNA. Ruby et al. have postulated that this motif is part of a 21U-RNA promoter. We conclude that piRNAs might be transcribed as individual transcripts in C. elegans and perhaps other species. Alternatively, these motifs might also be involved post-transcriptionally in piRNA biogenesis. Having linked a conserved motif to piRNAs in C. elegans it will be important to search for such motifs in other species. Perhaps such a motif might be found only for a subset of “primary” piRNAs in Drosophila or vertebrates.
No Evidence for Ping-Pong in C. elegans
Recently, a compelling model for piRNA production in Drosophila named the Ping-Pong amplification loop has been put forward (Brennecke et al., 2007; Gunawardane et al., 2007). In this model Piwi/Aub is bound by a primary piRNA with a 5′ uridine. Target slicing by this complex followed by 3′ end processing results in the generation of a secondary piRNA that is antisense to the primary piRNA, overlaps by ten complementary nucleotides, and has an adenine at position 10. The loop is completed by AGO3, the third Piwi protein of Drosophila, which binds the secondary piRNA and given a suitable template, in turn generates piRNAs with 5′ uridines. This model is supported by the finding that Piwi/Aub bound piRNAs have a strong bias for 5′ uridine and AGO3 bound piRNAs a strong bias for adenine at position 10 (Brennecke et al., 2007; Gunawardane et al., 2007). To test the Ping-Pong hypothesis in C. elegans, we analyzed the overlap among 21U-RNAs and between 21U-RNAs and other C. elegans small RNAs (Figure 6A). We found few cases of overlap and no adenine peak at position 10 (Figure S6). We conclude that there is currently no evidence for a role of Ping-Pong to generate piRNAs or to couple piRNAs to endogenous siRNA pathways in C. elegans.
Piwi and piRNAs Act Upstream of Secondary siRNA Pathways in Tc3 Silencing
We suggest that C. elegans and Drosophila share primary piRNAs, but while amplification of small RNAs in Drosophila might be achieved through a Ping-Pong amplification loop, in C. elegans amplification might be achieved through secondary siRNAs and RdRPs (Figure 6B). We demonstrate that Tc3 transposon silencing, and associated siRNAs depend on Piwi and MUT-7. However, we cannot directly implicate a specific piRNA in Tc3 transposon silencing. One attractive candidate might be 21UR-139, which maps to the Tc3 transposase ORF and might act in cis. A second 21U-RNA also maps to the Tc3 locus (Batista et al., 2008). Both are sense relative to the transposase gene. We propose a speculative model to explain how sense piRNAs might be required for antisense siRNA production at the Tc3 locus. Piwi loaded with sense piRNAs might stimulate RdRP activity using Tc3 antisense transcripts as templates, in a manner analogous to siRNAs in RNAi amplification (Baulcombe, 2007). The sense transcripts would be targeted by antisense siRNAs, and act as a template for a second round of small RNA-stimulated RdRP activity (Figure 6B). Such a loop would maintain high levels of Tc3 siRNAs. In the case of the Tc3 transposase this might lead to PTGS against the mRNA. This is in agreement with the low number of siRNAs mapping to the intron as compared to the two exons of the Tc3 transposase (Figure 5B). In the case of the Tc3 TIR, this might involve chromatin-mediated TGS. If the TIR-associated siRNAs are functionally distinct from the siRNAs mapping to the Tc3 transposase ORF, these two pathways might be separable. Indeed, Batista et al. (2008) observed that prg-1 mutants lack only TIR-associated siRNAs, but not siRNAs associated with the transposase ORF (Batista et al., 2008). Together, these data suggest that PRG-1 and PRG-2 might have functionally distinct roles in Tc3 silencing. This is particularly intriguing as PRG-1, but not PRG-2 is required for piRNA accumulation (Figure 2A).
The amplification loop we propose is sequence independent, given an initiating 21U-RNA. Therefore the link between piRNA and siRNA pathways might not be restricted to Tc3. Indeed, we observe a positional bias for antisense siRNAs close to 21U-RNAs to map downstream of the 21U-RNA locus (Figure 6A). piRNA and siRNA pathways might be linked in other species too. Drosophila and vertebrates have lost secondary siRNAs and RdRPs, which are found in plants, yeast and C. elegans. However, recently endogenous siRNAs in mouse oocytes were shown to map to the same loci as piRNAs (Tam et al., 2008).
Other Roles of Piwi in C. elegans?
