Abstract
Free full text

Hedgehog Secretion and Signal Transduction in Vertebrates
Abstract
Signaling by the Hedgehog (Hh) family of secreted proteins is essential for proper embryonic patterning and development. Dysregulation of Hh signaling is associated with a variety of human diseases ranging from developmental disorders such as holoprosencephaly to certain forms of cancer, including medulloblastoma and basal cell carcinoma. Genetic studies in flies and mice have shaped our understanding of Hh signaling and revealed that nearly all core components of the pathway are highly conserved. Although many aspects of the Drosophila Hh pathway are conserved in vertebrates, mechanistic differences between the two species have begun to emerge. Perhaps the most striking divergence in vertebrate Hh signaling is its dependence on the primary cilium, a vestigial organelle that is largely absent in flies. This minireview will provide an overview of Hh signaling and present recent insights into vertebrate Hh secretion, receptor binding, and signal transduction.
Introduction
Originally discovered for its role in Drosophila embryonic patterning, the Hedgehog (Hh) pathway is among a handful of signaling pathways governing the development of multicellular organisms. Hh signaling is essential for the development of nearly every organ system in vertebrates, from patterning the neural tube and limbs to regulating lung morphogenesis and hair follicle formation (1). Although the Drosophila genome encodes a single hh gene, vertebrates harbor between three (Sonic hedgehog (Shh), Desert hedgehog (Dhh), and Indian hedgehog (Ihh) in birds and mammals) and six (Shh, Dhh, and Ihh plus Tiggywinkle hedgehog (Twhh), Echidna hedgehog (Ehh), and Qiqihar hedgehog (Qhh) in fish) homologs, differing primarily in tissue distribution (2). In vertebrates, Shh is expressed throughout the developing nervous system and in many epithelial tissues, Ihh functions primarily in bone development, and Dhh expression is limited to the peripheral nervous system and reproductive organs (1). As a result of its widespread expression, much of what is known about vertebrate Hh signaling stems from work on Shh. All Hh ligands undergo a similar series of processing events that result in the covalent attachment of two lipid moieties and are essential for proper signaling activity and tissue distribution (Fig. 1). Secreted Hh ligands interact with Patched (Ptc)-coreceptor complexes on the surface of responding cells, relieving Ptc-mediated inhibition of the signal transducer Smoothened (Smo) (see Fig. 4). Activated Smo prevents the processing of full-length Gli transcription factors (Gli-FL)2 into transcriptional repressors (Gli-R) so as to allow Gli-FL to activate the transcription of Hh target genes. Thus, the relative abundance of Gli transcriptional activators and inhibitors ultimately regulates the transcription of Hh target genes.

Hh processing and release. Hh precursor peptides 45 kDa in size undergo a cholesterol-dependent autocatalytic cleavage in the endoplasmic reticulum to generate a cholesterol-modified N-terminal fragment (Hh-N, denoted by N) and a 25-kDa C-terminal fragment (Hh-C, denoted by C). Hh-C is recognized by the lectins OS-9 and XTP3 and ubiquitylated by the ubiquitin ligase Hrd1 and its partner Sel1. Ubiquitylated Hh-C is moved into the cytosol by the p97 ATPase and subsequently degraded by the proteasome. Cholesterol-modified Hh-N enters the secretory pathway, where the acyltransferase Hhat catalyzes the covalent attachment of palmitate to the N-terminal cysteine. Dually lipidated Hh is targeted to the cell membrane, where cholesterol facilitates the assembly of multimeric Hh complexes possibly by tethering Hh to the membrane and promoting interactions with HS proteoglycans (HSPG). Prior to its release, N- and C-terminal peptides may be cleaved by membrane-proximal proteases such as those belonging to the ADAM family, resulting in the removal of both lipid moieties. The 12-pass transmembrane protein Disp facilitates the release of Hh multimers into the extracellular environment, although the mechanistic details of this process are not well understood. Ub, ubiquitin.
