Abstract
Free full text

From the CoverPNAS Plus
A subset of group A-like var genes encodes the malaria parasite ligands for binding to human brain endothelial cells
Abstract
Cerebral malaria is the most deadly manifestation of infection with Plasmodium falciparum. The pathology of cerebral malaria is characterized by the accumulation of infected erythrocytes (IEs) in the microvasculature of the brain caused by parasite adhesins on the surface of IEs binding to human receptors on microvascular endothelial cells. The parasite and host molecules involved in this interaction are unknown. We selected three P. falciparum strains (HB3, 3D7, and IT/FCR3) for binding to a human brain endothelial cell line (HBEC-5i). The whole transcriptome of isogenic pairs of selected and unselected parasites was analyzed using a variant surface antigen-supplemented microarray chip. After selection, the most highly and consistently up-regulated genes were a subset of group A-like var genes (HB3var3, 3D7_PFD0020c, ITvar7, and ITvar19) that showed 11- to >100-fold increased transcription levels. These var genes encode P. falciparum erythrocyte membrane protein (PfEMP)1 variants with distinct N-terminal domain types (domain cassette 8 or domain cassette 13). Antibodies to HB3var3 and PFD0020c recognized the surface of live IEs and blocked binding to HBEC-5i, thereby confirming the adhesive function of these variants. The clinical in vivo relevance of the HBEC-selected parasites was supported by significantly higher surface recognition of HBEC-selected parasites compared with unselected parasites by antibodies from young African children suffering cerebral malaria (Mann–Whitney test, P = 0.029) but not by antibodies from controls with uncomplicated malaria (Mann–Whitney test, P = 0.58). This work describes a binding phenotype for virulence-associated group A P. falciparum erythrocyte membrane protein 1 variants and identifies targets for interventions to treat or prevent cerebral malaria.
Cerebral malaria (CM) is the most serious outcome of a Plasmodium falciparum infection, leading to death in 10–20% of cases and to long-term neurological deficits in others (1). The hallmark of the disease is microvascular sequestration, a process in which P. falciparum–infected erythrocytes (IEs) cytoadhere to endothelial cells, leading to microvascular obstruction, acidosis, hypoxia, and release of inflammatory cytokines (reviewed in ref. 2). Currently there is no specific treatment for CM other than standard antimalarial drugs and supportive therapies such as fluid replacement (3). The molecular mechanisms underlying CM are not understood, partly because of the lack of an appropriate animal model (4). However, human brain microvascular endothelial cell (HBEC) lines, such as HBEC-5i (5), can be used to study malaria host–parasite interactions, providing an in vitro model for CM (6–9). Despite the existence of this in vitro model, the parasite ligand(s) and host-cell receptor(s) mediating the interaction between IEs and brain endothelial cells remain unknown.
Previous work has identified variant surface antigens (VSA) such as P. falciparum erythrocyte membrane protein 1 (PfEMP1) as the parasite ligands mediating adhesion to a variety of receptors on human cells (10, 11). The PfEMP1 family is encoded by 50–60 var genes per parasite isolate (12). Although the sequence of each var gene is unique, all variants start with an N-terminal segment (NTS) and are followed by a succession of Duffy binding-like (DBL) and cysteine-rich interdomain region (CIDR) domains. These domains can be categorized into subtypes by the presence of short, conserved amino acid motifs, the rest of the sequence being highly polymorphic (13). The var gene family is subdivided into three main subgroups, A, B, and C, based on semiconserved upstream sequences (12). All group A var genes are located near the telomeres, all group C var genes are near the centromeres, and group B var genes can be found in either location. The var gene groups have functional and clinical significance. Group B and C var genes encode PfEMP1 variants that bind CD36 (14, 15) and are linked to nonvirulent clinical disease, whereas group A var genes encode non–CD36-binding variants linked to severe clinical disease including CM (16–19). Some group A var genes encode PfEMP1 variants that bind to uninfected erythrocytes to form rosettes (20–25); however, the adhesion phenotype of the majority of group A variants is unknown currently.
Although, because of their position on the surface of IEs, PfEMP1 encoded by var genes and other VSA such as rifins and stevors are the major candidates for parasite adhesion ligands, it remains possible that other parasite adhesins remain to be discovered. We investigated the whole transcriptome of parasites selected for binding to HBEC-5i cells to identify the parasite ligands for adhesion. We postulated that the parasite ligand(s) necessary for binding to HBEC-5i would be expressed at a higher level in selected (adherent) than in unselected (nonadherent) parasites and tested the hypothesis using microarray technology (25).
Results
Selection of P. falciparum for Binding to HBEC-5i.
Preliminary experiments indicated that in vitro cultures of P. falciparum adhere poorly to HBEC-5i (Fig. 1A, Left). Four P. falciparum laboratory strains (HB3, 3D7, IT, and Dd2) were selected for binding to HBEC-5i cells by repeated panning (9). After five to seven rounds of selection, HBEC-5i–adherent lines were obtained from the HB3 (Fig. 1A, Right), 3D7, and IT strains. The P. falciparum strain Dd2 did not increase in adhesion to HBEC-5i cells even after five rounds of selection, suggesting that Dd2 lacks (or is unable to transcribe or transport) the necessary parasite ligand or that the lack of knobs in Dd2 prevents adhesion (26). HB3 was selected twice independently on HBEC-5i to provide a replicate for subsequent experiments (HB3-HBEC1 and 2). HB3 also was selected on TNF-activated HBEC-5i (HB3-HBEC-TNF) to investigate whether different parasite ligands would be selected on cytokine-stimulated and resting endothelial cells.
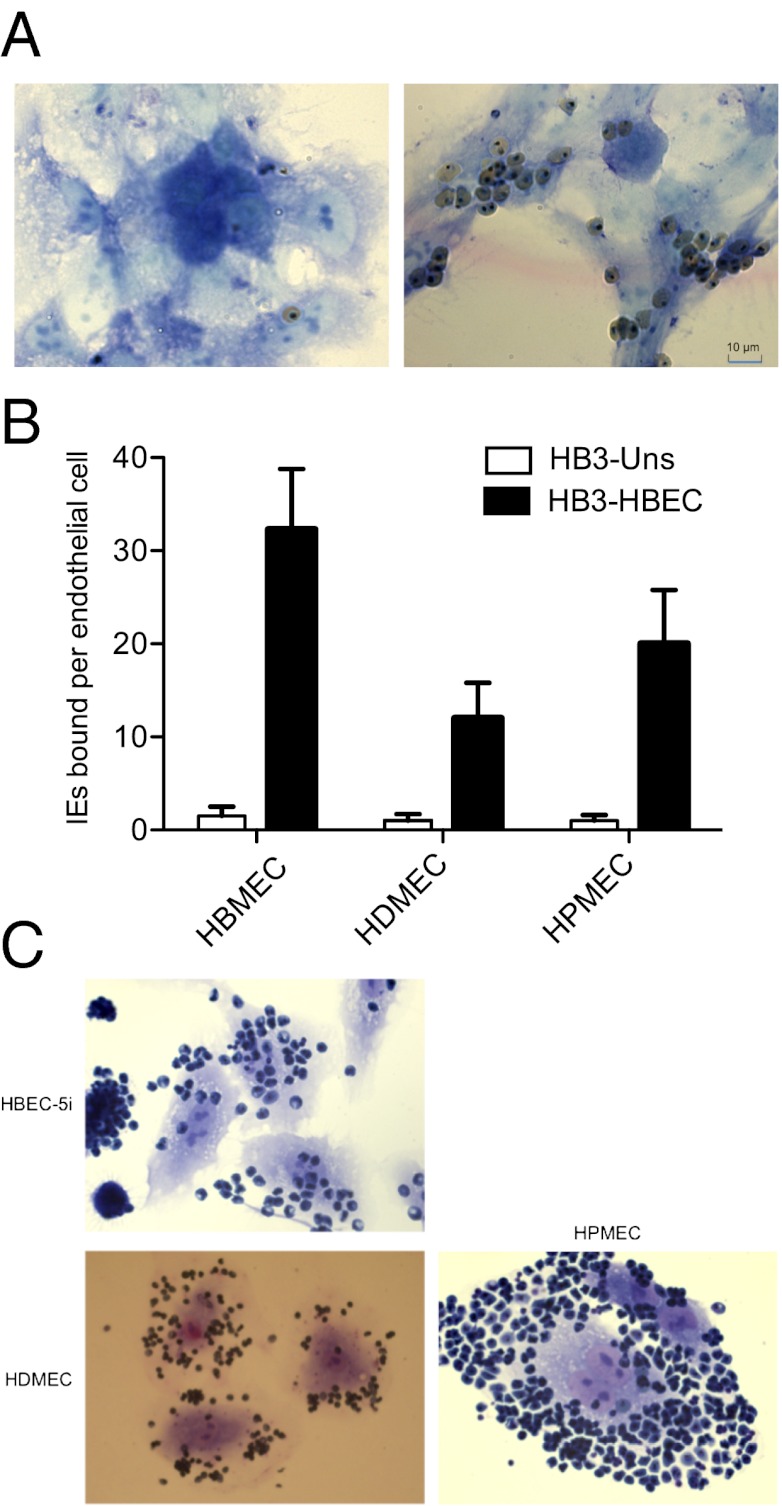
Selection of P. falciparum IEs for adhesion to HBMEC. (A) Unselected P. falciparum lines (e.g., HB3 shown here) show only minimal binding to HBEC-5i cells (Left). After five rounds of panning, adherent parasites are selected (Right). The cells were fixed with glutaraldehyde, stained with Giemsa, and visualized by microscopy at 1,000× magnification. (B) Binding of selected (HB3-HBEC) and unselected (HB3-Uns) parasite lines to primary endothelial cells from human brain (HBMEC), skin (HDMEC), and lung (HPMEC). Means + SE from triplicate wells are shown. (C) Rebinding of HBEC-5i–adherent IEs that were detached mechanically from HBEC-5i cells and then reincubated with HBEC-5i cells (positive control), HDMEC, or HPMEC.
