Abstract
Free full text

A Typical Hospital-Acquired Methicillin-Resistant Staphylococcus aureus Clone Is Widespread in the Community in the Gaza Strip
Abstract
Epidemiological data on community acquired methicillin-resistant-Staphylococcus aureus (CA-MRSA) carriage and infection in the Middle-East region is scarce with only few reports in the Israeli and Palestinian populations. As part of a Palestinian-Israeli collaborative research, we have conducted a cross-sectional survey of nasal S. aureus carriage in healthy children and their parents throughout the Gaza strip. Isolates were characterized for antibiotic susceptibility, mec gene presence, PFGE, spa type, SCCmec-type, presence of PVL genes and multi-locus-sequence-type (MLST). S. aureus was carried by 28.4% of the 379 screened children-parents pairs. MRSA was detected in 45% of S. aureus isolates, that is, in 12% of the study population. A single ST22-MRSA-IVa, spa t223, PVL-gene negative strain was detected in 64% of MRSA isolates. This strain is typically susceptible to all non-β-lactam antibiotics tested. The only predictor for MRSA carriage in children was having an MRSA carrier-parent (OR =
= 25.5, P
25.5, P =
= 0.0004). Carriage of the Gaza strain was not associated with prior hospitalization. The Gaza strain was closely related genetically to a local MSSA spa t223 strain and less so to EMRSA15, one of the pandemic hospital-acquired-MRSA clones, scarcely reported in the community. The rapid spread in the community may be due to population determinants or due to yet unknown advantageous features of this particular strain.
0.0004). Carriage of the Gaza strain was not associated with prior hospitalization. The Gaza strain was closely related genetically to a local MSSA spa t223 strain and less so to EMRSA15, one of the pandemic hospital-acquired-MRSA clones, scarcely reported in the community. The rapid spread in the community may be due to population determinants or due to yet unknown advantageous features of this particular strain.
Introduction
Staphylococcus aureus is a leading cause of human bacterial infection worldwide [1]. Nasal carriage is a major source of endogenous infection as well as of human to human transmission. Methicillin-Resistant S. aureus (MRSA) is a major problem in health-care facility settings (HA-MRSA). In the last two decades, community-acquired (CA-MRSA) infections have emerged as well, mainly in young healthy individuals [2]. CA-MRSA strains appear to be more virulent than HA-MRSA strains [3], [4], yet, they are more susceptible to non-β-lactam antibiotic groups [5].
While a few pandemic clones cause most HA-MRSA infections [1], a considerably diverse group of distinct CA-MRSA strains has been documented. Yet, three predominant CA-MRSA clones emerged: MRSA-ST8-IVa(USA300), causing a major epidemic in the USA , MRSA-ST30-IV in Asia and Oceania and MRSA-ST80-IV in Europe, causing mostly sporadic infections with a few local outbreaks [6].
Epidemiological data on CA-MRSA carriage and infection in the Middle-East region is scarce. Only rare cases were found in Israeli [7], [8], or Palestinian populations [9]. In 2009, we established the Palestinian-Israeli collaborative research (PICR) group, in order to explore important infectious disease issues in both Palestinian and Israeli communities.
The Gaza strip is a 360-km2 narrow area located on the Eastern coast of the Mediterranean Sea, bordering Egypt on the south, the Mediterranean on the west and Israel on the north and east. It is populated by ~1.5 million inhabitants. Annual population growth rate is 3.3%, and infant mortality rate is ~20/1000 (UNRWA data: http://unispal.un.org/UNISPAL.NSF/0/885BD85F 892778F28525772700503A4B).
In this study we report widespread CA-MRSA carriage of a single unique strain among children and their parents throughout different districts of the Gaza strip.
Methods
Institutional Review Board (IRB) and Patient Consent
IRB approval was given by the Sheba Medical Center's IRB as well as by a local ethics committee of the Health Ministry in the Gaza strip. Written informed consent was received for each participating individual before recruitment.
Study period & population
Between March and July 2009, healthy children younger than 5.5 years were randomly selected, using a simple random scheme, in 12 Gaza neighborhoods and villages in northern and central Gaza strip. Children and one of their parents were enrolled following parental signed informed consent. Participants were screened in their homes, nasal swabs were obtained and a questionnaire addressing demographic and medical history information was filled.