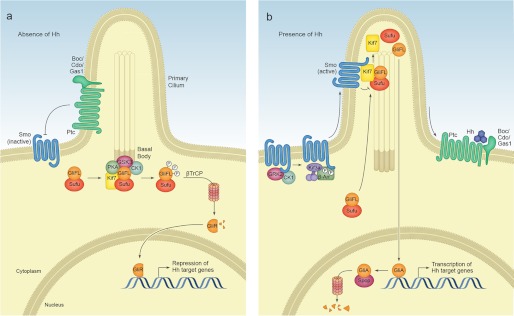
Vertebrate Hh signal transduction. a, in the absence of ligand, the 12-pass transmembrane protein Ptc localizes to the primary cilium base and maintains Smo in an inactive conformation. Gli-FL transcription factors complex with Sufu. Sufu sequesters Gli-FL in the cytosol and stabilizes the protein. Sufu and the kinesin-4 family member Kif7 promote the phosphorylation of C-terminal residues in Gli-FL by PKA, GSK3β, and CK1α, which may occur at the basal body of the primary cilium. Phosphorylated Gli-FL is recognized by the E3 ubiquitin ligase βTrCP, resulting in ubiquitylation and proteasomal degradation of C-terminal residues to generate a truncated N-terminal transcriptional repressor (Gli-R) that inhibits Hh target gene transcription. b, in the presence of ligand, Hh binding to Ptc causes Ptc to exit the cilium and relieves its inhibition of Smo. Smo is phosphorylated by CK1α and GRK2, inducing a conformational change and enabling β-arrestin (β-Arr)- and Kif3a-dependent transport into the cilium. Within the cilium, activated Smo promotes the disassembly of Sufu-Gli complexes. Kif7 also localizes to the cilium in the presence of Hh and likely assists Smo in this disassembly. Gli-FL accumulates in the tip of the cilium and is shuttled into the nucleus, perhaps on cytoplasmic microtubules. Within the nucleus, Gli-FL receives additional modifications that convert it to a labile transcriptional activator (Gli-A) that activates Hh target genes. Gli-A is subsequently degraded in a manner that requires the cullin-3-based adaptor Spop.
Although many aspects of Drosophila Hh signaling are conserved in vertebrates, vertebrate Hh signal transduction differs in its requirement for the primary cilium. Primary cilia are slim, microtubule-based, non-motile structures that project from the surface of nearly all vertebrate cells but are conspicuously absent in most Drosophila cell types (3). The assembly and maintenance of primary cilia require intraflagellar transport (IFT) proteins, and several members of the IFT family are essential for proper vertebrate Hh signaling (3, 4). Mutations in components of the kinesin-driven IFT-B complex, which mediates the anterograde transport of molecules from the base of the cilium to the tip, lead to a complete loss of Hh signaling (3). In contrast, mutations in members of the dynein-driven IFT-A complex, which controls retrograde transport, lead to aberrant Hh pathway activation (3). Nonetheless, it is not currently known whether IFT-A and IFT-B complexes interact directly with Hh pathway components to control their localization and activity or if, instead, these complexes facilitate Hh signaling simply by maintaining proper ciliary architecture. Indeed, recent genetic studies suggest that the primary cilium may function primarily as a scaffold for Hh signaling, arguing against a direct role for IFT proteins in regulating the movement of Hh pathway components (5).
In this minireview, we provide an overview of Hh production and cytosolic signaling in vertebrates (for excellent reviews of Drosophila Hh signaling, see Refs. 2 and 6). We discuss recent insights into ligand release, receptor binding, and signal transduction and attempt to incorporate these findings into existing models of Hh signaling. Additionally, we present remaining questions regarding Hh secretion and signal transduction that warrant further investigation.
Hedgehog Processing and Release
The signaling activity of Hh ligands is intimately linked to a complex sequence of post-translational modifications ultimately resulting in the covalent attachment of two lipid moieties, one at each terminus (Fig. 1). Following translation, the Hh precursor peptide (~45 kDa in size) translocates into the endoplasmic reticulum lumen, where it undergoes a cholesterol-dependent autocatalytic cleavage to generate a 19-kDa cholesterol-modified N-terminal peptide fragment and a 25-kDa C-terminal fragment (Fig. 1). This cleavage reaction occurs in two steps. In the first step, the free thiol of Cys-198 (human SHH) acts as a nucleophile, attacking the carbonyl carbon of the preceding glycine residue and generating a thioester intermediate (7–10). In the second step, this thioester intermediate is subjected to nucleophilic attack by the 3β-hydroxyl group of cholesterol, generating a cholesterol-modified N-terminal fragment (Hh-N) and displacing the C-terminal fragment (Hh-C). Although Cys-198 has long been recognized for its role in autocatalytic cleavage, a second conserved cysteine, Cys-363, is also required for cleavage, forming a disulfide bond with Cys-198 that likely facilitates protein folding and the reduction of which generates the reactive thiol required for cleavage (11). As such, mutating either cysteine residue prevents autoproteolysis of Hh precursors (11). Although processing-deficient full-length forms of Shh are able to illicit juxtacrine signaling in cell-based assays (12), the significance of this finding remains enigmatic, as Shh is found exclusively in its cleaved form during embryogenesis (13). Indeed, mutations disrupting the cleavage of full-length Hh peptides have been linked to developmental disorders such as holoprosencephaly (14, 15).