To assess the relevance of HBEC-5i selection for primary human tissues, HB3-HBEC parasites were tested for adhesion to primary endothelial cells derived from brain, dermal, and lung tissues (ScienCell). In all cases the selected parasites showed enhanced binding to primary endothelial cells (>10-fold greater adhesion of selected than unselected parasites) (Fig. 1B). Furthermore, HBEC-5i–binding IEs that were physically detached from HBEC-5i cells and then incubated with other endothelial cell lines showed strong binding (Fig. 1C), excluding the possibility that subpopulations of parasites bound to the different cell lines. These data show that the HBEC-5i–selected phenotype is relevant for primary human tissues and indicates that the host receptor is present on endothelia from diverse sources, consistent with sequestration of IEs in multiple endothelial beds in CM patients in vivo (27). Neither unselected nor HB3-HBEC–selected parasite lines bound above background levels to COS-7 cells, showing that the selected parasites do not bind nonspecifically to all cell types.
Adhesion Phenotype of HBEC-5i–Selected Parasites.
The other adhesion properties of the selected parasites were examined. The selected lines from all three strains did not form rosettes (SI Appendix, Fig. S1A), and platelet-mediated clumping was reduced significantly after selection for HBEC-5i binding (SI Appendix, Fig. S1B). HB3-HBEC–selected parasites showed significantly lower binding to CD36 and ICAM-1 than did HB3–unselected parasites (SI Appendix, Fig. S1C), and there was no increase in binding to any known P. falciparum adhesion receptor, including fractalkine, PECAM-1, P-selectin, E-selectin, VCAM-1, integrin αVβ3, thrombospondin, NCAM, fibronectin, heparin, chondroitin sulfate A, hyaluronic acid, gC1qR, or heparan sulfate (SI Appendix, Fig. S1C and Table S1). IT-HBEC–selected parasites also showed significantly lower binding to CD36 than did IT–unselected parasites, and the only minor but statistically significant increase was in binding to gC1qR (SI Appendix, Fig. S1D), although absolute levels of binding remained low. gC1qR has been implicated primarily as a platelet-mediated clumping receptor for P. falciparum, although it also is expressed on endothelial cells (28). IT-HBEC parasites had a clumping frequency of <2%; therefore it seems unlikely that gC1qR is a major receptor for these parasites. Adhesion-blocking antibodies to CD36 (29) and ICAM-1 (30) had no effect on binding of HBEC-selected parasites to HBEC-5i (SI Appendix, Fig. S1E), thus further excluding a role for these receptors in HBEC-5i binding. Therefore we were unable to identify the HBEC-5i receptor(s) used by the HBEC-selected parasite lines, and HBEC-5i binding may involve an unknown host endothelial receptor.
We also examined knob positivity in the selected and unselected parasite lines, because lack of knobs is known to influence parasite adhesion properties (26). We found that the percentage of knob-positive IEs in the selected and unselected lines of each strain did not differ significantly (mean percent of knob-positive IE (SE) for HB3-HBEC–selected, 62.5 (2.3) and –unselected, 65.0 (1.7), P = 0.41; IT-HBEC–selected, 69.3 (2.5) and –unselected, 68.3 (2.7), P = 0.79; 3D7-HBEC–selected, 54.0 (1.5) and –unselected, 55.5 (1.7), P = 0.53; unpaired t test in each case). Therefore, the presence or absence of knobs is unlikely to explain the differences in HBEC-5i adhesion between the selected and unselected parasite strains.
Whole-Transcriptome Analysis Identifies a Subset of Group A var Genes as Being Highly Transcribed in HBEC-5i–Selected Parasites.
We used a VSA-supplemented microarray chip to examine the whole transcriptome of selected and unselected parasites, with the aim of identifying candidate parasite adhesion ligands as gene(s) whose transcriptional levels increased markedly after selection (25). The VSA-supplemented chip contains oligonucleotide probes for all the VSA genes (var, rif, and stevor) from the HB3, IT, and Dd2 strains (25) added to the existing 3D7-based chip (31). Isogenic pairs of selected and unselected P. falciparum parasite cultures were synchronized (SI Appendix, Table S2), and RNA samples were collected every 8 h for 48 h. The selected and unselected parasites were at similar levels of maturity during the time-course experiment, as shown by Giemsa smear (SI Appendix, Fig. S2) and by strongly positive Pearson correlation coefficients when data from all oligonucleotide probes at each time point for selected and unselected parasites were compared (SI Appendix, Fig. S3; see exceptions for time point 3 of HB3-HBEC-TNF and time point 6 of IT-HBEC, whose data should be interpreted with caution).
The level of transcription of each gene in selected parasites was compared with that in unselected parasites and was expressed as a fold expression ratio. In HB3, the gene with the most markedly and consistently increased transcription in selected parasites was a group A var gene, HB3var3 (Fig. 2 A–D and G). There were five oligonucleotide probes on the microarray chip for this gene, and all five showed strongly increased hybridization signals in HB3-HBEC1 and HB3-HBEC2 compared with unselected HB3 (Fig. 2 B and C). Taking an average of all five probes, HB3var3 was up-regulated in selected parasites by up to 61-fold (Fig. 2G). (By “up-regulated” we mean that the amount of mRNA for a particular gene was increased in selected compared with unselected parasites.) Selection of HB3 parasites on TNF-activated HBEC-5i also resulted in a parasite population with increased transcription of HB3var3 (Fig. 2 D and G). Multiple other var genes were up-regulated at time point 3 in the parasites selected on TNF-activated HBEC-5i, but the upregulation of these genes may have been caused by poor synchronization between selected and unselected parasites at this time point (SI Appendix, Fig. S3).
