Abstract
Free full text

From the Cover
Detection of ultra-rare mutations by next-generation sequencing
Abstract
Next-generation DNA sequencing promises to revolutionize clinical medicine and basic research. However, while this technology has the capacity to generate hundreds of billions of nucleotides of DNA sequence in a single experiment, the error rate of ~1% results in hundreds of millions of sequencing mistakes. These scattered errors can be tolerated in some applications but become extremely problematic when “deep sequencing” genetically heterogeneous mixtures, such as tumors or mixed microbial populations. To overcome limitations in sequencing accuracy, we have developed a method termed Duplex Sequencing. This approach greatly reduces errors by independently tagging and sequencing each of the two strands of a DNA duplex. As the two strands are complementary, true mutations are found at the same position in both strands. In contrast, PCR or sequencing errors result in mutations in only one strand and can thus be discounted as technical error. We determine that Duplex Sequencing has a theoretical background error rate of less than one artifactual mutation per billion nucleotides sequenced. In addition, we establish that detection of mutations present in only one of the two strands of duplex DNA can be used to identify sites of DNA damage. We apply the method to directly assess the frequency and pattern of random mutations in mitochondrial DNA from human cells.
The advent of massively parallel DNA sequencing has ushered in a new era of genomic exploration by making simultaneous genotyping of hundreds of billions of base pairs possible at a small fraction of the time and cost of traditional Sanger methods (1). Unlike conventional techniques, which simply report the average genotype of an aggregate collection of molecules, next-generation sequencing technologies digitally tabulate the sequence of many individual DNA fragments, thus offering the unique ability to detect minor variants within heterogeneous mixtures. This concept of “deep sequencing” has been implemented in a variety of fields including metagenomics (2), paleogenomics (3), forensics (4), and human genetics (5) to disentangle subpopulations in complex biological samples. Clinical applications are rapidly being developed, such as prenatal screening for fetal aneuploidy (6), early detection of cancer (7), and monitoring its response to therapy (8) with nucleic acid-based serum biomarkers.
Although, in theory, DNA subpopulations of any size should be detectable when deep sequencing a sufficient number of molecules, a practical limit of detection is imposed by errors introduced during sample preparation and sequencing (9). PCR amplification of heterogeneous mixtures can result in population skewing due to differential amplification (10, 11), and polymerase mistakes generate point mutations resulting from base misincorporations and rearrangements due to template switching (10, 12). Combined with the additional errors that arise during cluster amplification, cycle sequencing, and image analysis, ~1% of bases are incorrectly identified, depending on the specific platform and sequence context (1). This background level of artifactual heterogeneity establishes a limit below which the presence of true rare variants is obscured (9).
A variety of improvements at the level of biochemistry (13, 14) and data processing (14–19) have been developed to improve sequencing accuracy. In addition, techniques whereby PCR duplicates arising from individual DNA fragments can be resolved on the basis of unique random shear points (20) or via exogenous tagging (21, 22) before amplification (23–28) have recently been reported. Because all amplicons derived from a particular starting molecule can be explicitly identified, any variation in the sequence or copy number of identically tagged sequencing reads can be discounted as technical error. This approach has been used to improve counting accuracy of DNA (25, 26, 28) and RNA templates (24, 25, 27, 29) and to correct base errors arising during PCR or sequencing (20, 23, 24, 26). For example, Kinde et al. (23) reported a reduction in error frequency of ~20-fold with a tagging method that is based on labeling single-stranded DNA fragments with a primer containing a 14-bp degenerate sequence. This approach allowed for an observed mutation frequency of ~0.001% mutations/bp in normal human genomic DNA. Nevertheless, a number of highly sensitive genetic assays have indicated that the true mutation frequency in normal cells is likely to be far lower, with estimates of per-nucleotide mutation frequencies generally ranging from 10−8 to 10−11 (30, 31). Thus, the majority of mutations seen in normal human genomic DNA by this method potentially still represent technical artifacts.
Prevailing next-generation sequencing platforms generate sequence data from single-stranded fragments of DNA. As a consequence, artifactual mutations introduced during the initial round of PCR amplification are undetectable as errors—even with tagging techniques—if the base change is propagated to all subsequent PCR duplicates. Multiple types of DNA damage are highly mutagenic and may lead to this scenario. Spontaneous DNA damage arising from normal metabolic processes results in thousands of damaging events per cell per day (32), and additional DNA damage is generated ex vivo during tissue processing and DNA extraction (33).