All of the signaling properties of Hh proteins reside within the N-terminal fragment. The C-terminal fragment undergoes endoplasmic reticulum-associated degradation, a process that requires the lectins OS-9 and XTP3, the ubiquitin ligase Hrd1 and its partner Sel1, and the p97 ATPase (Fig. 1) (11). Hh-N is subjected to a second covalent modification by Hh acyltransferase (Hhat)/Skinny hedgehog (Ski), which catalyzes the attachment of palmitate to the free amino group of the N-terminal cysteine (16–18). Thus, Hh-N has two covalently attached lipid moieties: cholesterol at its C-terminal end and palmitate at its N-terminal end.
One unique feature of Hh proteins is their capacity to travel very long distances, up to 300 μm in vertebrate limb, to reach their targets. The release and long-range signaling of cholesterol- and palmitate-modified Hh-N (hereafter referred to as Hh) require the activity of Dispatched (Disp), a 12-pass transmembrane protein belonging to the RND family of bacterial transporters (13, 19–21). Although mice and flies deficient in Disp synthesize Hh properly, Hh accumulates in producing cells, able to activate the pathway in neighboring cells but not competent for long-range signaling (19, 20, 22–24). Although the Hh-distributing function of murine Disp requires two presumptive proton-binding domains in transmembrane domains 4 and 10, little else is known about how Disp facilitates Hh secretion and long-range signaling (20). Recent studies of Drosophila imaginal discs indicate that Hh and Disp co-localize within endocytic vesicles and suggest that Disp may traffic Hh to the basolateral membrane, where it is released (24). Whether or not the trafficking function of Disp is coupled to its Hh-releasing function or if these two activities are distinct remains to be shown, and additional studies are needed to determine whether the trafficking function of Disp is conserved in vertebrates.
Lipid Modifications Regulate Activity and Distribution of Hedgehog
Genetic studies in flies and mice indicate that cholesterol and palmitate are essential for the proper activity and distribution of Hh ligands. The C-terminal cholesterol moiety is required for the formation of multimeric Hh complexes, which are thought to be the biologically relevant form of the morphogen (25–27). In cells expressing a truncated form of Hh that cannot be cholesterol-modified, Hh proteins are secreted as monomers in a Disp-independent manner (19, 23, 28). Although the process by which cholesterol mediates multimerization remains uncertain, one possibility is that by tethering Hh proteins to the membrane, the cholesterol moiety concentrates Hh within specific microdomains such as lipid rafts and promotes electrostatic interactions between Hh monomers (29–31). Cholesterol-mediated clustering may also promote interactions between Hh and other membrane-associated molecules such as heparin sulfate (HS) proteoglycans, whose HS moieties are known to interact with positively charged residues within a conserved Cardin-Weintraub motif present in all Hh proteins (Fig. 2) (26, 27, 30, 31). In Drosophila, the HS-containing glypicans Dally and Dally-like interact with both Hh and the hemolymph-derived lipoprotein lipophorin, leading to the formation of soluble lipoprotein complexes that mediate patterning in the wing imaginal disc (27, 32). Although the addition of HS is sufficient to induce dimerization of non-cholesterol-modified Shh in vitro, the composition of vertebrate Hh multimers remains uncharacterized (30).