S. aureus detection and antibiotic susceptibility testing
Swabs were collected from both anterior nares using a cotton-tipped polyester-swab placed in Amies transport-medium (Copan, Brescia, Italy). All swabs were transmitted to the central Maccabi-Healthcare-Services Laboratory within 24 hours, were plated on tryptic soy agar plates supplemented with 5% sheep blood (HyLabs, Rehovot, Israel) and incubated overnight at 35°C. S. aureus was identified by colony morphology, production of catalase, DNAse, and coagulase. Antibiotic susceptibilities were determined by the VITEK-2 system, using plate AST P536 (bioMe'rieux, Hazelwood, MO) for oxacillin, cefoxitine, erythromycin, clindamycin, inducible clindamycin, fusidic acid, gentamicin, trimethoprin-sulfamethoxazole, minocycline, ciprofloxacin and vancomycin. Ciprofloxacin and trimethoprin-sulfamethoxazole susceptibility were also assessed by disc-diffusion and E-test when VITEK-2 results were not available or when susceptibility by VITEK-2 was defined as resistant. Oxacillin resistance was also correlated with mecA gene presence by PCR.
Pulse field gel electrophoresis (PFGE)
Pulse field gel electrophoresis (PFGE) was performed on all MRSA isolates and on 40 randomly selected MSSA isolates. Briefly, SmaI digested DNA embedded in agarose plugs were subjected to PFGE analysis at 14°C in a CHEF-DR-III system (Bio-Rad) at, 6 V/cm for 19 h; initial pulse, 2 s; final pulse, 54 s; angle, 120°; in a 0.5×Tris-borate-EDTA buffer. PFGE profiles were analyzed by BioNumerics software v6.5 (Applied Maths, Sint-Martens Latem, Belgium) using the dice coefficient with a 1.5% position tolerance and 1% optimization value. Cluster analysis was performed by the un-weighted pair-group mean analysis (UPGMA). Similarity of 80% of Dice coefficients was defined as PFGE cluster.
Multilocus sequence typing (MLST)
At least one isolate from each PFGE pattern and at least two from each PFGE cluster were analyzed by MLST. Briefly, isolates were grown on blood agar overnight. Several colonies were re-suspended in 400 µl lysis solution (0.7 µl lysozyme 5 mg/ml, 7 µl lysostaphin 0.5 mg/ml, 4 µl Tris 1 M, 8 µl EDTA 0.5 M, 380 µl H2Ox2) and incubated at 37°C for 30 minutes followed by heating at 95°C for 10 minutes. PCR amplification was carried out according to the MLST website (http://www.mlst.net/). PCR products were sequenced and analyzed by the BioNumerics software. Clonal complex (CC) was determined using the program eBURST v3 based on related STs (http://eburst.mlst.net/).
Spa typing
PCR amplification was performed as described previously [10]. The PCR Products were purified with the Gene JET PCR DNA Purification kit (Fermentas, Burlington city, Ontario, Canada) and sequenced with the Big Dye Terminator v1.1 Cycle Sequencing kit (Applied Biosystems Warrington, UK).The sequencing products were purified with Big Dye XTerminator Purification kit (Applied Biosystems, Warrington, UK) and finally were sequenced using 3100 Avant Genetic Analyzer (ABI, Foster City, CA) and analyzed using the BioNumerics software.
SCCmec typing
SCCmec typing was performed using the multiplex-PCR assay described by Zhang et al [11] with some modifications. Briefly, DNA was extracted by rapid DNA extraction from a single colony suspended in 50 µl of sterile distilled water which was heated at 99°C for 10 min or by the ZR Fungal/Bacterial DNA Mini Prep (Zymo Research, Orange, CA). The cycling parameters were, 94°C for 5 min (or 15 min); 10 cycles of 94°C for 45 s, 60°C for 45 s, 72°C for 90 s; another 25 cycles of 94°C for 45 s, 52°C for 45 s, and 72°C for 90 s, and 72°C for 10 min. As not all SCCmec types and subtypes could be determined initially, ccr gene complex (types 1,2,3 and 5) and mec gene complex (class A, B and C) were assessed [11].
Panton Valentine Leukocidin
Detection of Panton Valentine Leukocidine (PVL) genes was performed using primers previously described [4]. This was performed for all MRSA and for the 40 MSSA isolates described above.
Statistical methods
Multivariate logistic regression models were used to assess predictors for S. aureus and MRSA carriage among parents and children. Covariates included: age, sex, recent hospitalization, recent antibiotic use, number of household members and being a pet owner (cat, dog, horse or other). Additional covariates assessed as potential predictors for child S. aureus carriage were day care attendance and parental S. aureus carriage; additional covariates in the parent S. aureus carriage model were child S. aureus carriage and frequent contact with elderly. MRSA carriage was assessed among S. aureus carriers. Covariates significant at p<0.2 in the univariate models were included in the multivariate analysis. To assess genetic diversity, the Simpson's index of diversity (SID) with a confidence interval of 95% was used [12]. The SID was estimated by the combination of the results obtained by PFGE, presence of PV- genes and SCCmec type. SAS 9.2 was used.