Limitations inherent to sequencing of single-stranded DNA can be overcome, however, as DNA naturally exists as a double-stranded entity, with one molecule reciprocally encoding the sequence information of its partner. Thus, it should be feasible to identify and correct nearly all forms of sequencing errors by comparing the sequence of individual tagged amplicons derived from one half of a double-stranded complex with those of the other half of the same molecule. Herein, we present an approach for tag-based error correction, termed Duplex Sequencing, which capitalizes on the redundant information stored in complexed double-stranded DNA. Our method has a theoretical background error rate of less than one artifactual error per 109 nucleotides sequenced and thus allows rare variants in heterogeneous populations to be detected with unprecedented sensitivity.
Results
To improve the sensitivity of variant detection by next-generation DNA sequencing, we designed an alternative approach to library preparation and analysis that we term Duplex Sequencing. The method entails tagging both strands of duplex DNA with a random, yet complementary double-stranded nucleotide sequence, which we refer to as a Duplex Tag. Double-stranded tag sequences are incorporated into standard Illumina sequencing adapters by first introducing a single-stranded randomized nucleotide sequence into one adapter strand and then extending the opposite strand with a DNA polymerase to yield a complementary, double-stranded tag (Fig. 1A). Following ligation of tagged adapters to sheared DNA, the individually labeled strands are PCR amplified from asymmetric primer sites on the adapter tails (Fig. 1B) and subjected to paired-end sequencing. Every PCR duplicate that arises from a single strand of DNA will carry the original strand’s tag sequence. Owing to the complementary nature of the Duplex Tags on the two strands, each strand in a DNA duplex pair generates a distinct, yet related, population of PCR duplicates. Comparing the sequence obtained from each of the two strands in a duplex facilitates differentiation of sequencing errors from true mutations: when an apparent mutation is, in fact, due to a PCR or sequencing error, the substitution will only be seen on a single strand. In contrast, with a true DNA mutation, complementary substitutions will be present on both strands.
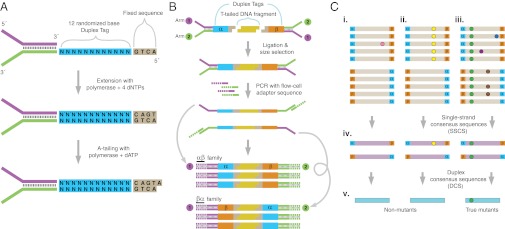
Overview of Duplex Sequencing. (A) Adapter synthesis. A double-stranded, randomized Duplex Tag sequence is appended to a sequencing adapter by copying a degenerate sequence in one strand of the adapter with DNA polymerase. Complete adapter A-tailing is ensured by extended incubation with polymerase and dATP. (B) Duplex Sequencing workflow. Sheared, T-tailed double-stranded DNA is ligated to A-tailed adapters. Because every adapter contains a Duplex Tag on each end, every DNA fragment becomes labeled with two distinct tag sequences (arbitrarily designated α and β in the single fragment shown). PCR amplification with primers containing Illumina flow-cell–compatible tails is carried out to generate families of PCR duplicates. Two types of PCR products are produced from each DNA fragment. Those derived from one strand will have the α tag sequence adjacent to flow cell sequence 1 and the β tag sequence adjacent to flow cell sequence 2. PCR products originating from the complementary strand are labeled reciprocally. (C) Error correction. (i–iii) Sequence reads sharing a unique set of tags are grouped into paired families with members having strand identifiers in either the αβ or βα orientation. Each family pair reflects the amplification of one double-stranded DNA fragment. (i) Mutations (colored spots) present in only one or a few family members represent sequencing mistakes or PCR-introduced errors occurring late in amplification. (ii) Mutations occurring in many or all members of one family in a pair arise from PCR errors during the first round of amplification such as might occur when copying across sites of mutagenic DNA damage. (iii) True mutations (green) present on both strands of a DNA fragment appear in all members of a family pair. Whereas artifactual mutations may co-occur in a family pair with a true mutation, all except those arising during the first round of PCR amplification can be independently identified and discounted when producing (iv) an error-corrected single-strand consensus sequence (SSCS). The sequences obtained from each of the two strands of an individual DNA duplex can then be compared to obtain (v) the duplex consensus sequence (DCS), which eliminates remaining errors that occurred during the first round of PCR.
During the PCR amplification step after tagging, many duplicate “families” of molecules are created, each of which arose from a single strand of an individual DNA molecule. After sequencing, members of each PCR family are identified and grouped by virtue of sharing an identical tag sequence (Fig. 1C). The sequences of uniquely tagged PCR duplicates are then compared to create a PCR consensus sequence. Only PCR families consisting of at least three duplicates and yielding the same sequence in at least 90% of the members at a given position are used to create the consensus sequence. This step filters out random errors introduced during sequencing or PCR to yield a set of sequences, each of which derives from an individual molecule of single-stranded DNA. We refer to these as single strand consensus sequences (SSCSs).