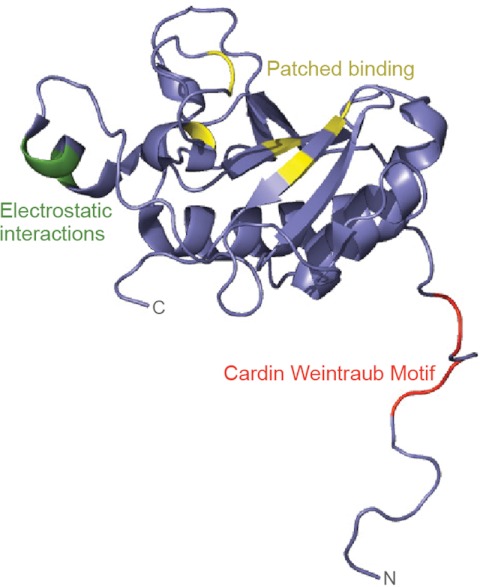
Regions of Shh important for receptor binding and multimerization. Shown is the structure of human SHH-N (non-cholesterol-modified N-terminal fragment; Protein Data Bank code 3M1N (99)). Residues in green (Glu-72, Arg-73, and Lys-75) mediate electrostatic interactions between Hh monomers and are required for multimerization (38). Arg-73 is the vertebrate equivalent of Drosophila Lys-132, the mutation of which results in decreased long-range signaling in the imaginal disc (26). Residues in yellow (His-133, His-134, His-140, His-180, and His-182) are important for Ptc binding (note that His-140 and His-182 coordinate with zinc). Residues in red (Lys-32, Arg-33, Arg-34, Lys-37, and Lys-38) form the Cardin-Weintraub motif and interact with HS. Note how the N terminus extends away from the globular domain of SHH-N; some of these residues may be cleaved in the formation of active Shh multimers (see text).
In addition to its role in multimerization, cholesterol also regulates the distribution of Hh ligands (23, 33, 34). Although there have been conflicting reports regarding how cholesterol affects Hh distribution, the majority of data are in agreement with a role for cholesterol in restricting the spread of Hh ligands (23, 33, 35, 36). Nonetheless, the mechanism by which cholesterol limits the distribution of Hh remains unclear, and the increased range of non-cholesterol-modified Hh ligands may be secondary to loss of multimerization or Disp-mediated release. Such an indirect role for cholesterol in regulating Hh distribution is supported by the finding that, in Drosophila, a cholesterol-modified form of Hh that cannot multimerize (due to a K132D mutation) has a restricted distribution and signaling range (Fig. 2) (26). Additionally, recent work in vertebrate cell lines suggests that the cholesterol moiety of Shh is removed by membrane-proximal proteases prior to its release (30). Taken together, these data indicate that the role of cholesterol in determining the range of Hh signaling may not be straightforward and warrants further investigation.
Whereas non-cholesterol-modified Hh ligands maintain some of their signaling capacity, loss of palmitoylation abolishes the signaling activity of Hh almost entirely (17, 18, 29, 37), indicating that palmitate is absolutely required for Hh signaling. Although the importance of palmitate has long been recognized, only recently have inroads been made in understanding why. Recent work in vitro suggests that palmitate facilitates the cleavage of N-terminal amino acids by membrane-proximal proteases such as ADAM (a disintegrin and metalloprotease) family members (38). Such cleavage is required for the formation of active Shh multimers, as these residues otherwise obstruct the Zn2+ coordination site on adjacent molecules, a region that likely interacts with Ptc and is known to regulate Shh stability and activity (Fig. 3) (39–42). Thus, in the absence of palmitoylation (due to mutation of the N-terminal Cys), Shh maintains the capacity to multimerize, but these multimers have significantly reduced signaling activity due to their inability to properly interact with Ptc (38). Although these data provide insight into the role of palmitoylation in Hh signaling, they also raise a number of questions regarding the production and secretion of Hh. For instance, how is the cleavage of lipid moieties coupled to Disp-mediated release? Are the lipid moieties of Drosophila Hh also cleaved? Future studies are needed to address these questions and to determine whether lipid moieties are also cleaved in vivo.
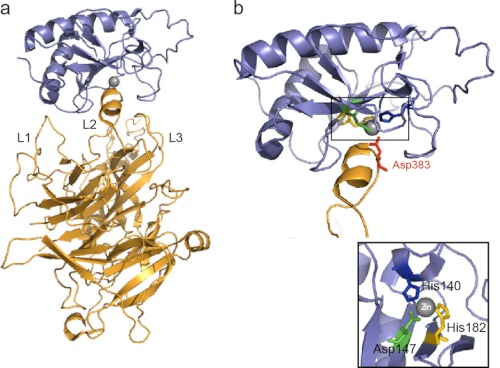
SHH-N receptor binding involves the Zn2+ coordination site. a, structure of human SHH-N in complex with HIP (Protein Data Bank code 3HO5 (39)). The L2 loop in the β-propeller domain of HIP interacts with SHH-N. b, HIP binds the pseudo-active site in SHH-N, and Asp-383 completes the tetrahedral coordination of Zn2+ in SHH-N. Inset, His-140, His-142, and Arg-147 of SHH-N coordinate Zn2+. Note that the Zn2+ coordination site is also required for binding to PTC, and PTC likely binds SHH in a manner similar to HIP (see text).
