Abstract
Free full text

Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens; a review of the considerable within-species diversity.
Abstract
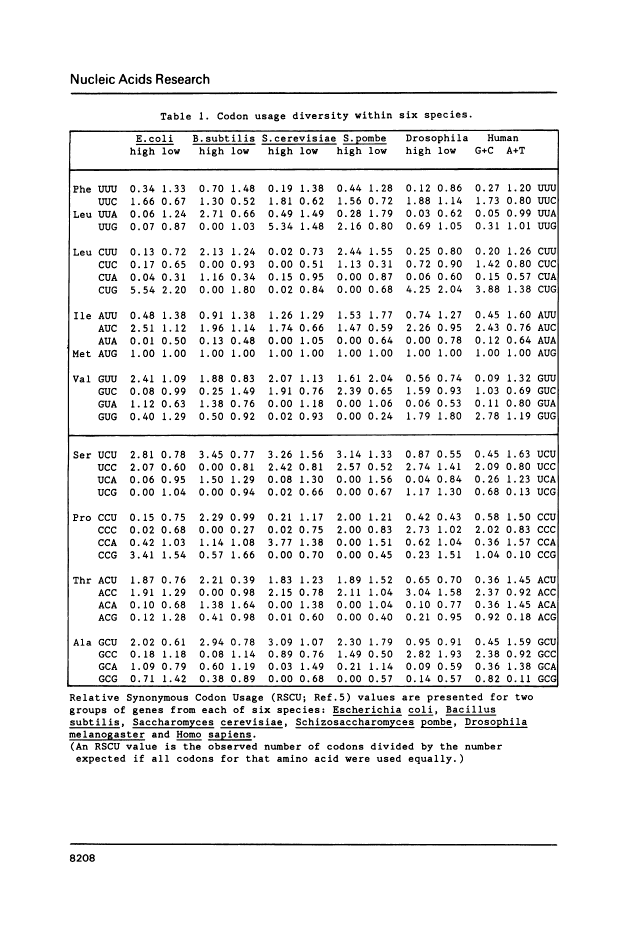
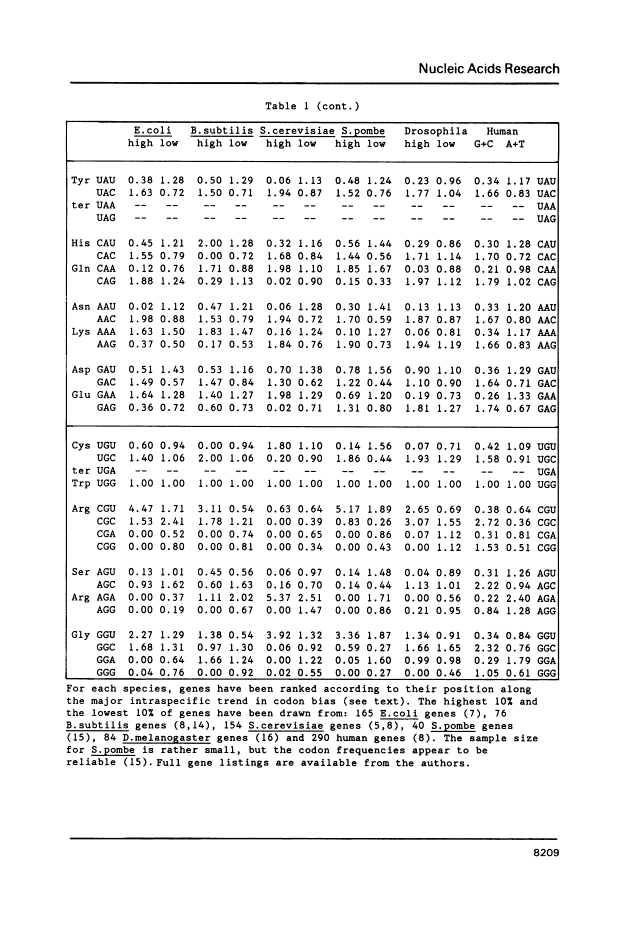
The genetic code is degenerate, but alternative synonymous codons are generally not used with equal frequency. Since the pioneering work of Grantham's group it has been apparent that genes from one species often share similarities in codon frequency; under the "genome hypothesis" there is a species-specific pattern to codon usage. However, it has become clear that in most species there are also considerable differences among genes. Multivariate analyses have revealed that in each species so far examined there is a single major trend in codon usage among genes, usually from highly biased to more nearly even usage of synonymous codons. Thus, to represent the codon usage pattern of an organism it is not sufficient to sum over all genes as this conceals the underlying heterogeneity. Rather, it is necessary to describe the trend among genes seen in that species. We illustrate these trends for six species where codon usage has been examined in detail, by presenting the pooled codon usage for the 10% of genes at either end of the major trend. Closely-related organisms have similar patterns of codon usage, and so the six species in Table 1 are representative of wider groups. For example, with respect to codon usage, Salmonella typhimurium closely resembles E. coli, while all mammalian species so far examined (principally mouse, rat and cow) largely resemble humans.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (375K), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Grantham R, Gautier C, Gouy M, Mercier R, Pavé A. Codon catalog usage and the genome hypothesis. Nucleic Acids Res. 1980 Jan 11;8(1):r49–r62. [Europe PMC free article] [Abstract] [Google Scholar]
- Grantham R, Gautier C, Gouy M, Jacobzone M, Mercier R. Codon catalog usage is a genome strategy modulated for gene expressivity. Nucleic Acids Res. 1981 Jan 10;9(1):r43–r74. [Europe PMC free article] [Abstract] [Google Scholar]
- Gouy M, Gautier C. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 1982 Nov 25;10(22):7055–7074. [Europe PMC free article] [Abstract] [Google Scholar]
- Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol. 1985 Jan;2(1):13–34. [Abstract] [Google Scholar]
- Sharp PM, Tuohy TM, Mosurski KR. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 1986 Jul 11;14(13):5125–5143. [Europe PMC free article] [Abstract] [Google Scholar]
- Aota S, Ikemura T. Diversity in G + C content at the third position of codons in vertebrate genes and its cause. Nucleic Acids Res. 1986 Aug 26;14(16):6345–6355. [Europe PMC free article] [Abstract] [Google Scholar]