Next, sequences belonging to the two complementary strands of each DNA duplex are identified by searching for complementary tag sequences among SSCS reads. Specifically, a 24-nucleotide tag sequence consists of two 12-nucleotide sequences at each end of the molecule that can be designated α and β. For a tag of form αβ in read 1, the opposite strand’s tag will be of form βα in read 2. Following partnering of the two strands, the sequences of the strands are compared. A sequence base at a given position is kept only if the read data from each of the two strands matches perfectly. A detailed illustration of the approach is provided in SI Materials and Methods. Comparing the sequences obtained from both strands eliminates errors introduced during the first round of PCR where an artifactual mutation may be propagated to all PCR duplicates of one strand and would not be removed by SSCS filtering alone. We refer to the resulting high-confidence sequences of individual DNA duplex molecules as duplex consensus sequences (DCSs).
Duplex Sequencing of M13 DNA.
To establish the sensitivity of Duplex Sequencing, we first applied the method to M13mp2 DNA, which is a substrate that has been used extensively in sensitive genetic mutation assays and has a well-established base substitution frequency of 3.0 × 10−6 (34). M13mp2 DNA was sheared and ligated to Duplex Sequencing adapters and subjected to deep sequencing on an Illumina HiSEq 2000 (Fig. 2A). Analysis of the data by standard methods (i.e., without consideration of the double-stranded tag sequences and with quality filtering for a Phred score of 30) resulted in an error frequency of 3.8 × 10−3, more than 1,000-fold higher than the true mutation frequency of M13mp2 DNA. Thus, >99.9% of the apparent mutations identified by standard sequencing are erroneous.
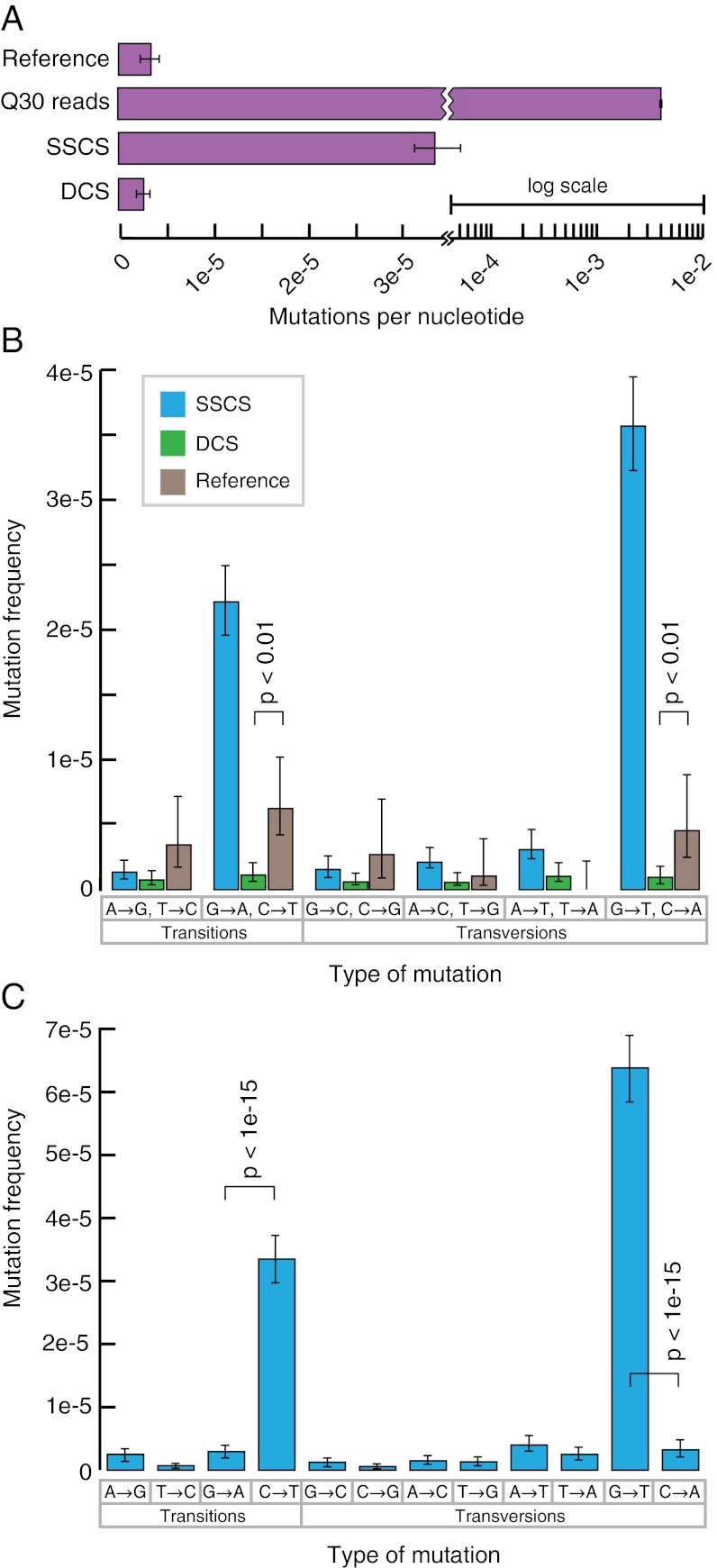
Duplex Sequencing of M13mp2 DNA. (A) Average mutation frequency of M13mp2 DNA as measured by a standard sequencing approach, SSCS, and DCS. Reference value of 3.0 × 10−6 is from ref. 34. Note that the axis is plotted on a split-log scale. (B) Single-strand consensus sequences (SSCSs) reveal a large excess of G→A/C→T and G→T/C→A mutations, whereas duplex consensus sequences (DCSs) yield a balanced spectrum. Mutation frequencies are grouped into reciprocal mispairs, as DCS analysis only scores mutations present in both strands of duplex DNA. All significant (P < 0.05) differences between DCS analysis and the literature reference values are noted. (C) Complementary types of mutations should occur at approximately equal frequencies within a DNA fragment population derived from duplex molecules. However, SSCS analysis yields a 15-fold excess of G→T mutations relative to C→A mutations and an 11-fold excess of C→T mutations relative to G→A mutations. All significant (P < 0.05) differences between paired reciprocal mutation frequencies are noted.