We demonstrate a role for Piwi in inhibiting Tc3 mobility. However, it seems unlikely that the defects in germline development observed in piwi mutants are solely due to Tc3 activity. It has been proposed that mobile element excision and excessive DNA breaks result in meiotic catastrophe in piwi mutants in Drosophila (Klattenhoff and Theurkauf, 2008). We cannot exclude the possibility that other mobile elements in addition to Tc3 are hyper-activated in piwi mutants in C. elegans, with similar consequences. Alternatively, Piwi/piRNAs might be involved in gene regulation during germline development in C. elegans. Surveying whole animal gene expression in wild-type and piwi mutant adult C. elegans using Affymetrix mRNA profiling we did not observe striking changes in gene expression (data not shown). However, changes might only be revealed by studying gene expression in germ cells. Indeed, a recent sudy suggests that the expression of a subset of mRNAs expressed during spermatogenesis is deregulated in male gonads isolated from prg-1 mutants (Wang and Reinke, 2008). The striking clustering of piRNAs on chromosome IV (Ruby et al., 2006) raises a number of interesting questions. Do piRNAs act in cis or do they act in trans, analogous to miRNA function? If piRNAs act in trans, what are the rules for piRNA:piRNA target interaction? Do piRNAs always act through siRNA pathways as we have demonstrated for Tc3? Having identified piRNAs in C. elegans we hope to address these questions using the powerful genetic tools that this organism offers.
EXPERIMENTAL PROCEDURES
C. elegans Strains and Culture
See Supplemental Data and Tables S3, S4 for strain information and culture conditions.
High-Throughput Sequencing and Data Analysis
We generated 5′-dependent libraries for N2 (2,719,949 reads), piwi(n4357; n4358) (764,960 reads), mut-7 (3,475,722 reads), dcr-1 (149,997 reads), and 5′-independent libraries for N2 (2,963,895 reads) and piwi(n4357; n4358) (3,017,027 reads) that were sequenced using the Illumina/Solexa platform. See Supplemental Data for additional information.
RNAi Experiments
RNAi experiments were carried out as reported previously (Fire et al., 1998; Timmons et al., 2001). Bacterial strains carrying plasmids expressing double-stranded RNA for the gene of interest were obtained from the Ahringer Laboratory genome-wide RNA library (Fraser et al., 2000; Kamath et al., 2003). All RNAi library inserts were confirmed by sequencing.
Transposon Excision Experiments
Transposon excision assays were carried out as described previously (Ketting et al., 1999).
ACKNOWLEDGMENTS
We thank Rob Shaw and Na An for strain management. We thank Andrew Hellman and Bob Horvitz for help generating prg-1 and prg-2 deletion strains. We thank Ericka Havecker, David Baulcombe, Leonie Kamminga, James Hadfield and Thomas Down for help with high-throughput sequencing. We thank Fuchou Tang for help with qRT-PCR assays. We thank Julie Ahringer, Jane Hubbard, Alla Grishok, Shohei Mitani and the Caenorhabditis Genetics Center (funded by the NIH National Center for Research Resources), University of Minnesota, Twin Cities, MN, USA for providing C. elegans strains. We thank Pedro Batista and Craig Mello for discussions on the manuscript and sharing of unpublished data. M.P.B. was supported by the Medical Research Council (MRC, UK). J.R.W. was supported by a Cancer Research UK studentship. L.D.G. was supported by an EPSRC fellowship (UK). S.T. is supported by Cancer Research UK and is a Royal Society-Wolfson Research Merit Award holder. This work was supported by a Cancer Research UK Programme Grant to E.A.M. (C13474).
Footnotes
Supplemental Data Supplemental data include a document with Supplemental Experimental Procedures, eight figures and five tables.