- Sharp PM, Li WH. Codon usage in regulatory genes in Escherichia coli does not reflect selection for 'rare' codons. Nucleic Acids Res. 1986 Oct 10;14(19):7737–7749. [Europe PMC free article] [Abstract] [Google Scholar]
- Aota S, Gojobori T, Ishibashi F, Maruyama T, Ikemura T. Codon usage tabulated from the GenBank Genetic Sequence Data. Nucleic Acids Res. 1988;16 (Suppl):r315–r402. [Europe PMC free article] [Abstract] [Google Scholar]
- Bennetzen JL, Hall BD. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [Abstract] [Google Scholar]
- Shields DC, Sharp PM. Synonymous codon usage in Bacillus subtilis reflects both translational selection and mutational biases. Nucleic Acids Res. 1987 Oct 12;15(19):8023–8040. [Europe PMC free article] [Abstract] [Google Scholar]
- Sharp PM, Li WH. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987 Feb 11;15(3):1281–1295. [Europe PMC free article] [Abstract] [Google Scholar]
Associated Data
Articles from Nucleic Acids Research are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/nar/16.17.8207
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC338553
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Evolution shapes and conserves genomic signatures in viruses.
Commun Biol, 7(1):1412, 30 Oct 2024
Cited by: 0 articles | PMID: 39478059 | PMCID: PMC11526014
A new era of mutation rate analyses: Concepts and methods.
Zool Res, 45(4):767-780, 01 Jul 2024
Cited by: 1 article | PMID: 38894520 | PMCID: PMC11298668
Review Free full text in Europe PMC
Complete mitogenome and intra-family comparative mitogenomics showed distinct position of Pama Croaker Otolithoides pama.
Sci Rep, 14(1):13820, 15 Jun 2024
Cited by: 0 articles | PMID: 38879694 | PMCID: PMC11180200
Codon usage bias and phylogenetic analysis of chloroplast genome in 36 gracilariaceae species.
Funct Integr Genomics, 24(2):45, 01 Mar 2024
Cited by: 1 article | PMID: 38429550
Comparative Analysis of Six Complete Plastomes of <i>Tripterospermum</i> spp.
Int J Mol Sci, 25(5):2534, 22 Feb 2024
Cited by: 0 articles | PMID: 38473781 | PMCID: PMC10931592
Go to all (362) article citations
Other citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Synonymous codon usage in Bacillus subtilis reflects both translational selection and mutational biases.
Nucleic Acids Res, 15(19):8023-8040, 01 Oct 1987
Cited by: 166 articles | PMID: 3118331 | PMCID: PMC306324
Codon usage bias is correlated with gene expression levels in the fission yeast Schizosaccharomyces pombe.
Genes Cells, 14(4):499-509, 01 Apr 2009
Cited by: 54 articles | PMID: 19335619
Selective differences among translation termination codons.
Gene, 63(1):141-145, 01 Jan 1988
Cited by: 36 articles | PMID: 3133285
Codon usage and tRNA content in unicellular and multicellular organisms.
Mol Biol Evol, 2(1):13-34, 01 Jan 1985
Cited by: 1055 articles | PMID: 3916708
Review