We generated SSCSs by using the unique tag affixed to each molecule to create a consensus of all PCR products that came from an individual molecule of single-stranded DNA. This resulted in a mutation frequency of 3.4 × 10−5, suggesting that ~99% of sequencing errors are corrected in SSCS reads. However, this mutation frequency is >10-fold higher than the reference value of 3.0 × 10−6, indicating that ~90% of the mutations identified by SSCSs are still artifacts.
Next, we further corrected errors by using the complementary tags to compare the DNA sequence arising from the two strands of each single molecule of duplex DNA to create DCSs. This approach resulted in a mutation frequency of 2.5 × 10−6, nearly identical to the frequency of 3.0 × 10−6 determined by well-established genetic methods (34). The number of nucleotides of DNA sequence obtained by a standard sequencing approach, and after SSCS and DCS analysis, may be found in Table S1.
DNA Damage Alters SSCS Mutation Spectrum.
We next examined the spectrum of mutations identified by both SSCS and DCS analysis relative to literature reference values (34) for the M13mp2 substrate (Fig. 2B). SSCS analysis revealed a large excess of G→A/C→T and G→T/C→A mutations relative to reference (P < 10−6, two-sample t test). In contrast, DCS analysis was in excellent agreement with the literature values with the exception of a decrease relative to reference of these same mutational events: G→A/C→T and G→T/C→A (P < 0.01). To probe the potential cause of these spectrum deviations, the SSCS data were filtered to consist of forward-mapping reads from read 1 (i.e., direct sequencing of the reference strand) and the reverse complement of reverse-mapping reads from read 1 (i.e., direct sequencing of the antireference strand.) True double-stranded mutations should result in an equal balance of complementary mutations observed on the reference and antireference strand. However, SSCS analysis revealed a large number of single-stranded G→T mutations, with a much smaller number of C→A mutations (Fig. 2C). A similar bias was seen with a large excess of C→T mutations relative to G→A mutations.
Base-specific mutagenic DNA damage is a likely explanation of these imbalances. Excess G→T mutations are consistent with the oxidative product 8-oxo-guanine (8-oxo-G) causing first round PCR errors and artifactual G→T mutations. DNA polymerases, including those commonly used in PCR, have a strong tendency to insert adenine opposite 8-oxo-G (35, 36), and misinsertion of A opposite 8-oxo-G would result in erroneous scoring of a G→T mutation. Likewise, the excess C→T mutations are consistent with spontaneous deamination of cytosine to uracil (37), a particularly common DNA damage event that results in insertion during PCR of adenine opposite uracil and erroneous scoring of a C→T mutation.