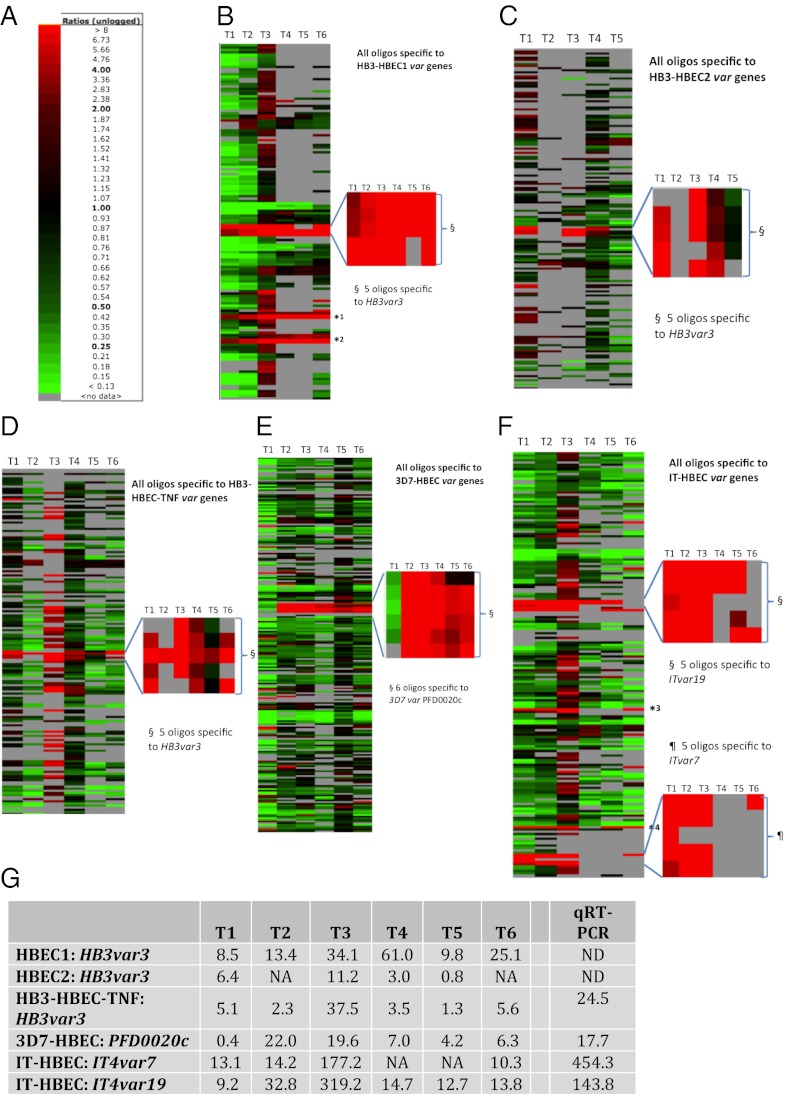
Microarray analysis indicates that a subset of group A var genes shows markedly increased transcription after selection for HBEC-5i binding. (A) Color scale showing the fold expression ratios of gene transcription in selected versus unselected parasites. (B–F) Var gene transcription throughout the 48 h asexual life cycle in HBEC-5i–selected and unselected P. falciparum strains HB3: (B) replicate 1, HB3-HBEC1; (C) replicate 2, HB3-HBEC2; (D) HB3 on TNF-stimulated cells;,HB3-HBEC-TNF; (E) 3D7-HBEC; and (F) IT-HBEC. Each row represents a different oligonucleotide probe, and each column is a time point. T1 = maximum parasite maturity, 8 h postinvasion; T2 = 16 h; T3 = 24 h; T4 = 32 h; T5 = 40 h; T6 = 48 h. Red rectangles indicate higher transcription in the selected than in the unselected population. Green rectangles indicate higher transcription in the unselected population. A gray rectangle indicates no var gene transcription detected above background level in at least one parasite population. For each selection, the most highly up-regulated var gene(s) has been zoomed out. All oligonucleotide probes are shown except for those with no transcription above background level at any time point. A few var genes have sections of sequence identical to other var genes within the same strain (62). For example, the second half of HB3var3 shows similarities with HB3var5 and HB3var46p. Therefore, some oligos are not unique to a single sequence. *1, oligos HB3var46p_2 and HB3var46p_3 match the HB3var3 gene. Oligos HB3var46p_0 and HB3var46p_1 are unique to HB3var46p and are not up-regulated. *2, oligos HB3var5_2 and HB3var5_3 match HB3var3. Oligos HB3var5_0 and HB3var5_1 are unique to HB3var5 and are not up-regulated. *3, oligos ITvar32_2, ITvar32_3, and *4, oligo ITvar6_0 - match ITvar19. However, other oligos specific to ITvar32 (ITvar32_0 and 1) and ITvar6 (ITvar6_1, 2 and 3) are not up-regulated. (G) Expression fold ratios (unlogged) of the most up-regulated var genes in each selection. Numbers represent the average of all oligonucleotide probes specific to that gene. The last column indicates real-time qPCR data obtained from RNA at late ring stage. NA, not available (i.e., no transcription above background level was detected in at least one parasite population); ND, not determined.
In 3D7, a single group A var gene, PFD0020c, showed the most marked and consistent increase in transcription after selection (Fig. 2 E and G). All six oligonucleotide probes to PFD0020c showed a similar pattern. In the IT strain, two var genes, ITvar7 (group A) and ITvar19 (group B), showed highly increased transcription after selection (Fig. 2F). The five probes specific to each of these two genes showed, on average, an up-regulation of 10- to more than 100-fold (Fig. 2G).
When the five selections are considered together, the microarray data for the var gene family are striking. In all five selections, one or two var genes show markedly increased transcription in the selected parasites, but multiple other var genes are down-regulated. At time point 3 (16–24 h postinvasion), there is an 11- to 320-fold increase in HB3var3, PFD0020C, ITvar9, and ITvar19 in the selected populations (Fig. 2G). No other var gene was up-regulated by more than twofold across more than one time point.
Quantitative real-time PCR (qPCR) with specific primers to the var genes of each parasite strain confirmed the up-regulation detected by microarray (Fig. 2G) and did not detect any other up-regulated var genes. Expression profiling by RT-PCR with universal primers to the var gene DBLα region (32, 33) also identified the same set of up-regulated var genes (SI Appendix, Fig. S4 A–D), confirming that all three methods are comparable. RT-PCR experiments on HB3 parasites selected for binding to human dermal microvascular endothelial cells (HDMEC) and human pulmonary microvascular endothelial cells (HPMEC) showed that the group A var gene up-regulated with HBEC-5i (HB3var3) also was up-regulated following selection on other endothelial types (SI Appendix, Fig. S4E).
Rif Genes Located Head-to-Head with the var Gene Ligand Candidates Are Up-Regulated.
In the rif family, three genes showed increased transcription by threefold or more in at least one time point in all three HB3 selections (Fig. 3A). Only one of these genes, HB3RifA_081, showed a substantial, eight- to 100-fold increase (Fig. 3B). Interestingly, this rif gene is located next to HB3var3, on the opposite strand, and is transcribed in the opposite direction. Thus, the selection of HB3 parasites on HBEC-5i, with or without prior activation with TNF, led to the increased transcription of HB3var3 and its “head-to-head” HB3_RifA_081 gene. No stevor gene was up-regulated by threefold or more in all three HB3-derived selections.
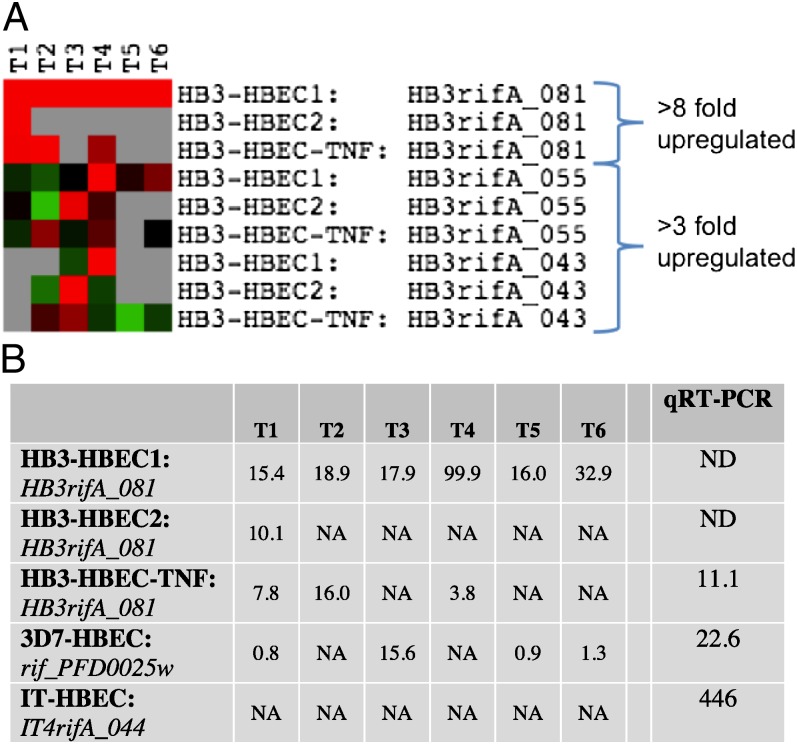
The rif genes located head-to-head with the up-regulated var genes are up-regulated also. (A) HB3 rif genes up-regulated by at least threefold in all HB3-derived strains. Because of the small size of rif genes (1–2 kbp), there was only one oligonucleotide probe per gene on the microarray chip. Color scale is as in Fig. 2. (B) Expression fold ratios of the rif genes located head-to-head with the most up-regulated var genes in each selection. The last column indicates real-time qPCR data, obtained from RNA at late ring stage. NA, not available (i.e., no transcription above background level was detected in at least one parasite population); ND, not determined.