Results
Study population
A total of 379 pairs of children and parents were enrolled ( Table 1 ). The median children's age was 1.8 years (range: 3 weeks to 5.5 years); the median parents' age was 32 (range: 19.5–58 years). Among children both genders were equally represented (49.3% vs. 50.7%, male:female). Of the parents, 68.9% were mothers, among which 94% reported to be housewives. Of the fathers, 37% reported to be unemployed. Most of the screened children (93%) had siblings, with a median number of household members of seven (range:2–25). Only 12 (3.4%) children reported attending day care centers. All participants were reported to be healthy and none of them reported a skin infection on the day of screening.
Table 1
| MRSA carriage | S. aureus carriage | n (total) | Variable | ||
| aOR$(95% CI);p | n (%**) | aOR* (95% CI);p | n (%) | ||
| 50 (46.7) | 107(28.2) | 379 | All | ||
| Age (months) | |||||
| Ref | 6 (75.0) | Ref | 8 (26.7) | 30 | <6 |
| 0.40(0.10–1.53);0.18 | 5 (33.3) | 0.89(0.32–2.46);0.81 | 15 (25.0) | 60 | 6–11 |
| 0.35(0.11–1.18);0.09 | 8 (32.0) | 0.79(0.31–2.04);0.63 | 25 (21.7) | 115 | 12–23 |
| 1.14(0.36–3.54);0.83 | 14 (53.8) | 1.46(0.56–3.82);0.44 | 26 (34.7) | 75 | 24–35 |
| 0.96(0.32–2.93);0.95 | 17 (51.5) | 1.36(0.53–3.52);0.52 | 33 (33.3) | 99 | 36–66 |
| sex | |||||
| Ref | 24 (46.2) | Ref | 52 (27.8) | 187 | Male |
| 1.28(0.67–2.44);0.45 | 26 (47.3) | 1.05(0.66–1.68);0.84 | 55 (28.6) | 192 | Female |
| Number of household members | |||||
| 2 (40.0) | Ref | 5 (18.5) | 27 | 2–3 | |
| 30(51.7) | 1.49(0.52–4.31);0.46 | 58 (27.1) | 214 | 4–8 | |
| 18(40.9) | 1.86(0.63–5.52);0.26 | 44 (32.1) | 137 | 9+ | |
| Attend Day care | |||||
| Ref | 101 (27.6) | 366 | No | ||
| 1.95(0.61–6.27);0.26 | 6 (46.2) | 13 | Yes | ||
| Parental S. aureus/MRSA carriage+ | |||||
| Ref | 27 (40.3) | Ref | 67 (25.0) | 268 | No |
| 6.30(3.01–13.18);<0.001 | 22(56.4) | 1.73(1.05–2.86);0.03 | 39 (36.1) | 108 | Yes |
| Cat owner | |||||
| Ref | 86 (26.3) | 327 | No | ||
| 2.11(1.11–4.00);0.02 | 21 (40.4) | 52 | Yes | ||
S. aureus and MRSA carriage among children (Table 1)
Of the children, 107/379 (28.2%) were S. aureus carriers. MRSA was detected in 50 children (46.7% of S. aureus carriers and 13.2% of all children). Predictors for S. aureus carriage in children were having a S. aureus carrier parent (aOR =
= 1.73, 95% CI 1.05–2.86;P
1.73, 95% CI 1.05–2.86;P =
= 0.03) and being a cat owner (aOR
0.03) and being a cat owner (aOR =
= 2.11, 95% CI 1.11–4.00;P
2.11, 95% CI 1.11–4.00;P =
= 0.02). The only predictor for MRSA carriage in children was having a MRSA carrier parent (aOR
0.02). The only predictor for MRSA carriage in children was having a MRSA carrier parent (aOR =
= 6.3, 95% CI 3.01–13.18;P
6.3, 95% CI 3.01–13.18;P =
= <0.001).
<0.001).