Dual Roles of Patched in Hedgehog Reception and Pathway Inhibition
The Hh receptor Ptc is a 12-pass transmembrane protein with homology to the RND family of bacterial transporter proteins. Reception of Hh by Ptc is enhanced by the presence of additional Hh-binding proteins on the cell surface. These presumptive coreceptors include a family of immunoglobulin- and fibronectin type III (FnIII)-containing integral membrane proteins (Ihog and Boi in Drosophila and Cdo and Boc in vertebrates) and the vertebrate-specific cell surface protein Gas1 (43–45). Although removal of a single coreceptor leads to a modest, tissue-specific reduction in Hh pathway activity, removal of two or three coreceptors from Drosophila or mice, respectively, leads to a complete loss of signaling, underscoring the importance of these coreceptors in Hh pathway transduction (43, 45, 46).
In addition to Boc, Cdo, and Gas1, vertebrates harbor a fourth Hh-binding protein, Hip, which has no downstream signaling function and likely acts as a decoy receptor by competing with Ptc for Hh binding (39, 47). Analysis of the crystal structure of Hip in complex with Shh revealed that Asp-383 of Hip displaces water and completes the tetrahedral coordination of Zn2+ in the Shh pseudo-active site (Fig. 3) (39, 40). Sequence comparisons of Hip and Ptc revealed that Ptc contains a similar sequence of amino acids capable of binding Shh and competing with Hip for Shh binding, providing novel insight into Hh-receptor interactions (39). Given that Drosophila Hh lacks a Zn2+ coordination site and is unable to directly bind Ptc, these data also suggest that Hh-Ptc interactions differ between flies and vertebrates (44). This possible divergence is further supported by the finding that Drosophila Hh binds the second FnIII repeat in Ihog, whereas vertebrate Hh proteins bind a third, non-orthologous FnIII repeat in Cdo (48). Thus, despite the conserved function of Ptc and coreceptors in Hh signaling, the mode of binding between Hh and these receptor complexes does not appear to be conserved.
In addition to serving as the Hh receptor, Ptc functions as a potent negative regulator of the Hh pathway by inhibiting the seven-pass transmembrane protein Smo. In the absence of Hh, Ptc localizes to the primary cilium and maintains Smo in an inactive conformation, preventing Smo from entering the cilium (49). Although early studies suggested that Ptc could directly bind to and inhibit Smo (50), subsequent work revealed that Ptc-mediated inhibition is non-stoichiometric, making direct inhibition unlikely (51). The mechanism by which Ptc inhibits Smo remains enigmatic. Sequence similarities between Ptc and the RND family of bacterial transporter proteins have led many to hypothesize that Ptc may regulate the flux molecules that activate or inhibit Smo, a theory that is supported by the susceptibility of Smo to modulation by small molecules such as the steroidal alkaloid cyclopamine (52–54). Given that Ptc is enriched around the base of the primary cilium, where vertebrate Hh signaling likely occurs, Ptc might locally control the abundance of Smo inhibitors or activators (49). Although a number of Smo agonists and antagonists have been identified, to date, none have been shown to be regulated by Ptc. Recent work in Drosophila suggests that Ptc may inhibit Hh signaling by regulating the synthesis of phosphatidylinositol 4-phosphate (PI4P), revealing that increased and decreased levels PI4P lead to Hh pathway activation and repression, respectively (55). Importantly, by showing that cells deficient in Ptc have increased PI4P levels, this work provides the first evidence of an endogenous Hh activator that is regulated by Ptc. Nonetheless, future studies are needed to determine how Ptc regulates PI4P synthesis and to verify that PI4P activates the pathway at the level of Smo rather than acting farther downstream.