In 3D7-HBEC, the most highly up-regulated rif gene was PFD0025w (Fig. 3B and SI Appendix, Fig. S5A). Again, this rif gene is located head-to-head with the most highly transcribed var gene, PFD0020c. Several other rif genes and four stevor genes showed more than threefold up-regulation in at least one time point (SI Appendix, Fig. S5A). These data are each based on a single probe (because of the small size of rif and stevor genes) and a single experiment; therefore further replicates would be required to determine whether these increases are consistent. In IT-HBEC, five rif genes and one stevor gene showed more than threefold up-regulation in a single time point (SI Appendix, Fig S5B). No expression data were available for IT4rifA_044, the rif gene found head to head with ITvar7. This lack of data may mean that this gene is not transcribed above background levels in one of the parasite populations or could be the result of a technical problem, such as misprinting of the oligonucleotide probe. The other up-regulated var gene in IT parasites, ITvar19, does not have a head-to-head rif gene, because these head-to-head arrangements are found only with group A var genes.
To confirm the up-regulation of the specific head-to-head rif genes in the HB3 and 3D7 strains, and to examine further the transcription of IT4rifA_044, the rif gene found head-to-head with ITvar7, we carried out real-time qPCR with rif-specific primers (34). This evaluation confirmed the up-regulation of HB3_RifA_081 in HB3-selected parasites and PFD0025w in 3D7-selected parasites (Fig. 3B). Furthermore, the data showed a 446-fold increase in IT4rifA_044 in IT-selected parasites (Fig. 3B).
Taken together, these data indicate that selection for binding to HBEC-5i leads to increased transcription of specific group A var genes and that the rif genes located head-to-head with these var genes are up-regulated also. Therefore, the genes HB3rifA_081, PFD0025w, and IT4rifA_044 also should be considered as encoding potential parasite ligands for HBEC-5i binding, given the known location of rifins on the surface of IEs (35).
Some Exported Proteins Show Increased Transcription in HBEC-Selected Parasites.
The four var genes identified above and their accompanying rif genes are by far the most highly and consistently up-regulated genes after selection. However, other non-VSA genes could be involved in cytoadherence to HBEC-5i. The vast majority of non-VSA genes have a sequence that is virtually identical from one strain to another; therefore the three strains (HB3, 3D7, and IT) can be considered as replicates of each other (36). Using the arbitrary cutoff of a twofold increase in at least one time point, 15 non-VSA genes were found to be up-regulated, and 58 non-VSA genes were down-regulated in all five selections (SI Appendix, Figs. S6 and S7). No specific functional pathways were statistically significantly up-regulated by functional enrichment analysis. Furthermore, no non-VSA genes showed increases in transcription comparable in intensity or consistency to the up-regulated var genes (SI Appendix, Fig. S6 and Fig. 2). Only one gene was up-regulated by at least threefold in all five selections: PF14_0472, a member of the Plasmodium helical interspersed subtelomeric (PHISTa) family (37). Six other up-regulated genes encode proteins predicted or proven to be exported: P. falciparum erythrocyte membrane protein 3 (PfEMP3) (38), ring-IE surface antigen (RESA-1) (39), membrane associated histine-rich protein 1 (MAHRP-1) (40), and three others with a Pexel motif: PF14_0740, PF11_0035, and PFF0055w (37). These six genes and PF14_0472 are up-regulated at time point 3 (16–24 h postinvasion) in almost all selections, indicating that they could be translated in time for cytoadherence at the pigmented trophozoite stage.
Of the 58 down-regulated genes, 11 were down-regulated by at least threefold in at least one time point (SI Appendix, Fig. S7). Intriguingly, 41 of the 58 down-regulated genes encode proteins proven or predicted to be involved with merozoite invasion. Indeed, a functional enrichment analysis revealed the merozoite invasion pathway to be highly enriched (P ~0) (SI Appendix, Table S3). In other words, the selection of parasites for binding to HBEC-5i resulted in a statistically highly significant decrease in transcription of invasion-related genes. This decrease in transcription occurred during time points 1–5 but not at time point 6 (schizogony) when invasion genes primarily are expected to be transcribed and translated. Thus, the actual amounts of invasion proteins necessary for merozoites to invade red blood cells may remain the same in HBEC-5i–selected parasites.
Taken together, examination of the entire transcriptome of three P. falciparum strains selected for adhesion to HBEC-5i identifies a subset of group A var genes and their head-to-head rif genes as the most likely candidates for adhesion ligands because of their markedly increased transcription after selection.
Up-Regulated Var Genes Show Similarities in PfEMP1 Architecture.
Examination of the var genes up-regulated in HBEC-5i–selected parasites reveals that they encode PfEMP1 variants that are diverse in sequence (32–46% amino acid identity between each pair) (SI Appendix, Table S4). However, the up-regulated variants do show similarities in PfEMP1 architecture (Fig. 4). As expected for group A var genes, HB3var3, PFD0020c, and ITvar7, all encode variants with a DBLα1 domain at the N-terminus, whereas the group B ITvar19 variant has a DBLα2 domain (Fig. 4). Despite its group B upstream region, the ITvar19-encoded variant has some group A-like features: It is large (with seven extracellular domains, compared with four for a typical group B variant) and has a CIDRα1 domain (usually found in group A variants). The presence of the CIDRα1 domain, which is uncommon (present in <10% of all var genes) is the most obvious distinguishing feature of the four adhesion ligand candidates. DBLα1 and CIDRα1 domains recently have been classified into subgroups on the basis of sequence homology (41). Both HB3var3 and ITvar7 have the N-terminal domain pair DBLα1.7/CIDR1α1.4, also known as “domain cassette (DC) 13.” [Domain cassettes are sets of PfEMP1 domains that usually occur together (41).] Of seven P. falciparum strains whose var gene repertoires have been fully sequenced and classified, most strains have only one variant with DC13, and these variants are all group A var genes that show PfEMP1 architecture similar to that of HB3var3 and ITvar7 (41). Thus, DC13-type variants, including HB3var3 and ITvar7, may represent functionally similar alleles in different parasite strains. PFD0020c and ITvar19 have different DBLα types, but they share very similar CIDRα1.1 domains with 74% amino acid identity (but only 39–43% identity with CIDRα1.4 of HB3var3 and ITvar7) (SI Appendix, Table S4). Other sequenced strains usually have only one variant with a CIDRα1.1 domain, and most of these strains are group B PfEMP1 variants with PfEMP1 architecture similar to that in ITvar19, represented by DC8 (41). PFD0020c has three of the four domains usually found in DC8, differing only in the first DBL domain (Fig. 4).
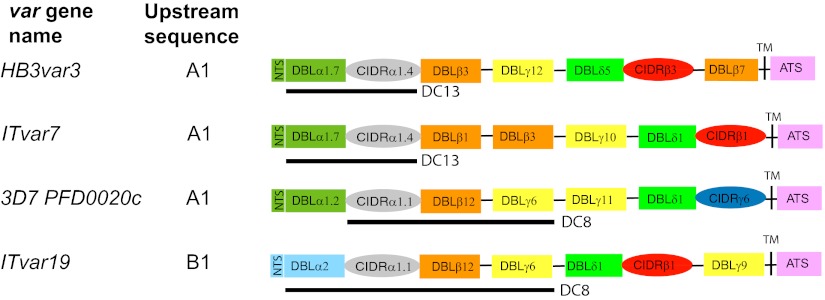
PfEMP1 architecture of variants up-regulated after HBEC-5i selection. ATS, acidic terminal segment; CIDR, cysteine-rich interdomain region; DBL, Duffy binding-like domain; DC13, domain cassette 13; DC8, domain cassette 8; NTS, N-terminal segment. Pairwise amino acid identities between domains are shown in SI Appendix, Tables S4 and S5.
The HB3var3 variant is identical to HB3var5 from the DBLγ domain to the end of the extracellular region. The fact that HB3var3 but not HB3var5 was up-regulated after selection for binding to HBEC-5i suggests that the functional binding region includes the first three domains (NTS-DBLα-CIDRα-DBLβ) that differ in HB3var3 and HB3var5.
Antibodies Against HB3var3 Inhibit Adhesion of HB3-HBEC to HBEC-5i.
The transcriptional patterns shown above combined with the known adhesion properties of PfEMP1 make the PfEMP1 variants encoded by HB3var3, PFD0020c, ITvar7, and ITvar19 the most likely candidate parasite adhesion ligands for HBEC-5i binding. To test the hypothesis that this subset of group A-like var genes encodes the parasite ligands for adhesion to HBEC-5i, we raised antibodies to specific PfEMP1 domains.
The NTS-DBLα1 and the NTS-DBLα1-CIDRα1 (hereafter called the “di-domain”) from HB3var3 were expressed as recombinant proteins in Escherichia coli. For NTS-DBLα1, the recombinant protein was mostly of the expected molecular weight (54.1 kDa) and showed a shift upon reduction, indicating the presence of disulfide bonds in the recombinant protein (Fig. 5A, Left). For the di-domain, much of the protein was degraded, although some protein at the expected molecular weight (88.2 kDa), and which showed a shift upon reduction, was obtained (Fig. 5A, Right).