S. aureus and MRSA carriage among parents (Table 2)
Table 2
| MRSA carriage | S. aureus carriage | n (total) | Variable | ||
| aOR$ (95% CI);p | n, (%**) | aOR*(95% CI);p | n (%) | ||
| 44 (40.7) | 108 (28.5) | 379 | All | ||
| 1.00(0.95–1.04);0.87+ | 0.96(0.93–0.99);0.02+ | Age (years) | |||
| Ref | 7 (31.8) | Ref | 22 (35.5) | 62 | <25 |
| 0.88(0.33–2.35);0.80 | 20 (37.0) | 0.85(0.44–1.65);0.63 | 54 (32.9) | 164 | 25–34 |
| 1.05(0.38–2.91);0.92 | 15 (53.6) | 0.48(0.23–0.99);0.05 | 28 (21.9) | 128 | 35–44 |
| 0.92(0.15–5.74);0.93 | 2 (66.7) | 0.45(0.11–1.94);0.29 | 3 (23.1) | 13 | 45+ |
| Sex | |||||
| Ref | 10 (43.5) | Ref | 23 (21.3) | 108 | Male |
| 1.33(0.60–2.94);0.48 | 33 (40.2) | 1.48(0.84–2.62);0.18 | 82 (34.2) | 240 | Female |
| Hospitalization in the previous 6 months | |||||
| Ref | 107 (30.1) | 356 | No | ||
| 0.08(0.01–0.64);0.02 | 1 (4.30) | 23 | Yes | ||
| Pet owner | |||||
| Ref | 76 (26.2) | 290 | No | ||
| 1.27(0.73–2.21);0.39 | 32 (36.4) | 88 | Yes | ||
| Child S. aureus/MRSA carriage ++ | |||||
| 2 (11.8) | Ref | 69 (25.4) | 272/17 | No | |
| 6.38(3.05–13.32);<0.0001 | 17 (77.3) | 1.78(1.05–3.04);0.03 | 39 (36.4) | 107/22 | Yes |
Of the parents 108/379 (28.5%) were S. aureus carriers, and 44 (40.7% of carriers and 11.6% of all parents) were MRSA carriers. The variables significantly associated with S. aureus carriage among parents were younger age (aOR 0.96, 95%CI 0.93–0.99 per year; p =
= 0.02), no previous hospitalization (aOR 0.08, 95%CI 0.01–0.64;p
0.02), no previous hospitalization (aOR 0.08, 95%CI 0.01–0.64;p =
= 0.02) and S. aureus carriage by the child (aOR 1.78, 95%CI 1.05–3.04;p
0.02) and S. aureus carriage by the child (aOR 1.78, 95%CI 1.05–3.04;p =
= 0.03). The only predictor for MRSA carriage among parents was having a MRSA carrier child (aOR 6.38, 95%CI 3.05–13.32; p<0.001).
0.03). The only predictor for MRSA carriage among parents was having a MRSA carrier child (aOR 6.38, 95%CI 3.05–13.32; p<0.001).
Characteristics of the MRSA clones
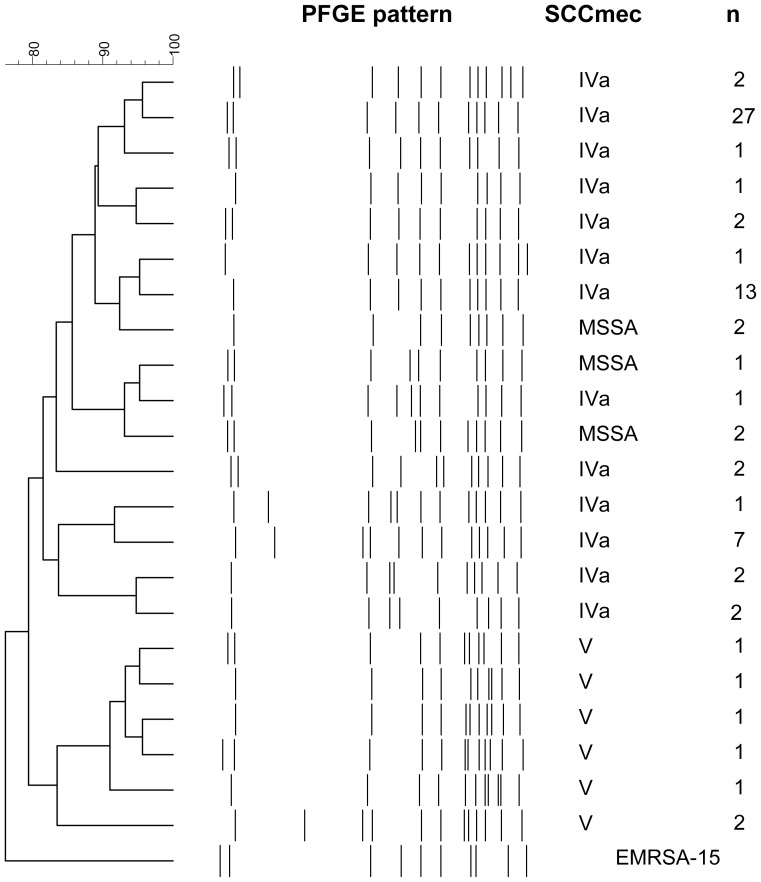
The 94 MRSA isolates identified were further characterized by PFGE, SCCmec typing, spa typing and presence of PVL genes. Twenty-seven isolates were subject to MLST typing, 15 of these were ST22. Characterization of the isolates is presented in
Table 3
. Overall, a low genetic diversity of MRSA clones, as determined by combining the results obtained by PFGE, SCCmec typing and presence of genes encoding PVL was observed (SID 0.58, 95%CI 047–0.70). The predominant clonal complex, CC22 was found in 70 strains, 74.5% of all MRSA isolates. The genetic diversity of MRSA belonging to this clonal complex was relatively low (SID 0.28, 95%CI 0.15–0.41). Moreover, a single strain, as determined by the Simpson's index (ST22-MRSA-IVa, negative for PVL -genes and of a single PFGE cluster) was detected in 60/94 (64%) cases and defined here as the Gaza strain. Other clonal complexes detected were mainly CC88 (n =
= 7, 7.4%) and CC80 (n
7, 7.4%) and CC80 (n =
= 5, 5.3%).