To determine whether the excess G→T mutations seen in SSCSs might reflect oxidative DNA damage at guanine nucleotides, before sequencing library preparation we incubated M13mp2 DNA with the free radical generator hydrogen peroxide in the presence of iron, a protocol that induces DNA damage (38). This treatment resulted in a substantial further increase in G→T mutations by SSCS analysis (Fig. 3A), consistent with PCR errors at sites of DNA damage as the likely mechanism of this biased mutation spectrum. In contrast, induction of oxidative damage did not alter the mutation spectrum seen with DCS analysis (Fig. 3B), indicating that duplex consensus sequences are not similarly susceptible to DNA damage artifacts.
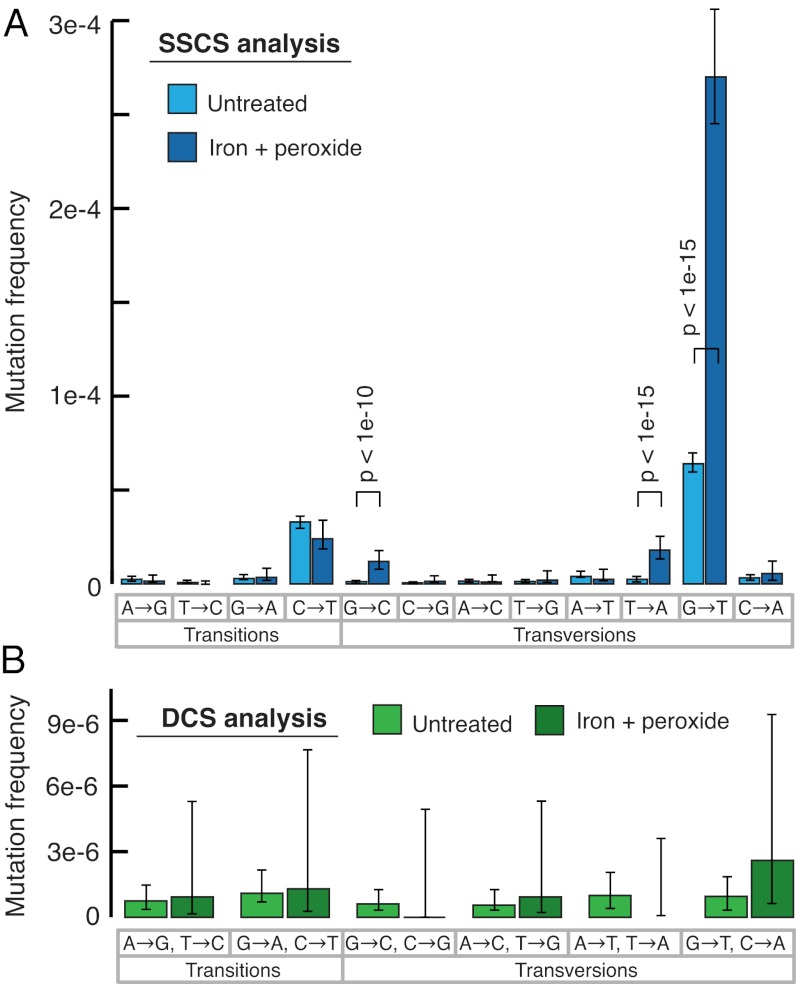
Effect of DNA damage on mutation spectrum. DNA damage was induced by incubating purified M13mp2 DNA with hydrogen peroxide and FeSO4. (A) SSCS analysis reveals a further elevation from baseline of G→T mutations, indicating these events to be the artifactual consequence of nucleotide oxidation. All significant (P < 0.05) changes from baseline mutation frequencies are noted. (B) Induced DNA damage had no effect on the overall frequency or spectrum of DCS mutations.
Furthermore, relative to the literature reference values, DCS analysis results in a lower frequency of G→T/C→A and C→T/G→A mutations (Fig. 2B), which are the same mutations elevated in SSCS analysis as a probable result of DNA damage. Notably, the M13mp2 LacZ assay, from which reference values have been derived, is dependent upon bacterial replication of a single molecule of M13mp2 DNA. Thus, the presence of oxidative damage within this substrate could cause an analogous first-round replication error by Escherichia coli, converting a single-stranded damage event into a fixed, double-stranded mutation during replication. The slight reduction in the frequency of these two types of mutations measured by DCS analysis may, therefore, reflect the absence of damage-induced errors that are scored by the in vivo LacZ assay.
Mutant Recovery.