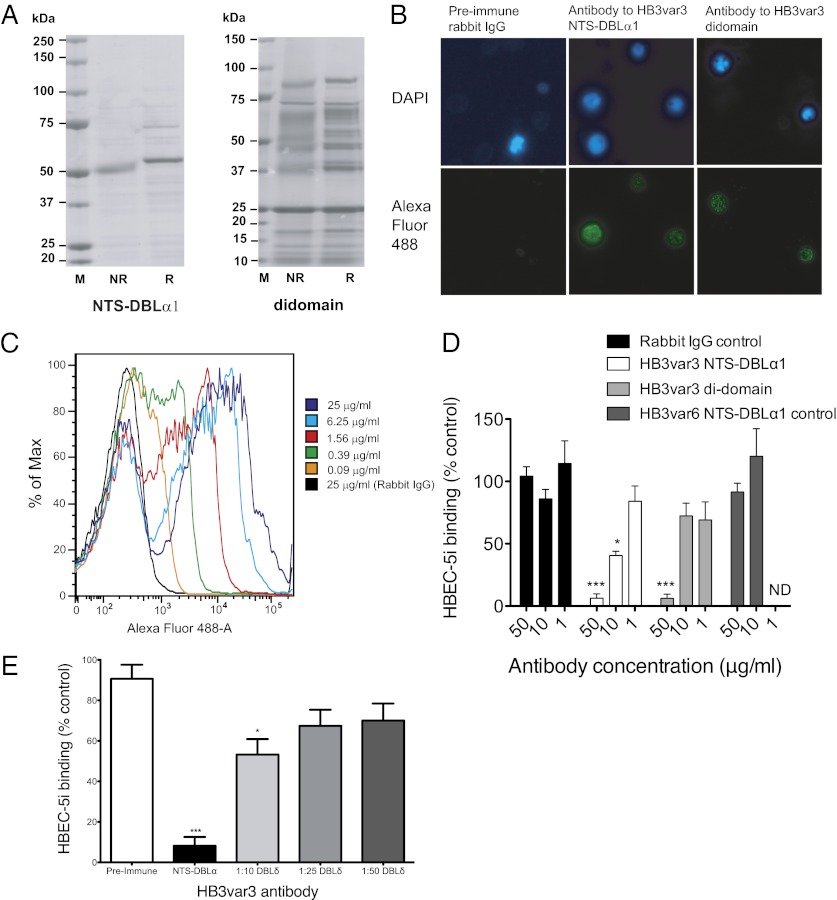
Antibodies against HB3var3 inhibit adhesion of HB3-HBEC to HBEC-5i cells. (A) SDS/PAGE showing recombinant NTS-DBLα1 and di-domain (NTS-DBLα1-CIDRα1) domains from HB3var3 expressed in E. coli. (B) Immunofluorescence assay with antisera to HB3var3 (1/100 dilution) showing punctate surface fluorescence on live IEs. IEs are shown by DAPI staining of the parasite nuclei. PfEMP1 antibody was detected with highly cross-absorbed goat anti-rabbit IgG Alexa Fluor 488 (1/1,000). (C) Titration of anti-HB3var3 antibodies (purified total IgG) against live HB3-HBEC IEs by flow cytometry. (D) Inhibition of HB3-HBEC binding to HBEC-5i cells by antibodies to HB3var3 NTS-DBLα1 and di-domain. Means and SD from quadruplicate wells of a representative experiment are shown. Binding is compared with control wells with no added IgG. Negative controls are nonimmunized rabbit IgG and IgG raised to the NTS-DBLα1 domain of a rosette-mediating group A PfEMP1 variant (HB3var6). A significant decrease in adhesion was seen in the presence of HB3var3 antibodies (P < 0.0001, ANOVA). ***P < 0.001, *P < 0.05, Tukey’s multiple comparison test. (E) Inhibition of HB3-HBEC binding to HBEC-5i cells by antibodies to HB3var3 DBLδ5. Mean and SE from three independent experiments are shown. Binding is compared with control wells with no added IgG. The negative control is preimmune serum from the rabbit used to raise DBLδ5 antibodies. A significant decrease in adhesion was seen in the presence of HB3var3 antibodies (P < 0.0001, ANOVA). *P < 0.05, ***P < 0.001, Tukey’s multiple comparison test.
Antisera to HB3var3 NTS-DBLα1 and the di-domain showed punctate surface fluorescence of live HB3-HBEC IEs by immunofluorescence assay (IFA) (Fig. 5B) typical of PfEMP1 antibodies (23). Rabbit preimmune sera and nonimmunized rabbit control sera were negative by IFA. Total IgG were purified from the antiserum and used at fourfold dilutions in flow cytometry to determine the end titer (here defined as the lowest concentration of total IgG to stain in >50% of live IEs). The HB3var3 NTS-DBLα1 antibodies gave surface reactivity down to 0.39 μg/mL of total IgG (Fig. 5C), and the di-domain antibodies gave surface reactivity down to 1.56 μg/mL of total IgG. The HB3var3 NTS-DBLα1 antibodies were tested at 400 μg/mL for live IE surface reactivity with other P. falciparum strains selected for a range of different adhesion phenotypes (24). The antibodies did not show surface reactivity with HB3-unselected parasites, 3D7-HBEC, 3D7-unselected, IT-HBEC, IT-unselected, six P. falciparum rosetting strains, or 12 clinical isolates (data are shown in figures 4b and 6a of ref. 24, in which the non-Ros group A control is HB3var3 NTS-DBLα1 antibody). Therefore, the HB3var3 NTS-DBLα1 antibodies are highly variant- and strain-specific and show surface reactivity only with the homologous parasite line HB3-HBEC expressing the HB3var3 PfEMP1 variant.
To determine whether the HB3var3 PfEMP1 variant is the parasite ligand for binding to HBEC-5i, the effect of HB3var3 antibodies on adhesion was examined. Both NTS-DBLα1 and di-domain antibodies inhibited binding to HBEC-5i compared with control nonimmunized rabbit IgG (Fig. 5D). Polyclonal rabbit IgG raised against a PfEMP1 variant not involved in HBEC-binding (against the NTS-DBLα1 domain of the rosette-mediating HB3var6 group A PfEMP1 variant) (24) showed no inhibition of binding (Fig. 5D). We also tested antiserum raised against another recombinant domain of the HB3var3 variant (DBLδ5) produced in baculovirus (42). Previous work has shown that antibodies to multiple PfEMP1 domains are able to block adhesion in the case of rosetting parasites (23). The HB3var3 DBLδ5 antiserum showed punctate surface fluorescence with live IEs of HB3-HBEC parasites at 1/25 and 1/100 dilution, whereas the preimmune serum was negative at the same concentrations. The antiserum was negative by IFA against HB3-unselected, 3D7-HBEC, 3D7-unselected, IT-HBEC, and IT-unselected parasites. The antiserum to HB3var3 DBLδ5 showed specific inhibition of adhesion of HB3-HBEC IEs to HBEC-5i, although it was not as effective as NTS-DBLα1 antibody (Fig. 5E). Therefore, three different antibody preparations against two distinct domains of the HB3var3 PfEMP1 variant inhibited binding of HB3-HBEC to HBEC5i, confirming that HB3var3 is a parasite adhesion ligand and making it extremely unlikely that antibody cross-reactivity with other molecules on the surface of IEs is responsible for the data shown. None of the antibodies caused agglutination of IEs at the concentrations used in the adhesion inhibition assays.
Antibodies to two distinct domains of the PfEMP1 variant up-regulated in 3D7 parasites, PFD0020c, also were tested for adhesion inhibition. Antibodies to the NTS-DBLα1 domain of PFD0020c (produced in E. coli) gave punctate surface fluorescence of 3D7-HBEC live IEs down to 6.25 μg/mL of total IgG, whereas antibodies to DBLγ6 (produced in Baculovirus) gave surface reactivity down to 100 μg/mL Both antibodies were variant- and strain-specific and did not show surface reactivity with the IT- or HB3-HBEC–selected or –unselected parasite lines when tested at 400 μg/mL The NTS-DBLα1 antibodies inhibited binding of 3D7-HBEC to HBEC-5i by 52% compared with rabbit IgG control at 50 μg/mL of total IgG, whereas the DBLγ6 antibodies inhibited binding by 64% compared with rabbit IgG control at 250 μg/mL of total IgG (P < 0.001, paired t test). Therefore, for PFD0020c, two independent antibody preparations to separate domains show variant- and strain-specific surface reactivity and inhibit adhesion of 3D7-HBEC to HBEC-5i, providing evidence that the PFD0020c PfEMP1 variant is the parasite adhesion ligand.