5, 5.3%).
Table 3
| CC* | MLST* | SCCmec | PVL-genes | n | Antibiotic susceptibility profile | ||||||
| cip | min | tri | Gen | fus | cli | ery | |||||
CC22 (n = = 71) 71) | ST22 (69) | IVa (62) | neg (60) | 37 | S | S | S | S | S | S | S |
| 23 | S | S | S | S | S | R | R | ||||
| pos (2) | 2 | S | S | S | S | S | S | S | |||
| V (7) | neg | 4 | S | S | S | S | S | S | S | ||
| 2 | S | S | S | R | S | S | S | ||||
| 1 | S | S | S | I | S | S | S | ||||
| ST1784 (2) | V | neg | 2 | S | S | S | S | S | S | S | |
CC88 (n = = 7) 7) | ST78 | IVa | neg | 5 | S | S | S | S | S | R | R |
| 1 | R | S | S | S | S | R | R | ||||
| 1 | S | S | S | S | S | S | S | ||||
CC80 (n = = 5) 5) | ST80 | IVa | pos | 4 | S | S | S | S | I | S | S |
| IV not a–d | pos | 1 | S | S | S | S | I | S | S | ||
CC5 (n = = 3) 3) | ST5 (1) | V | neg | 1 | S | S | S | S | S | R | R |
| ST1785 (2) | IVa | neg | 2 | S | R | S | S | S | R | R | |
CC30 (n = = 2) 2) | ST30 | IVc | pos | 1 | S | S | S | S | S | R | R |
| ST1734 | IVc | neg | 1 | S | S | S | S | S | S | S | |
CC913 (n = = 1) 1) | ST913 | IVa | neg | 1 | S | S | S | S | S | R | R |
Others (n = = 5) 5) | neg | 5 | |||||||||
While the SCCmec type of the Gaza strain was IVa, nine closely related isolates (by PFGE) carried SCCmec type V; seven of these also belonged to ST22 and two isolates belonged to a novel single locus variant of ST22: ST1784.
Of all 94 MRSA isolates, eight (8.5%) were positive for PVL-genes; Five PVL-gene positive isolates belonged to a single ST80-MRSA-IVstrain, a single ST30MRSA-IVc PVL-gene positive isolate was detected, and two isolates of the Gaza strain PFGE cluster were PVL-gene positive. The spa type of the Gaza strain as well as that of the closely related strains (the ST22-MRSA-IVa- PVL-gene positive, as well as the ST22-MRSA-V strains) were all spa type t223.
The Gaza strain consisted of 13 closely related PFGE patterns with two predominant patterns, all with similarity of more than 81% as determined by the Dice coefficients. This strain is genetically related to the epidemic HA-MRSA-ST22 clone, EMRSA-15, with 76% PFGE pattern similarity ( Figure 1 ).
Antibiotic susceptibility
All MRSA isolates were vancomycin, trimethoprim/sulfametoxazole and ciprofloxacin susceptible excluding a single isolate of ST78-MRSA which was ciprofloxacin resistant and had inducible resistance to clindamycin. Interestingly, 27% of the Gaza strain isolates were initially reported as trimethoprim/sulfametoxazole resistant by VITEK-2. Due to previous reports of inaccuracy in detecting susceptibility by the automatic systems [13], we also tested these strains using the disk diffusion and E-test methods and finally defined them as susceptible. Erythromycin resistance was observed in 37.9% of cases and all had inducible clindamycin resistance. Aminoglycoside resistance was detected in 3/94 isolates. Most CC22 isolates carrying SCCmec IV were susceptible to all non-β-lactam classes. Similarly, most CC22 isolates carrying SCCmec type V were susceptible to all non-β-lactam classes, however 3/9 (33%) were non-susceptible to gentamicin.