To further validate the capability of DCS analysis to detect rare mutations, we constructed a series of M13mp2 variants containing specific single base substitutions and mixed the variants together at known ratios. The final mixture was then sequenced with Duplex Sequencing adapters. With conventional analysis of the sequencing data (i.e., without consideration of the tag sequences and filtering for a read quality score of 30), variants present at a level of <1/100 could not be accurately identified because artifactual mutations occurring at a background frequency of about 1/100 obscured the presence of less abundant true mutations (Fig. S1). In contrast, when the data are analyzed as duplex consensus sequences with ~20,000-fold final depth, accurate recovery of mutant sequences was possible down to the lowest tested level of one mutant molecule per 10,000 wild-type molecules.
Duplex Sequencing of Human Mitochondrial DNA.
Having established the methodology for Duplex Sequencing with M13mp2 DNA, which is a substrate for which the mutation frequency and spectrum are fairly well established, we next wished to apply the approach to a human DNA sample. Thus, we isolated mitochondrial DNA from human brain tissue and sequenced the DNA after ligation of Duplex Sequencing adapters. A standard sequencing approach with quality filtering for a Phred score of 30 resulted in a mutation frequency of 2.7 × 10−3, and SSCS analysis yielded a mutation frequency of 1.5 × 10−4. In contrast, DCS analysis revealed a much lower overall mutation frequency of 3.5 × 10−5 (Fig. 4A). The frequency of mutations in mitochondrial DNA has previously been difficult to measure directly due in part to sources of error in existing assays that can result in either overestimation or underestimation of the true value. An additional confounder has been that most approaches are limited to interrogation of mutations within a small fraction of the genome (39). The method of single-molecule PCR, which has been proposed as an accurate method of measuring mitochondrial mutation frequency (39) and is considered resistant to damage-induced background errors (40), has resulted in a reported mitochondrial mutation frequency in human colonic mucosa of 5.9 × 10−5 ± 3.2 × 10−5 (39), which is in excellent agreement with our result. Likewise, mitochondrial DNA sequence divergence rates in human pedigrees are consistent with a mitochondrial mutation frequency of 3–5 × 10−5 (41, 42).
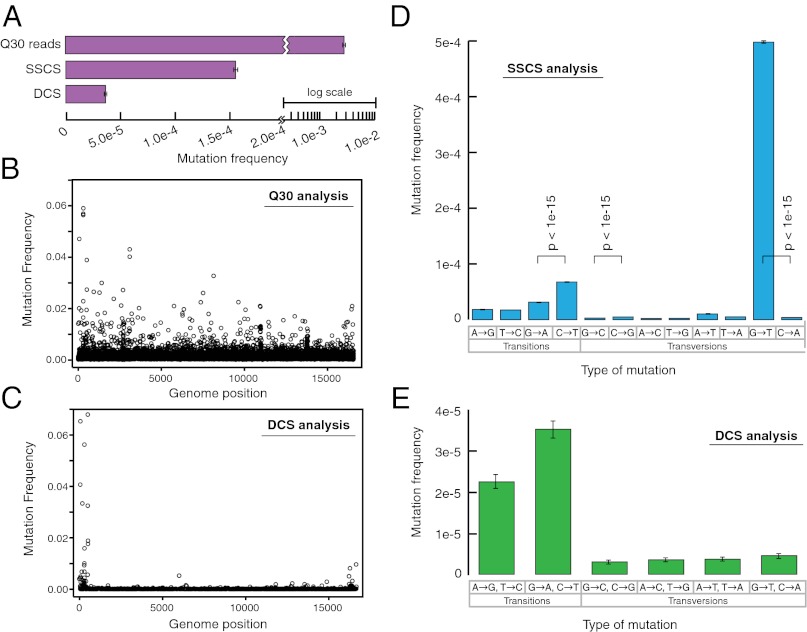
Duplex Sequencing of human mitochondrial DNA. (A) Overall mutation frequency as measured by a standard sequencing approach, SSCS, and DCS. (B) Pattern of mutation in human mitochondrial DNA by a standard sequencing approach. The mutation frequency (vertical axis) is plotted for every position in the ~16-kb mitochondrial genome. Due to the substantial background of technical error, no obvious mutational pattern is discernible by this method. (C) DCS analysis eliminates sequencing artifacts and reveals the true distribution of mitochondrial mutations to include a striking excess adjacent to the mtDNA origin of replication. (D) SSCS analysis yields a large excess of G→T mutations relative to complementary C→A mutations, consistent with artifacts from damaged-induced 8-oxo-G lesions during PCR. All significant (P < 0.05) differences between paired reciprocal mutation frequencies are noted. (E) DCS analysis removes the SSCS strand bias and reveals the true mtDNA mutational spectrum to be characterized by an excess of transitions.