In Vivo Significance of the HBEC-5i–Binding Subset of Group A PfEMP1 Variants.
No animal model adequately reflects the pathology and clinical features of severe P. falciparum malaria in humans; therefore, it is not possible to test directly whether HBEC-5i–selected parasites cause CM. As an alternative approach to examine the in vivo significance of the HBEC-5i–binding variants, we investigated whether African children suffering from CM develop antibodies that recognize the HBEC-selected parasite lines. Plasma samples from acutely ill or convalescent Kenyan children with CM or uncomplicated malaria (UM) were used to assess surface recognition of live IEs from selected and unselected parasites by flow cytometry. Acute plasma samples collected from both clinical groups at the time of hospital admission (predominantly reflecting prior exposure) showed no significant difference in their reactivity with selected or unselected parasite lines (Fig. 6 A and B). However, plasma samples from convalescent patients with CM (reflecting antibody responses to the variant causing the clinical episode) showed significantly greater recognition of the HBEC-selected lines than of the unselected lines (Fig. 6C). In contrast, plasma samples from convalescent patients with UM did not show differential reactivity with HBEC-selected lines compared with -unselected lines (Fig. 6D), although surface reactivity against both HBEC-selected and -unselected lines increased compared with the samples from acutely ill patients (Fig. 6 B and D). When each parasite strain was considered individually, all three strains showed a trend toward increased recognition of HBEC-selected compared with -unselected parasites in sera from convalescent patients with CM, but the increase was statistically significant only in the 3D7 strain (SI Appendix, Fig. S8).
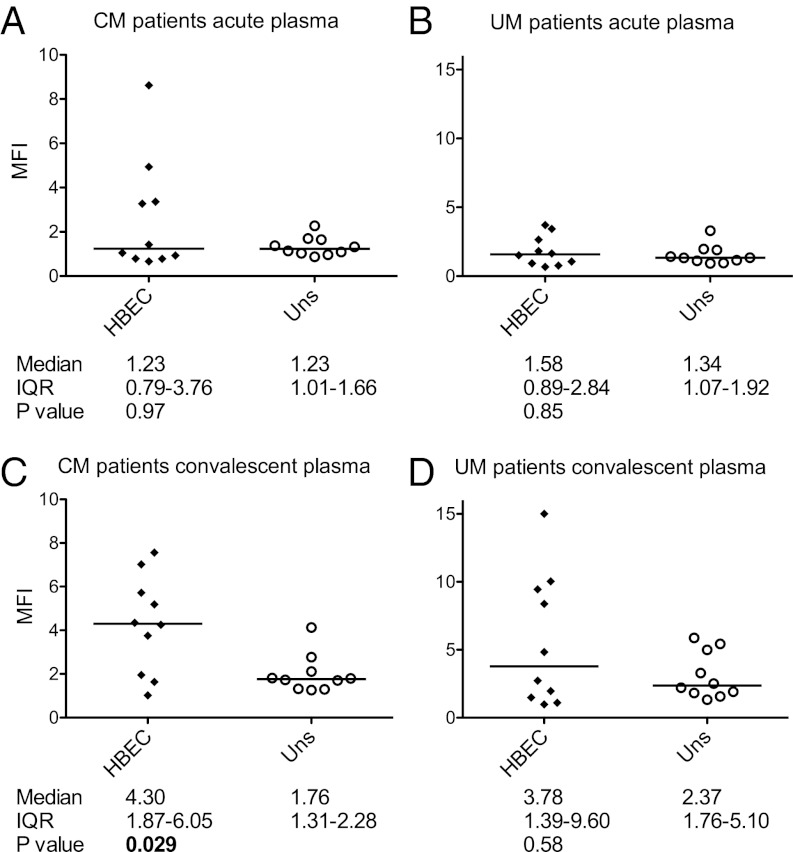
Recognition of HBEC-selected and -unselected parasites lines by antibodies from African malaria patients. Acute and convalescent plasma samples were collected from 10 patients with CM and 10 age- and date of admission-matched patients with UM. Surface recognition of HBEC-selected and -unselected (uns) parasite lines was tested by flow cytometry. The mean fluorescence intensity (MFI) of the uninfected erythrocyte population was subtracted from the MFI of the IE population to give the specific MFI of the IE population shown on the y axis. Each data point represents plasma from one patient. The median of the values from the three individual strains was used to represent the specific recognition of HBEC-selected or -unselected parasites by each plasma sample (the individual strains are shown in SI Appendix, Fig. S8). (A) Acute plasma from patients with CM. (B) Acute plasma from patients with UM. (C) Convalescent plasma from patients with CM. (D) Convalescent plasma from patients with UM. IQR, interquartile range.
Discussion
In this work we found that a distinct subset of group A-like var genes was highly transcribed in three P. falciparum strains selected for adhesion to an HBEC line (Fig. 2). The up-regulated genes encode PfEMP1 variants with similar domain structures exemplified by the presence of DC8 (3D7_PFD0020c and ITvar19) or DC13 (HB3var3 and ITvar7) (Fig. 4). In their companion articles, Avril et al. (43) provide independent confirmation of the up-regulation of DC8 genes following selection on brain endothelial cells, and Lavstsen et al. (44) show DC8 and DC13 genes to be highly differentially transcribed in patients with severe malaria compared with UM controls, confirming the clinical importance of DC8- and DC13-expressing parasites. DC8 genes were differentially transcribed in all clinical forms of severe malaria (CM, severe malarial anemia, and respiratory distress), consistent with our findings and those of Avril et al. (43) that parasites selected for binding to HBEC-5i also could adhere to other primary endothelial cell types, giving the potential to sequester at multiple sites throughout the body in addition to the brain. Taken together, these three studies identify parasites expressing DC8 and DC13 as having high potential for cytoadherence to endothelial cells in the brain and elsewhere and suggest that DC8 and DC13 PfEMP1 variants may be important targets for interventions to treat or prevent severe malaria.
To confirm the role of DC8 and DC13 PfEMP1 variants in binding to HBEC-5i, we raised antibodies to domains of HB3var3 and 3D7_PFD0020. In each case antibodies to two distinct domains showed variant- and strain-specific reactivity with the surface of live IEs and blocked binding of IEs to HBEC-5i (Fig. 5). These data strongly support the identification of the HB3var3 and PFD0020c PfEMP1 variants as parasite adhesion ligands for HBEC-5i binding.
Our examination of the whole transcriptome of three parasite strains showed that the only other highly up-regulated genes after selection for HBEC-5i binding were the rif genes located head-to-head with the up-regulated group A var genes (Fig. 3). Given the known location of rifins on the surface of IEs (35), it is possible that the rifins could contribute to host cell binding; further experiments will be required to test this possibility. An alternative hypothesis is that the rif genes are up-regulated because their transcriptional pattern is linked to the adjacent head-to-head var gene. The coregulation of group A var genes and their adjacent rif genes has been reported previously (34, 45) and also was seen in our proof-of-concept experiment for the VSA-supplemented microarray with rosetting parasites (25).
The microarray approach has the potential to reveal non-VSA genes that could be involved directly or indirectly in cytoadherence, for example by contributing to the trafficking of PfEMP1 to the IE surface. We found only 15 non-VSA genes that were up-regulated by twofold or more in all selections; among these genes were several encoding proteins predicted to be exported into the infected red blood cell cytoplasm (SI Appendix, Fig. S6). Two of these genes, MAHRP-1 and PF14_0752 (PHISTa), also were modestly up-regulated in IT parasites selected for rosetting (25). An indirect role for MAHRP-1 in cytoadherence already had been demonstrated, because deletion of the gene leads to depletion of surface-exposed PfEMP1 (46). PF14_0752 is part of the PHISTa subfamily, whose members are specific to P. falciparum (37). PF14_0752 was one of the most highly up-regulated genes in CD36-selected parasites (47), in field isolates (48), and in 3D7 gametocytes (49). Recently a PHISTc family member was shown to bind to the intracellular domain of PfEMP1 (50); however the function of other PHIST family members is unknown and awaits further research.
One of the attractions of the microarray approach is its potential to reveal previously unrecognized parasite adhesion proteins. At first sight, to conclude that the major adhesion ligands are PfEMP1 variants, with a possible contribution from rifins, could be considered disappointing. Similarly, our previous microarray study of rosetting parasites confirmed the marked transcriptional up-regulation of the known parasite rosetting ligand, the PfEMP1 variant encoded by ITvar9 (20), and did not identify any strong alternative candidates (25). However, we suggest that these data, rather than being disappointing, are important precisely because they provide a convincing demonstration of the importance of PfEMP1 in parasite adhesion that is based on global transcriptional data rather than relying solely on the examination of individual genes and gene families that has characterized previous work.