REFERENCES
- Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol. 2003;13:807–818. [Abstract] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. [Abstract] [Google Scholar]
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11:1017–1027. [Abstract] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. [Abstract] [Google Scholar]
- Barton MK, Schedl TB, Kimble J. Gain-of-function mutations of fem-3, a sex-determination gene in Caenorhabditis elegans. Genetics. 1987;115:107–119. [Europe PMC free article] [Abstract] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb J, Fahlgren N, Kasschau KD, Chiang R, Chaves DA, Gu W, Duan S, Conte D, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for germline development in C. elegans. Cell. 2008 (under review) [Europe PMC free article] [Abstract] [Google Scholar]
- Baulcombe DC. Molecular biology. Amplified silencing. Science. 2007;315:199–200. [Abstract] [Google Scholar]
- Beanan MJ, Strome S. Characterization of a germ-line proliferation mutation in C. elegans. Development. 1992;116:755–766. [Abstract] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. [Abstract] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc’his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–514. [Abstract] [Google Scholar]
- Carmell MA, Xuan Z, Zhang MQ, Hannon GJ. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16:2733–2742. [Abstract] [Google Scholar]
- Cerutti L, Mian N, Bateman A. Domains in gene silencing and cell differentiation proteins: the novel PAZ domain and redefinition of the Piwi domain. Trends Biochem Sci. 2000;25:481–482. [Abstract] [Google Scholar]
- Chen Y, Pane A, Schüpbach T. Cutoff and aubergine mutations result in retrotransposon upregulation and checkpoint activation in Drosophila. Curr Biol. 2007;17:637–642. [Europe PMC free article] [Abstract] [Google Scholar]
- Consortium C.e.S. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. [Abstract] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12:3715–3727. [Europe PMC free article] [Abstract] [Google Scholar]
- Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–514. [Abstract] [Google Scholar]
- Deng W, Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2:819–830. [Abstract] [Google Scholar]
- Desset S, Meignin C, Dastugue B, Vaury C. COM, a heterochromatic locus governing the control of independent endogenous retroviruses from Drosophila melanogaster. Genetics. 2003;164:501–509. [Europe PMC free article] [Abstract] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. [Abstract] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. [Abstract] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, et al. Endogenous siRNAs Derived from Transposons and mRNAs in Drosophila Somatic Cells. Science. 2008 [Europe PMC free article] [Abstract] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. [Abstract] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. [Abstract] [Google Scholar]
- Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. [Europe PMC free article] [Abstract] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. [Abstract] [Google Scholar]
- Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol. 2007;17:1265–1272. [Abstract] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. [Abstract] [Google Scholar]
- Joshua-Tor L. The Argonautes. Cold Spring Harb Symp Quant Biol. 2006;71:67–72. [Abstract] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. [Abstract] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. [Europe PMC free article] [Abstract] [Google Scholar]
- Ketting RF, Haverkamp TH, van Luenen HG, Plasterk RH. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell. 1999;99:133–141. [Abstract] [Google Scholar]
- Kim JK, Gabel HW, Kamath RS, Tewari M, Pasquinelli A, Rual JF, Kennedy S, Dybbs M, Bertin N, Kaplan JM, et al. Functional genomic analysis of RNA interference in C. elegans. Science. 2005;308:1164–1167. [Abstract] [Google Scholar]
- Kimble J, Edgar L, Hirsh D. Specification of male development in Caenorhabditis elegans: the fem genes. Dev Biol. 1984;105:234–239. [Abstract] [Google Scholar]
- Kirino Y, Mourelatos Z. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat Struct Mol Biol. 2007;14:347–348. [Abstract] [Google Scholar]
- Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA, Theurkauf WE. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev Cell. 2007;12:45–55. [Abstract] [Google Scholar]
- Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135:3–9. [Abstract] [Google Scholar]
- Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. [Europe PMC free article] [Abstract] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. [Abstract] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Yomogida K, Kuroiwa A, Tadokoro Y, Fujita Y, Sato M, Matsuda Y, Nakano T. Two mouse piwi-related genes: miwi and mili. Mech Dev. 2001;108:121–133. [Abstract] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. [Abstract] [Google Scholar]
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. [Abstract] [Google Scholar]
- Lee RC, Ambros VR. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. [Abstract] [Google Scholar]
- Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol. 2005;15:1501–1507. [Abstract] [Google Scholar]
- Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. [Europe PMC free article] [Abstract] [Google Scholar]
- Lin H, Spradling AC. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. [Abstract] [Google Scholar]
- Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell. 2002;110:689–699. [Abstract] [Google Scholar]
- Moerman DG, Waterston RH. Spontaneous unstable unc-22 IV mutations in C. elegans var. Bergerac. Genetics. 1984;108:859–877. [Europe PMC free article] [Abstract] [Google Scholar]
- Nelson GA, Lew KK, Ward S. Intersex, a temperature-sensitive mutant of the nematode Caenorhabditis elegans. Dev Biol. 1978;66:386–409. [Abstract] [Google Scholar]