Predictors for carriage
We could not identify any predictors for carriage of ST22-MRSA-IVa; it was isolated from all villages and neighborhoods that were sampled in this survey and from both children (n =
= 34, 55%) and parents (28, 45%). Only 7 (11.3%) of the carriers of this strain had a history of previous hospitalization (similar to MSSA and non-S. aureus carriers).
34, 55%) and parents (28, 45%). Only 7 (11.3%) of the carriers of this strain had a history of previous hospitalization (similar to MSSA and non-S. aureus carriers).
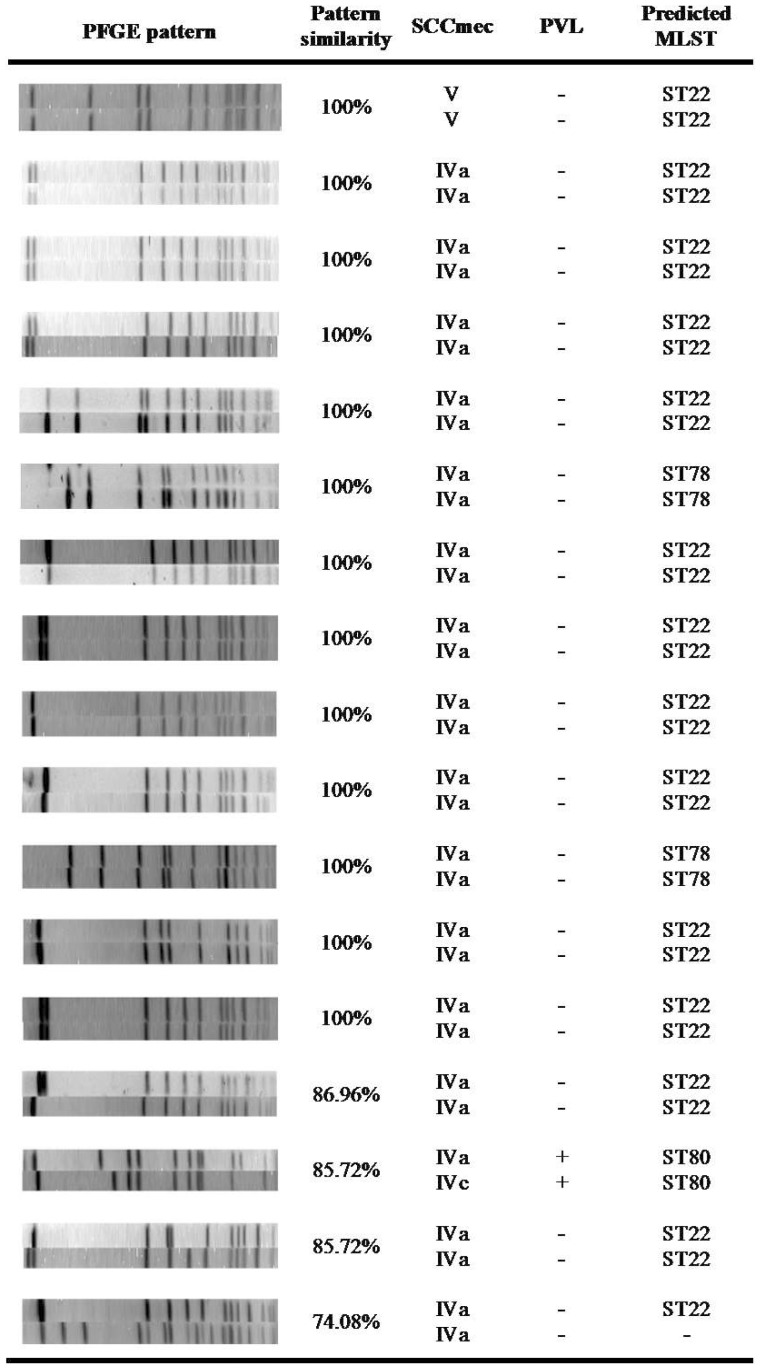
Since the only predictor for MRSA carriage by a child was having a MRSA-carrier parent (88.9% versus 39.6% carriage rates in children of MRSA-carrier parents vs. non-MRSA carriers (p<0.0001), we assessed the genetic relatedness of MRSA isolates from parents and their children. In seventeen child-parent couples (36.1% of all MRSA carriers), both the child and his/her parent were MRSA carriers ( Figure 2 ). 13/17 (82.35%) carried an identical strain.