The demonstration here of up-regulation of a subset of group A-like var genes in HBEC-5i–selected parasites fits well with a body of previous work on var gene transcription in P. falciparum clinical isolates as well as the companion article by Lavstsen et al. (44). The majority of studies addressing the relationship between var genes and clinical disease have found an association between group A var gene transcription and severe malaria or CM (16–19, 51). Previously, the only known binding phenotype for group A PfEMP1 variants was rosetting (20–23), with two group A domain cassettes (DC11 and DC16) being linked to rosette formation (24). DC16 was not associated with severe malaria in the study by Lavstsen et al. (44) and DC11 was not tested. DC11 is linked to IgM-positive rosetting (24), which probably is the most clinically important rosetting phenotype (52). Further research will be required to study the association between DC11 variants and severe malaria.
ICAM-1 often is cited as an endothelial receptor involved in CM (53); however, its precise role remains uncertain, because several studies show no significant increase in ICAM-1 binding in parasite isolates from patients with CM as compared with controls (54–56). Furthermore, a human ICAM-1 polymorphism that occurs at high frequency in Africa (57) and reduces the ability of P. falciparum to bind to ICAM-1 (53, 58) does not protect against CM or severe malaria (59). In the work shown here, although ICAM-1 is expressed on the surface of HBEC-5i, it is not the receptor used by the HBEC-selected parasites because (i) in spot-binding assays HBEC-selected parasites did not show increased binding to recombinant ICAM-1 compared with -unselected parasites (SI Appendix, Fig. S1 C and D); (ii) ICAM-1 antibodies did not block adhesion of selected parasites to HBEC-5i (SI Appendix, Fig S1E); and (iii) IT-HBEC parasites express ITvar7 and ITvar19, which have been shown to be ICAM-1 nonbinders (60). The endothelial receptor(s) used by the HBEC-5i–selected parasites are currently unknown.
In conclusion, we show that the P. falciparum ligands for adhesion to HBEC-5i are a subset of group A-like PfEMP1 variants (DC8 and DC13 type). The clinical relevance of these variants was shown by recognition of HBEC-selected lines by sera from children recovering from CM, thus validating the interaction with HBEC-5i as an in vitro model for CM sequestration. The DC8 and DC13 PfEMP1 variants represent potential targets for interventions to prevent or treat CM.
Materials and Methods
Please see SI Appendix, Text for the materials and methods related to the adhesion properties of selected and unselected parasite lines; microarray hybridizations; real-time qPCR; the generation of polyclonal antibodies to HB3var3 NTS-DBLα1, HB3var3 di-domain, and 3D7 PFD0020c NTS-DBLα1; the generation of polyclonal antibodies to HB3var3 DBLδ5 and 3D7 PFD0020c DBLγ6; and flow cytometry with plasma from African children.
Parasite and Human Endothelial Cell Culture.
P. falciparum strains HB3, 3D7, IT/FCR3, and Dd2 were cultured in supplemented RPMI medium as described (61). The HBEC-5i cell line is an immortalized endothelial line described previously (5). HBEC-5i cells were grown in DMEM/F-12 medium (Sigma) supplemented with 2 mM l-glutamine, endothelial cell growth supplement (5 mL of 100× per 500 mL of medium, hereafter referred as “incomplete medium”) (ScienCell), and 10% vol/vol heat-inactivated bovine serum. Parasite strains were selected by panning as described in detail previously (9). For HB3-HBEC-TNF, HBEC-5i cells were activated 24 h before selection by addition of TNFα at 50 μg/mL. HBMEC, HDMEC, and HPMEC (all from ScienCell) were cultured as for HBEC-5i cells, except for the use of endothelial cell medium (ScienCell) and bovine serum at 5% vol/vol, following the manufacturer’s instructions.
Binding Assays on Endothelial Cells.
The day before the assay, endothelial cells were plated on fibronectin-coated eight-well chamber slides (BioCoat 354628; BD) at 104 cells per well. For each assay, at least two replicate wells per slide were used. Parasite cultures were washed twice with incomplete medium. Two hundred microliters of parasite culture (pigmented trophozoites at 5–10% parasitemia and 2% hematocrit) in incomplete DMEM/1% BSA were added per well. The slide was incubated for 75 min at 37 °C with gentle resuspension of cells halfway through the incubation. The chamber then was removed according to the manufacturer’s instructions. The slide was washed in a 100-mm Petri dish (351029; BD) filled with incomplete DMEM. The slide was held still using tweezers, and the dish was rocked gently. This step was repeated four times, each time in a new dish with fresh medium. The slide was checked under an inverted microscope to ensure there were no or few unbound cells remaining. It then was fixed and stained with 1% glutaraldehyde for at least 1 h and was stained with 5% vol/vol Giemsa for 20 min. IEs bound to at least 100 isolated cells were counted by light microscopy using the 100× objective. For rebinding assays, after the washing step, bound IEs were detached by flushing the cell surface with incomplete DMEM using a Pasteur pipette at least 10 times. IEs were placed in a 15-mL tube and centrifuged at 300 × g for 5min. They were resuspended with 200 μL of incomplete DMEM per well and then were reincubated on endothelial cells as above.
IFA and Flow Cytometry with Antibodies to PfEMP1.
IFA with rabbit preimmune and immune sera and live IEs were carried out as described (23). For each antigen, the rabbit serum giving the brightest IFA signal with the lowest background was chosen for purification of total IgG. Staining for flow cytometry was carried out as for IFA except that all incubations included 1.25 μg/mL Hoechst (instead of DAPI) to stain parasite nuclei. After the secondary incubation (1:1,000 of a highly cross-absorbed Alexa Fluor 488 goat anti-rabbit IgG) (Invitrogen) and washes, cells were fixed in 0.5% paraformaldehyde on ice for 20 min and washed twice in FACS buffer (PBS/0.1% BSA/0.1% sodium azide). Cells were resuspended in 500 μL of FACS buffer and analyzed on a Becton-Dickinson LSRII flow cytometer by counting 500,000 events.
Antibody Inhibition of Binding to HBEC-5i Cells.
Binding assays were carried out as described above, with a preincubation step in which the culture suspension (5–10% parasitemia and 2% hematocrit in incomplete DMEM/1% BSA) was incubated for 20 min with antibodies to PfEMP1 or controls (concentrations are shown in figure legends). Two hundred microliters of this suspension were added per well on the endothelial cell culture slide, and the assay was completed as described above. Binding was compared with control wells with no added IgG. All antibodies were tested in at least three independent experiments with at least triplicate wells per antibody in each experiment.
Statistical Analysis.
Graphs and statistical analysis were carried out using GraphPad Prism software (GraphPad).
Acknowledgments
This work was funded by the Wellcome Trust through a 4-y doctoral studentship (to A.C.), a Senior Fellowship in Basic Biomedical Science (to J.A.R), Grant 084226, a Strategic Award (Grant 084538 to C.A.), and Wellcome Trust Programme Grants 084535 and 077092 (to P.C.B.). C.W.W. and L.T. were supported by Danish Medical Research Council Grant 271-08-0540. This paper was submitted with the permission of the director of the Kenya Medical Research Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.H. is a guest editor invited by the Editorial Board.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE32211).
See Commentary on page 10130.
See Author Summary on page 10156 (volume 109, number 26).
This article contains supporting information online at www.pnas.org/lookup/suppl/10.1073/pnas.1120461109/-/DCSupplemental.
References
Author Summary
Author Summary
Cerebral malaria is the most deadly outcome of infection with the protozoan parasite Plasmodium falciparum (1). The pathology of cerebral malaria is characterized by the accumulation of parasitized red blood cells in small blood vessels in the brain. This accumulation occurs because of the binding of parasite adhesion molecules (ligands) on the surface of infected red blood cells to human receptors on brain microvascular endothelial cells; however, the parasite and host molecules involved in this interaction are unknown. We investigated the parasite ligands used for adhesion to human brain endothelial cells to gain fundamental insights into the host–parasite interactions that lead to cerebral malaria. We identified the parasite adhesion ligands as a restricted subset of variant surface antigen molecules encoded by genes called “Group A var genes.” These findings represent an advance in our understanding of cerebral malaria and identify potential targets for therapies and disease prevention.