Transcriptional Repression in Absence of Hedgehog
The zinc finger-containing Gli transcription factors are the principal effectors of canonical Hh signaling. Depending on the availability of Hh ligands, Gli proteins function either as transcriptional activators or repressors. In the absence of Hh, Gli-FL is proteolytically processed to yield a truncated N-terminal transcriptional repressor (Gli-R) (Fig. 4a). Whereas Drosophila harbors a single Gli family member, Cubitus interruptus (Ci), vertebrates have three, Gli1–Gli3. Of these, Gli2 and Gli3 function as both transcriptional activators and repressors, whereas Gli1 is a target of Hh signaling and exists only as an activator.
Although many aspects of vertebrate Gli-R formation remain enigmatic, Suppressor of Fused (Sufu), the kinesin Kif7, and the primary cilium are required for efficient processing of Gli-FL into Gli-R (Fig. 4a) (3, 56–59). Sufu stabilizes Gli2-FL and Gli3-FL and sequesters both proteins in the cytosol, thus preventing their nuclear translocation and activation (6, 60–62). Sufu also promotes the phosphorylation of C-terminal residues in Gli-FL by protein kinase A (PKA), which primes Gli-FL for further phosphorylation by glycogen synthase kinase 3β (GSK3β) and casein kinase 1α (CK1α) (63, 64). Phosphorylated Gli-FL is recognized by the E3 ubiquitin ligase βTrCP, leading to the ubiquitylation and degradation of C-terminal peptides to generate Gli-R (63–66). In contrast to its relatively minor role in Drosophila, Sufu is absolutely required for proper development and is essential for Gli-R formation in vertebrates (56, 67). Mice deficient in Sufu die around embryonic day 9.5 with significantly reduced levels of both full-length and repressor forms of Gli and features of aberrant Hh activation that resemble loss of Ptc (56, 67). In the absence of Sufu, Gli-FL enters the nucleus and is converted into a labile transcriptional activator (Gli-A) that is quickly degraded within the nucleus in a manner that depends upon the cullin-3-based ubiquitin ligase adaptor Spop (62, 68–70). Indeed, Sufu and Spop have been shown to compete for Gli-FL binding, and loss of Spop from Sufu−/− cells leads to a significant recovery of Gli-FL levels (62). Together, these data indicate that Sufu regulates Gli-R formation by stabilizing Gli-FL in the cytosol and preventing Spop-dependent degradation in the nucleus. In addition to its role in Gli processing, Sufu may also inhibit the transcription of Hh target genes through its interaction with SAP18, a component of the mSin3-histone deacetylase repressor complex (71). However, this processing-independent role for Sufu was recently challenged (69), and additional data are needed to clarify the function of nuclear Sufu in Hh pathway inhibition.
In addition to Sufu, the kinesin-4 family member Kif7 also appears to be required for optimal Gli processing (57–59, 72). Mice deficient in Kif7 have increased levels of Gli-FL and decreased levels of Gli-R and exhibit features of pathway derepression such as polydactyly (57–59). Although the mechanism by which Kif7 promotes Gli processing remains unclear, one possibility is that, like its Drosophila homolog Costal2 (Cos2), Kif7 recruits PKA, GSK3β, and CK1α to phosphorylate Gli-FL (Fig. 4a). Although Kif7 has been shown to interact with Gli, additional data are needed to determine whether the scaffolding function of Kif7 is conserved in vertebrates.
Studies both in vivo and in vitro indicate that the primary cilium is required for efficient processing of Gli-FL into Gli-R (3). Interestingly, the role of Sufu in Gli-R production appears to be independent of cilia, as cells lacking both primary cilia and Sufu exhibit aberrant Hh pathway activity akin to Sufu−/− cells (69, 73). By contrast, the role of Kif7 in Gli processing is cilium-dependent, as mice lacking both cilia and Kif7 resemble cilia mutants (57). Although the exact function of the cilium in Gli processing remains enigmatic, the cilium may serve as a platform for Gli-processing machinery. Indeed, Kif7, PKA, GSK3β, and CK1α are present in the primary cilia and/or basal body in the absence of Hh signaling (57–59, 74–76). Although Sufu cannot localize to the cilium on its own, it is likely recruited there by Gli, as low levels of both Sufu and Gli can be observed in the cilium even in the absence of Hh signaling (60, 61). Thus, although Sufu-Gli complexes form throughout the cytosol, they may be directed to the cilium by Gli for efficient processing in a Kif7- and kinase-dependent manner.