Characteristics of the MSSA clones
Since the origin of the predominant MRSA clone could have originated from a widespread MSSA clone, we assessed the genetic relatedness of 40 randomly chosen MSSA ( Figure 3 ) isolates. Three main clones were identified; ST291in 8 (20.5%) individuals, ST1278 in 7 (17.9%) individuals and ST15 in 7 (17.9%) individuals. A single ST22 strain was detected in five individuals (12.8%), 3 children and 2 unrelated parents. This strain was closely related to the ST22-MRSA-IVa clone (over 80% similarity by PFGE), also of spa t223 ( Figure 1 ). None of the ST22-MSSA isolates carried the PVL genes. A single ST121-MSSA isolate was positive for genes encoding PVL.
Discussion
We report widespread CA-MRSA carriage of a single strain in the Gaza strip. The Gaza strain belongs to CC22 and is genetically related to the epidemic hospital EMRSA15 clone.
In this study, nearly 30% of healthy children and parents carried S. aureus, similar to reports from other geographical regions [14]. Yet, MRSA carriage rate was dramatically higher (12.4% of the study population). This prevalence rate is much greater than any report from the region [7], [8], including a report on CA-MRSA in the West Bank [9]. Predictors for S. aureus carriage were living in large families, owning pet cats and S. aureus carriage by another family member. Yet, the only predictor for MRSA carriage was MRSA carriage by another family member.
The most common ST22-MRSA clone worldwide is the epidemic HA-MRSA EMRSA-15, which emerged in the UK in 1991. Since then, it has become one of the dominant strains in that region and is currently considered one of the global HA-MRSA pandemic clones [15]. While EMRSA-15 is regarded as a typical HA-MRSA strain, it was recently reported to be carried asymptomatically by a few (5/879) healthy individuals without associated health-care risk factors [16] [23]. Another recent study reported EMRSA-15 in urban Portuguese public buses [17]. Yet, a few key features differentiate EMRSA-15 from the Gaza strain; While EMRSA-15 is known to be ciprofloxacin resistant [15] the Gaza strain is ciprofloxacin susceptible, EMRSA-15 contains SCCmec-IVh (vs. SCCmec-IVa) and its common spa types are t032 and t022 [18] (vs. t223). The quinolone-susceptibility difference may be due to infrequent quinolone use in the Gaza strip [IA personal communication] and does not necessarily imply evolutionary difference between the Gaza strain and EMRSA-15.
Another hospital acquired ST22 clone, referred to as the Barnim epidemic-MRSA strain, considered ancestral to EMRSA-15 in Germany, contains SCCmec-IVa. This clone emerged in three hospitals in north Berlin in 1996 and spread to other healthcare facilities throughout Germany. Like EMRSA-15, the Barnim strain is ciprofloxacin resistant and has not yet been reported in the community [19], [20].
Interestingly, EMRSA-15 has been reported to be a common clone isolated from small companion animals, specifically dogs and cats, in the community [21]. We found that owning a cat was associated with increased S. aureus carriage, but not particularly MRSA carriage.
The origin of the Gaza clone could either be an HA-MRSA that spread into the community, or alternatively a local ST22-MSSA that evolved into a novel CA-MRSA clone. While previous hospitalization was relatively frequent (13.7%) it was not a predictor for carrying this strain or MRSA in general. Furthermore, our data is suggestive of local evolution rather than import of EMRSA-15 as implied by the high genetic relatedness of the Gaza strain to the ST22-MSSA-spa-t223 strains in the region. Further thorough genomic studies of these strains would be required to conclusively determine its evolution.
The ST22-MRSA-V clone, which is closely related to the predominant strain; with ~80% similarity by PFGE and of the same spa type, was detected in only 7/94 (7.44%) vs. 61/94 (64.89%) of ST22-MRSA-IVa isolates. ST22-MRSA-V may have evolved from the same ancestor or alternatively could have evolved from the Gaza-strain, by acquiring SCCmec-V cassette after losing the IVa cassette, as has been suggested for ST8-MRSA strains [22].
The striking fact that a single strain is attributed to over 60% of all MRSA isolates may suggest that specific yet unknown features of this strain confer advantage over competing MRSA and MSSA strains. The relatively lower proportion of ST22 among MSSA isolates as compared to that among MRSA is supportive of this idea. However, we cannot rule out the possibility that the overcrowding in this region facilitated spread of a coincidental, typically nosocomial strain in the community.
Furthermore, intrafamilial MRSA transmission has been implicated to play an important role in MRSA transmission [23]. Given the large number of household members and the fact that only two family-members were sampled, the intrafamilial MRSA transmission reported in this study was probably under-estimated. It is well known that crowded living conditions contribute to CA-MRSA transmission, as reported among athletes, prisoners and military trainees [24], [25], [26] as well as among minorities and developing populations [27].