When the distribution of mutations throughout the mitochondrial genome is considered, the quality filtered reads (analyzed without consideration of the tags) have many artifactual errors, such that identification of mutational hotspots is difficult or impossible (Fig. 4B). DCS analysis removed these artifacts (Fig. 4C) and revealed striking hypermutability of the region of replication initiation (D loop), which is consistent with prior estimates of mutational patterns in mitochondrial DNA based upon sequence variation at this region within the population (43).
SSCS analysis produced a strong mutational bias, with a 130-fold excess of G→T relative to C→A mutations (Fig. 4D), consistent with oxidative damage of the DNA leading to first-round PCR mutations as a significant source of background error. A high level of oxidative damage is expected in mitochondrial DNA, due to extensive exposure of mitochondria to free radical species generated as a byproduct of metabolism (44). DCS analysis (Fig. 4E) removed the mutational bias and revealed that transition mutations are the predominant replication errors in mitochondrial DNA. The DCS mutation spectrum is in accord with prior estimates of deamination events (45) and T-dGTP mispairing by the mitochondrial DNA polymerase (46) as primary mutational forces in mitochondrial DNA. Furthermore, the mutation spectrum of our mitochondrial data are consistent with previous reports of heteroplasmic mutations in human brain showing an increased load of A→G/T→C and G→A/C→T transitions, relative to transversions (47, 48). A similar spectral bias has also been reported in mice (45, 49) and in population studies of Drosophila melanogaster (50).
Discussion
The accuracy of standard approaches to next-generation sequencing is constrained by a general reliance on analysis of single-stranded DNA, which makes certain technical sources of single-stranded errors fundamentally limiting. The complementary strands of native duplex DNA harbor redundant sequence information and here we have demonstrated an approach for error correction, termed Duplex Sequencing, which capitalizes on this biochemical redundancy to greatly lower the error rate of sequencing.
The most sensitive approach previously reported for improving accuracy of next-generation sequencing involves use of a random tag sequence in a PCR primer (23). In this technique, PCR duplicates are generated from a single strand of DNA, and the sequences of tag-identified duplicates are compared such that mutations are scored only when present in multiple duplicates. This method, conceptually analogous to our approach of SSCS analysis, results in ~20-fold improvement in accuracy relative to standard Illumina sequencing, but is presumably susceptible to the same sort of artifactual, largely damage-mediated, first-round PCR errors we observed in SSCS.
Notably, because SSCS is prone to damage-induced PCR errors, SSCS analysis can be used as a tool for detection of sites and patterns of DNA damage occurring in vivo. For example, the occurrence of G→T mutations in SSCS analysis in excess of reciprocal C→A mutations can be used as a marker for the extent of oxidative DNA damage in a sample. The ability to detect damage by SSCS could be further enhanced by using different DNA polymerases in the initial rounds of PCR, which have a proclivity to catalyze specific misinsertions opposite defined types of damage (51, 52).
In contrast to the damage sensitivity of single-strand consensus sequences, for DNA damage to result in an artifactual mutation in DCS, mutagenic lesions (or spontaneous, recurrent first-round PCR errors) would need to occur at the same nucleotide position on both strands of a molecule of duplex DNA and result in complementary errors. Thus, the background error frequency of our method may be calculated as (probability of error on one strand) × (probability of error on other strand) × (probability that both errors are complementary).
Based on the SSCS background error frequency of 3.4 × 10−5 from the M13mp2 DNA sequencing experiment, the error frequency of DCS can be approximated as: (3.4 × 10−5) × (3.4 × 10−5) × 1/3 = 3.8 × 10−10. This calculated error frequency represents a 10 million-fold improvement over the 3.8 × 10−3 value we obtained by standard methods. Of note, the calculation simplistically assumes that all mutational events are equally likely by multiplying by the factor one-third (because any given nucleotide can mutate to any one of three other nucleotides). In reality, the strong mutational bias observed in SSCSs indicates that reciprocal mispairs are not equally probable and, hence, the background of DCS is expected to be lower than this estimate.
In addition to their application for high sensitivity detection of rare DNA variants, the degenerate tags in our Duplex Sequencing adapters can also be used for single-molecule counting to precisely determine absolute DNA or RNA copy numbers (25, 29). Because tagging occurs before amplification, the relative abundance of variants in a population can be accurately assessed given that proportional representation is not subject to skewing by amplification biases. As with their use for error correction, because the degenerate tags are present in the adapters, there are no additional steps required during library preparation, which is in contrast to many existing methods of tag-based counting.