Although Gli2 and Gli3 both undergo partial proteolytic degradation in the absence of Hh, the processing of Gli3 is significantly more efficient than that of Gli2 (77). Consequently, Gli3-R serves as the principal transcriptional repressor of Hh signaling in the absence of ligand, whereas Gli2-A functions as the predominant transcriptional activator when Hh is present (78). The increased efficiency of Gli3 processing is due in large part to the sequence of a 200-residue processing determinant domain in its C terminus (79). Together with an appropriate degron and the zinc finger domain, the processing determinant domain forms a three-part signal that is essential for efficient Gli3 processing (80). But what happens to Gli2-FL in the absence of Hh? Like Gli3, the C terminus of Gli2 is phosphorylated by PKA in the absence of Hh. Although this phosphorylation leads to a limited amount of processing, it may also destabilize Gli2-FL, leading to complete degradation by the proteasome (77, 81). Such a processing-independent role of PKA in Hh pathway inhibition is supported by recent genetic data showing that mice lacking both catalytic subunits of PKA (Prkaca−/−;Prkacb−/−) die midgestation with a completely ventralized neural tube, a defect that cannot be explained by loss of Gli processing alone and that suggests a increase in Gli activation (75, 82). Given that PKA may also regulate the entry of Sufu-Gli complexes into the cilium, additional studies are required to clarify the mechanism(s) by which PKA inhibits Gli activation and to determine to what extent Gli2 phosphorylation inhibits pathway activation (61, 75, 83).
Smoothened and Gli Activation in Presence of Hedgehog
In the presence of Hh, Ptc relieves its inhibition of Smo and allows Smo to become activated. Despite significant sequence differences, many aspects of Drosophila Smo activation are conserved in vertebrates. In Drosophila, phosphorylation of C-terminal residues by PKA, CK1α, and G protein-coupled receptor kinase 2 (GRK2) cause Smo to adopt an open conformation and promote its accumulation on the membrane (84–89). Although the C terminus of vertebrate Smo differs significantly compared with Drosophila and lacks PKA phosphorylation sites, recent data indicate that vertebrate Smo is also phosphorylated in response to Hh signaling (76, 90, 91). CK1α and GRK2 phosphorylate the C-terminal tail of vertebrate Smo, inducing conformational changes and facilitating its lateral translocation into the primary cilium (Fig. 4b) (76). The movement of Smo into the cilium is dependent upon β-arrestins and the kinesin-2 motor subunit Kif3a, both of which are recruited to Smo following its phosphorylation by CK1α and GRK2 (76, 90, 92, 93).
Activated Smo both inhibits Gli processing and promotes additional ill-defined modifications that convert Gli-FL proteins into transcriptional activators. Although the details of this process remain somewhat enigmatic, activated Smo likely promotes the disassembly of Sufu-Gli complexes that accumulate in the cilium following pathway activation (Fig. 4b) (60–62, 94). Kif7 may also promote Sufu-Gli disassembly, as it localizes to the cilium in response to Hh and interacts with overexpressed Smo in tissue culture cells (58). Indeed, such a positive role of Kif7 in Hh signaling is consistent with the finding that mice deficient in Kif7 exhibit features of decreased Hh pathway activity such as reduced Ptc expression in the notochord and floor plate (57, 58). Nonetheless, additional studies are needed to determine whether Kif7-Smo interactions are dependent on Smo phosphorylation, as they are for Drosophila Cos2 (95, 96). The disassembly of Sufu-Gli complexes allows Gli-FL to enter the nucleus, where it is converted to its activator form (Gli-A) (61). The translocation of Gli requires cytoplasmic microtubules, as microtubule-destabilizing agents such as nocodazole have been shown to inhibit its nuclear accumulation and activity (60, 97). Although the details of Gli activation remain nebulous, they may involve phosphorylation, as Gli2 and Gli3 appear to be phosphorylated within the nucleus in response to Hh (60). Given that the nucleus is also the site of Spop-mediated degradation, however, it is difficult to ascertain whether this phosphorylation is coupled to Gli activation or degradation (62, 69). Gli proteins might also be deacetylated in response to Hh stimulation, as HDAC1 overexpression in tissue culture cells leads to Gli1 deacetylation (98). Activated Gli promotes the transcription of genes involved in differentiation, proliferation, and cell survival as well as several negative regulators of the pathway such as Ptc and Hip to down-regulate pathway activity.