Yet, the fact that a single MRSA strain became so predominant, while total S. aureus carriage rate was not different from that in other regions, suggests the Gaza strain has advantages over other local MRSA strains. Further studies to determine the unique characteristics of this strain are needed.
Acknowledgments
We greatly acknowledge the Gaza team for their hard work and dedication to the project. We thank Bracha Shainberg for her support and acknowledge Assaf Rockney, Sharon Mikhailov, Yulia Roytman, Dana Elhadad and the Maccabi Healthcare Services microbiology team for their technical assistance. Gill Smollen for technical advice. Aylana Reiss-Mandel for English editing and proofreading of the manuscript. We acknowledge H. deLancastre for sending us the EMRSA-15 strain, used here as a reference strain.
The data were presented, in part, at the 51st Interscience Conference on Antimicrobial Agents and Chemotherapy (ICAAC), Chicago, September 2011.
The PICR investigators (2009): PI: Regev-Yochay Gili (MD) Co-PIs: Abullaish Izzeldin (MD), Daana Mohanned (MD), Hamdan Ayob (MD), Jaar Fuad (MD), Kassem Samir (MD), Malley Richard (MD), Rahav Galia (MD), Raz Meir (MD), Rodity Avraham (MD), Roizin Hector (MD), Stern Ora (MD), Thalji Amin (MD), Varon Miriam (RN), Yakirevitch Luba (MD).
Funding Statement
This study was funded by the Gertner Institute for Epidemiology Research and Health Policy 2009 and by Maccabi Healthcare Services Research Institute RC-24010. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
Articles from PLOS ONE are provided here courtesy of PLOS
Full text links
Read article at publisher's site: https://doi.org/10.1371/journal.pone.0042864
Read article for free, from open access legal sources, via Unpaywall:
https://journals.plos.org/plosone/article/file?id=10.1371/journal.pone.0042864&type=printable
Citations & impact
Impact metrics
Citations of article over time
Article citations
Global epidemiology of asymptomatic colonisation of methicillin-resistant <i>Staphylococcus aureus</i> in the upper respiratory tract of young children: a systematic review and meta-analysis.
Arch Dis Child, 109(4):267-274, 19 Mar 2024
Cited by: 1 article | PMID: 38296611 | PMCID: PMC10958260
Review Free full text in Europe PMC
Resistance to aminoglycoside and quinolone drugs among Klebsiella pneumoniae clinical isolates from northern Jordan.
Heliyon, 10(1):e23368, 06 Dec 2023
Cited by: 1 article | PMID: 38163217 | PMCID: PMC10757003
Novel multidrug-resistant sublineages of Staphylococcus aureus clonal complex 22 discovered in India.
mSphere, 8(5):e0018523, 12 Sep 2023
Cited by: 0 articles | PMID: 37698417 | PMCID: PMC10597471
Prevalence of mecA and Panton-Valentine Leukocidin Genes in Staphylococcus aureus Clinical Isolates from Gaza Strip Hospitals.
Microorganisms, 11(5):1155, 28 Apr 2023
Cited by: 0 articles | PMID: 37317129 | PMCID: PMC10221152
Characterisation of Methicillin-Resistant Staphylococcus aureus from Alexandria, Egypt.
Antibiotics (Basel), 12(1):78, 01 Jan 2023
Cited by: 3 articles | PMID: 36671279 | PMCID: PMC9855118
Go to all (43) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Genomic epidemiology of meticillin-resistant Staphylococcus aureus ST22 widespread in communities of the Gaza Strip, 2009.
Euro Surveill, 23(34), 01 Aug 2018
Cited by: 12 articles | PMID: 30153881 | PMCID: PMC6113745
[Infectivity-resistotype-genotype clustering of methicillin-resistant Staphylococcus aureus strains in the Central Blacksea Region of Turkey].
Mikrobiyol Bul, 48(1):14-27, 01 Jan 2014
Cited by: 4 articles | PMID: 24506712
Dissemination of the Methicillin-resistant Staphylococcus aureus Pediatric Clone (ST5-T002-IV-PVL+) as a Major Cause of Community-associated Staphylococcal Infections in Bedouin Children, Southern Israel.
Pediatr Infect Dis J, 38(3):230-235, 01 Mar 2019
Cited by: 6 articles | PMID: 29912846
Community-associated meticillin-resistant Staphylococcus aureus in children in Taiwan, 2000s.
Int J Antimicrob Agents, 38(1):2-8, 11 Mar 2011
Cited by: 65 articles | PMID: 21397461
Review