To investigate host–parasite interactions involved in cerebral malaria, we used Plasmodium falciparum culture-adapted parasite strains and a human brain endothelial cell line, HBEC-5i (2). Initial parasite populations bound poorly to HBEC-5i, showing that these parasites did not show the adhesion phenotype involved in cerebral malaria. However, repeated rounds of selection for HBEC-5i–binding cells yielded highly adherent parasite populations showing “cerebral malaria-type” adhesion properties. We then were able to analyze gene transcription in the highly adherent (selected, cerebral malaria-type) parasites compared with the nonadherent (unselected, “non–cerebral malaria-type”) parasites. We looked for genes that showed markedly increased transcription in the cerebral malaria-type parasites obtained after selection to identify candidate adhesion ligands. Only one or two genes out of the whole genome showed a striking increase in transcription after selection for cerebral malaria-type adhesion properties, allowing straightforward identification of the parasite adhesion ligands.
We successfully selected three P. falciparum strains—HB3, 3D7, and IT/FCR3—for HBEC-5i binding and analyzed global gene expression throughout the 48-h blood-stage life cycle of selected and unselected pairs from each strain using microarray technology (Fig. P1) (3). The only genes that were up-regulated markedly and consistently after selection for HBEC-5i binding were one or two var genes encoding specific members of the P. falciparum erythrocyte membrane protein one (PfEMP1) family. PfEMP1 is a parasite-derived variant antigen expressed on the surface of infected red blood cells that plays a major role in malaria pathology and immune evasion. The genome of every P. falciparum isolate contains about 60 var genes that encode highly polymorphic PfEMP1 variants. The var gene repertoires of different P. falciparum isolates are largely nonoverlapping, resulting in huge diversity in the PfEMP1 family in natural P. falciparum populations. Only one var gene is expressed at a time in an infected red cell, and switching of expression to a different var gene results in antigenic variation of malaria parasites. The up-regulated var genes in the HBEC-5i–selected parasites fall into a subset called “Group A,” characterized by a conserved upstream sequence and location in the subtelomeric regions of the chromosomes. The up-regulated Group A-like PfEMP1 variants from the three different HBEC-5i–selected P. falciparum strains were variable in amino acid sequence, as expected for PfEMP1, but showed strong similarities in domain composition. PfEMP1 variants are composed of cysteine-rich extracellular domains of various types, and sets of domains that commonly occur together are called “domain cassettes” (DC). The up-regulated PfEMP1 variants in HBEC-5i–selected parasites contained either DC8 or DC13. We raised antibodies against the up-regulated PfEMP1 variants that were able to block adhesion of infected erythrocytes to HBEC-5i, confirming the adhesive function of the DC8 and DC13 PfEMP1 variants.
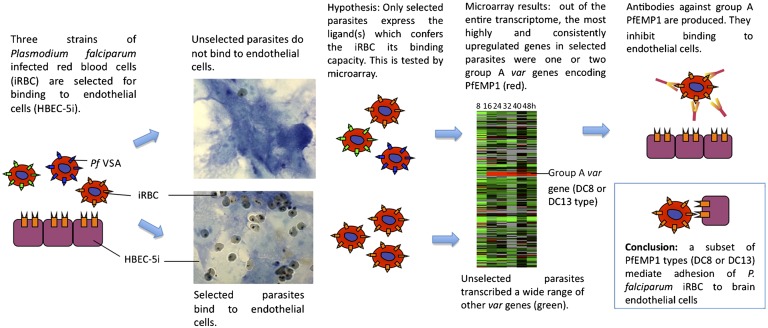
Group A-like PfEMP1 variants mediate adhesion of infected red blood cells to HBEC-5i. P. falciparum-infected red blood cells (iRBC) were selected for binding to a layer of HBEC-5i, mimicking the sequestration that occurs in cerebral malaria cases. The entire transcriptomes of both unselected (nonbinding) and selected (binding) populations of parasites were analyzed by microarray. In the three P. falciparum strains tested, HB3, 3D7, and IT/FCR3, Group A-like var genes were the most up-regulated genes in the selected populations. The up-regulated genes encode a subset of Group A PfEMP1 variants containing the specific domains DC8 and/or DC13. Antibodies raised against the up-regulated PfEMP1 variants blocked the adhesion of iRBC to HBEC-5i, indicating that the DC8 and DC13 type of Group A PfEMP1 variants are the parasite ligands mediating adhesion to human brain endothelial cells. Pf VSA, P. falciparum variant surface antigen.
At present, there are no suitable animal models for human cerebral malaria; therefore, the in vivo significance of in vitro findings is difficult to test. We investigated the clinical significance of our data by examining antibody recognition of the selected and unselected parasite lines by plasma samples from African children recovering from cerebral malaria. This investigation revealed enhanced recognition of the HBEC-5i–selected parasites compared with unselected parasites, highlighting the significance of these strains and associated expressed proteins in cerebral malaria.
In summary, we have shown that the P. falciparum ligands for adhesion to HBEC-5i are a subset of Group A-like PfEMP1 variants characterized by DC8 or DC13. Furthermore, we validated the interaction between P. falciparum-infected red cells and HBEC-5i as an in vitro model for cerebral malaria, which will serve as a useful tool for testing adhesion-blocking drug and vaccine candidates. Furthermore, the strong support provided in our study for the involvement of PfEMP1 in cell adhesion interactions associated with life-threatening P. falciparum infection suggests that the identified variants could be used as possible targets for preventative therapies and treatments for cerebral malaria.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.1120461109
Read article for free, from open access legal sources, via Unpaywall:
https://www.pnas.org/content/pnas/109/26/E1772.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Hyper-diverse antigenic variation and resilience to transmission-reducing intervention in falciparum malaria.
Nat Commun, 15(1):7343, 26 Aug 2024
Cited by: 1 article | PMID: 39187488 | PMCID: PMC11347654
Detection of naturally acquired, strain-transcending antibodies against rosetting Plasmodium falciparum strains in humans.
Infect Immun, 92(7):e0001524, 06 Jun 2024
Cited by: 0 articles | PMID: 38842304 | PMCID: PMC11238554
A human pluripotent stem cell-derived in vitro model of the blood-brain barrier in cerebral malaria.
Fluids Barriers CNS, 21(1):38, 01 May 2024
Cited by: 0 articles | PMID: 38693577 | PMCID: PMC11064301
A novel computational pipeline for var gene expression augments the discovery of changes in the Plasmodium falciparum transcriptome during transition from in vivo to short-term in vitro culture.
Elife, 12:RP87726, 25 Jan 2024
Cited by: 1 article | PMID: 38270586 | PMCID: PMC10945709
Pathogenetic mechanisms and treatment targets in cerebral malaria.
Nat Rev Neurol, 19(11):688-709, 19 Oct 2023
Cited by: 5 articles | PMID: 37857843
Review
Go to all (137) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Selection of Plasmodium falciparum parasites for cytoadhesion to human brain endothelial cells.
J Vis Exp, (59):e3122, 03 Jan 2012
Cited by: 15 articles | PMID: 22230803 | PMCID: PMC3369769
A restricted subset of var genes mediates adherence of Plasmodium falciparum-infected erythrocytes to brain endothelial cells.
Proc Natl Acad Sci U S A, 109(26):E1782-90, 22 May 2012
Cited by: 124 articles | PMID: 22619321 | PMCID: PMC3387091
Rosetting Plasmodium falciparum-infected erythrocytes bind to human brain microvascular endothelial cells in vitro, demonstrating a dual adhesion phenotype mediated by distinct P. falciparum erythrocyte membrane protein 1 domains.
Infect Immun, 82(3):949-959, 16 Dec 2013
Cited by: 34 articles | PMID: 24343658 | PMCID: PMC3958005
Modulation of malaria virulence by determinants of Plasmodium falciparum erythrocyte membrane protein-1 display.
Curr Opin Hematol, 13(3):124-130, 01 May 2006
Cited by: 17 articles | PMID: 16567953
Review
Funding
Funders who supported this work.
Wellcome Trust (5)
Measuring the impact of naturally acquired immunity on the expression of Plasmodium falciparum variant surface antigens.
Dr Peter Bull, University of Oxford
Grant ID: 084535
Centre for Immunity, Infection and Evolution.
Prof Keith Matthews, University of Edinburgh
Grant ID: 095831
Investigation into Plasmodium falciparum rosetting mechanisms and development of rosette-disrupting interventions to treat or prevent severe malaria.
Dr Rowe, University of Edinburgh
Grant ID: 084226
Developing research capacity and leadership in East Africa from the KEMRI- Wellcome Trust Research Programme, Kenya
Professor Kevin Marsh, Kenya Medical Research Institute (KEMRI)
Grant ID: 084538
Director's discretionary recruitment fund and contribution toward Kilifi District Hospital
Professor Kevin Marsh, University of Oxford
Grant ID: 077092