- Ohara T, Sakaguchi Y, Suzuki T, Ueda H, Miyauchi K, Suzuki T. The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat Struct Mol Biol. 2007;14:349–350. [Abstract] [Google Scholar]
- Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. [Abstract] [Google Scholar]
- Pane A, Wehr K, Schüpbach T. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell. 2007;12:851–862. [Europe PMC free article] [Abstract] [Google Scholar]
- Pepper AS, Killian DJ, Hubbard EJ. Genetic analysis of Caenorhabditis elegans glp-1 mutants suggests receptor interaction or competition. Genetics. 2003;163:115–132. [Europe PMC free article] [Abstract] [Google Scholar]
- Prud’homme N, Gans M, Masson M, Terzian C, Bucheton A. Flamenco, a gene controlling the gypsy retrovirus of Drosophila melanogaster. Genetics. 1995;139:697–711. [Europe PMC free article] [Abstract] [Google Scholar]
- Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sánchez Alvarado A. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005;310:1327–1330. [Abstract] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. [Abstract] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214–2222. [Europe PMC free article] [Abstract] [Google Scholar]
- Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi-interacting RNAs at their 3′ ends. Genes Dev. 2007;21:1603–1608. [Europe PMC free article] [Abstract] [Google Scholar]
- Savitsky M, Kwon D, Georgiev P, Kalmykova A, Gvozdev V. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 2006;20:345–354. [Europe PMC free article] [Abstract] [Google Scholar]
- Sijen T, Plasterk RH. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature. 2003;426:310–314. [Abstract] [Google Scholar]
- Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. [Abstract] [Google Scholar]
- Simmer F, Tijsterman M, Parrish S, Koushika SP, Nonet ML, Fire A, Ahringer J, Plasterk RH. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol. 2002;12:1317–1319. [Abstract] [Google Scholar]
- Smardon A, Spoerke JM, Stacey SC, Klein ME, Mackin N, Maine EM. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr Biol. 2000;10:169–178. [Abstract] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. [Abstract] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008 [Europe PMC free article] [Abstract] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. [Abstract] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. [Abstract] [Google Scholar]
- van Luenen HG, Colloms SD, Plasterk RH. Mobilization of quiet, endogenous Tc3 transposons of Caenorhabditis elegans by forced expression of Tc3 transposase. EMBO J. 1993;12:2513–2520. [Europe PMC free article] [Abstract] [Google Scholar]
- van Luenen HG, Colloms SD, Plasterk RH. The mechanism of transposition of Tc3 in C. elegans. Cell. 1994;79:293–301. [Abstract] [Google Scholar]
- Vos JC, De Baere I, Plasterk RH. Transposase is the only nematode protein required for in vitro transposition of Tc1. Genes Dev. 1996;10:755–761. [Abstract] [Google Scholar]
- Vos JC, van Luenen HG, Plasterk RH. Characterization of the Caenorhabditis elegans Tc1 transposase in vivo and in vitro. Genes Dev. 1993;7:1244–1253. [Abstract] [Google Scholar]
- Wang G, Reinke V. A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr Bio. 2008;18 in press, 10.1016/j.cub.2008.05.009. [Europe PMC free article] [Abstract] [Google Scholar]
- Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006;20:1732–1743. [Europe PMC free article] [Abstract] [Google Scholar]
- Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.molcel.2008.06.003
Read article for free, from open access legal sources, via Unpaywall:
http://www.cell.com/article/S1097276508003924/pdf
Subscription required at www.molecule.org
http://www.molecule.org/cgi/content/reprint/31/1/79
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Pachytene piRNAs control discrete meiotic events during spermatogenesis and restrict gene expression in space and time.
Sci Adv, 10(40):eadp0466, 02 Oct 2024
Cited by: 0 articles | PMID: 39356768 | PMCID: PMC11446278
Germ granule compartments coordinate specialized small RNA production.
Nat Commun, 15(1):5799, 10 Jul 2024
Cited by: 0 articles | PMID: 38987544 | PMCID: PMC11236994
RNAi-dependent expression of sperm genes in ADL chemosensory neurons is required for olfactory responses in <i>Caenorhabditis elegans</i>.
Front Mol Biosci, 11:1396587, 11 Jul 2024
Cited by: 0 articles | PMID: 39055986 | PMCID: PMC11269235
Interaction between a J-domain co-chaperone and a specific Argonaute protein contributes to microRNA function in animals.
Nucleic Acids Res, 52(11):6253-6268, 01 Jun 2024
Cited by: 0 articles | PMID: 38613392 | PMCID: PMC11194074
Catalytic residues of microRNA Argonautes play a modest role in microRNA star strand destabilization in C. elegans.
Nucleic Acids Res, 52(9):4985-5001, 01 May 2024
Cited by: 2 articles | PMID: 38471816 | PMCID: PMC11109956
Go to all (275) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans.
Mol Cell, 31(1):67-78, 19 Jun 2008
Cited by: 385 articles | PMID: 18571452 | PMCID: PMC2570341
C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts.
Cell, 150(1):78-87, 25 Jun 2012
Cited by: 260 articles | PMID: 22738724 | PMCID: PMC3410639
Maternal piRNAs Are Essential for Germline Development following De Novo Establishment of Endo-siRNAs in Caenorhabditis elegans.
Dev Cell, 34(4):448-456, 13 Aug 2015
Cited by: 65 articles | PMID: 26279485
Roles of piRNAs in transposon and pseudogene regulation of germline mRNAs and lncRNAs.
Genome Biol, 22(1):27, 08 Jan 2021
Cited by: 46 articles | PMID: 33419460 | PMCID: PMC7792047
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Cancer Research UK (1)
Grant ID: C13474
Medical Research Council
Wellcome Trust (1)
Core support for the Wellcome Trust Cancer Research UK Gurdon Institute.
Professor Daniel St Johnston, University of Cambridge
Grant ID: 092096