Conclusions and Perspectives
Over the past 2 decades, mouse and fly genetics have been instrumental in identifying components of the Hh pathway and elucidating their functions, revealing a high degree of conservation between the two species. However, the discovery that vertebrate Hh signaling requires the primary cilium has significantly changed how the pathway is studied and made it somewhat more difficult to draw comparisons between vertebrates and flies. Despite these challenges, significant progress has been made in defining vertebrate Hh signal transduction. Nonetheless, several questions regarding vertebrate Hh secretion and signal transduction remain unanswered. The mechanistic details of Disp-mediated secretion remain elusive, as does the composition of secreted Hh multimers. The mechanism by which Ptc inhibits Smo continues to be a mystery, and a detailed understanding of how activated Smo promotes Gli activation is lacking. Additional studies are needed to examine the role of Kif7 in Gli processing and activation as well as determine to what extent the motor function of Kif7 is important for Hh signaling. Perhaps most intriguing are the questions of how and why the primary cilium plays such an essential role in vertebrate Hh signal transduction. As cell and developmental biologists continue to adapt to the challenges inherent in the study of vertebrate Hh signaling, the answers to these and other questions will undoubtedly be revealed.
Acknowledgments
Footnotes
2The abbreviations used are:
- Gli-FL
- full-length Gli
- Gli-R
- Gli repressor
- Gli-A
- Gli activator
- IFT
- intraflagellar transport
- HS
- heparin sulfate
- FnIII
- fibronectin type III
- PI4P
- phosphatidylinositol 4-phosphate
- PKA
- protein kinase A
- GSK3β
- glycogen synthase kinase 3β
- CK1α
- casein kinase 1α
- GRK2
- G protein-coupled receptor kinase 2
- βTrCP
- β-transducin repeat-containing E3 ubiquitin protein ligase.
REFERENCES
Articles from The Journal of Biological Chemistry are provided here courtesy of American Society for Biochemistry and Molecular Biology
Full text links
Read article at publisher's site: https://doi.org/10.1074/jbc.r112.356006
Read article for free, from open access legal sources, via Unpaywall:
http://www.jbc.org/article/S0021925820500379/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1074/jbc.r112.356006
Article citations
Hepatic transcript profiling in beef cattle: Effects of rumen-protected niacin supplementation.
PLoS One, 18(8):e0289409, 03 Aug 2023
Cited by: 0 articles | PMID: 37535643 | PMCID: PMC10399858
Gestational iron deficiency affects the ratio between interneuron subtypes in the postnatal cerebral cortex in mice.
Development, 150(20):dev201068, 03 Mar 2023
Cited by: 3 articles | PMID: 36805633 | PMCID: PMC10110419
Targeting the Hedgehog Pathway in Rhabdomyosarcoma.
Cancers (Basel), 15(3):727, 24 Jan 2023
Cited by: 3 articles | PMID: 36765685 | PMCID: PMC9913695
Review Free full text in Europe PMC
Understanding the Roles of the Hedgehog Signaling Pathway during T-Cell Lymphopoiesis and in T-Cell Acute Lymphoblastic Leukemia (T-ALL).
Int J Mol Sci, 24(3):2962, 03 Feb 2023
Cited by: 4 articles | PMID: 36769284 | PMCID: PMC9917970
Review Free full text in Europe PMC
GRAMD1/ASTER-mediated cholesterol transport promotes Smoothened cholesterylation at the endoplasmic reticulum.
EMBO J, 42(3):e111513, 16 Dec 2022
Cited by: 5 articles | PMID: 36524353 | PMCID: PMC9890235
Go to all (78) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Protein structures in PDBe (2)
-
(1 citation)
PDBe - 3HO5View structure
-
(1 citation)
PDBe - 3M1NView structure
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The primary cilium as a Hedgehog signal transduction machine.
Methods Cell Biol, 94:199-222, 23 Dec 2009
Cited by: 109 articles | PMID: 20362092 | PMCID: PMC2867239
Biochemical mechanisms of vertebrate hedgehog signaling.
Development, 146(10):dev166892, 15 May 2019
Cited by: 129 articles | PMID: 31092502 | PMCID: PMC6550017
Review Free full text in Europe PMC
Hedgehog signaling.
Curr Top Dev Biol, 149:1-58, 06 May 2022
Cited by: 38 articles | PMID: 35606054
Review
Hedgehog signalling.
Development, 143(3):367-372, 01 Feb 2016
Cited by: 130 articles | PMID: 26839340
Review