In principle, Duplex Sequencing could be performed on the Illumina or similar platforms without the use of Duplex Tags, but instead by using the randomly sheared ends of the DNA fragments as unique identifiers (20): specifically, for a given DNA sequence seen in sequencing read 1 with 5′ sheared end sequence α and 3′ sheared end sequence β used as a tag of form αβ, the partner strand will occur as a matching sequence in read 2 tagged with 5′ shear point β and 3′ shear point α. In practice, this approach will be limited by the finite number of possible shear points that overlap any given DNA position, and thus, will not be scalable to sequencing DNA at great depth at any given position. However, Duplex Sequencing analysis based on shear points alone may have a role for retrospective confirmation that specific mutations of interest are true mutations that were indeed present in the starting sample (i.e., present in both DNA strands), as opposed to technical artifacts. Overall, however, Duplex Sequencing is most generally applicable when randomized, complementary double-stranded tags are used. We used a 24-nucleotide tag in the current work, which yields up to 424 = 2.8 × 1014 distinct tag sequences. Combining information regarding the shear points of DNA with the tag sequence would allow a shorter tag to be used, thus minimizing loss of sequencing capacity owing to that used for sequencing of the tag sequence itself.
Once Duplex Tag-containing adapters are synthesized by a straightforward series of enzymatic steps, they can be substituted for standard sequencing adapters without any significant deviations from the normal workflow of sample preparation for Illumina sequencing. Moreover, Duplex Sequencing can be generalized to essentially any sequencing platform: a double-stranded tag can be incorporated into other existing adapters or for sequencing approaches that do not require adapters, a double-stranded tag can be ligated onto a duplex DNA sample before sequencing. In the setting of this simple compatibility with existing workflows, the ability of Duplex Sequencing to radically lower the error rate of sequencing, and the ability of the method to enable precise molecular counting and inference of sites of DNA damage, our Duplex Sequencing approach offers a powerful next-generation sequencing tool for diverse areas of medicine and biology.
Materials and Methods
Adapter Synthesis.
Duplex Tag adapters were synthesized from two oligonucleotides with noncomplementary Y-shaped tails. A randomized single-stranded 12-nucleotide sequence present in one of the oligonucleotides was rendered double stranded by copying with DNA polymerase. After reaction cleanup, adapter A-tailing was performed by incubation with DNA polymerase and dATP. Further details are provided in SI Materials and Methods.
Sequencing Library Preparation.
Double-stranded DNA was sheared and end repaired by standard protocols, followed by T-tailing with DNA polymerase and dTTP. T-tailed DNA was ligated to A-tailed Duplex Tag adapters, followed by PCR amplification for 18–20 cycles and sequencing on an Illumina HiSEq 2000. Further details are provided in SI Materials and Methods.
Data Processing.
Reads were filtered for those containing a properly located tag sequence, and the 12-nucleotide tags present on each end of the paired reads were computationally combined to form a single 24-nucleotide tag for each read. Reads containing identical tag sequences were grouped together to form SSCS reads. Next, partner strands among SSCS reads were identified by virtue of the complementary tag sequences. The SSCS reads corresponding to both of the two strands of individual molecules of duplex DNA were then compared to form DCS reads. Resultant sequence positions were considered only when information from both DNA strands was in perfect agreement. Further details are provided in SI Materials and Methods.
Acknowledgments
We thank Elizabeth Swisher and Thomas Montine for providing human tissue samples and Tom Walsh, Anne Thornton, Ming Lee, and Bryan Paeper for assistance with DNA sequencing. This work was supported by National Institutes of Health Grants R01 CA115802, R01 CA102029, and P01 AG0751 (to L.A.L.); T32 AG000057 (to S.R.K.); F30 AG033485 (to J.J.S.); and F30 AG039173 (to J.B.H.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
See Commentary on page 14289.
This article contains supporting information online at www.pnas.org/lookup/suppl/10.1073/pnas.1208715109/-/DCSupplemental.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.1208715109
Read article for free, from open access legal sources, via Unpaywall:
https://www.pnas.org/content/pnas/109/36/14508.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1073/pnas.1208715109
Article citations
Longitudinal ultra-sensitive mutation burden sequencing for precise minimal residual disease assessment in AML.
Nat Commun, 15(1):9853, 14 Nov 2024
Cited by: 0 articles | PMID: 39543210 | PMCID: PMC11564880
Measurable residual FLT3 tyrosine kinase domain mutations before allogeneic transplant for acute myeloid leukemia.
Bone Marrow Transplant, 18 Oct 2024
Cited by: 0 articles | PMID: 39424958
Somatic mutations in autoinflammatory and autoimmune disease.
Nat Rev Rheumatol, 20(11):683-698, 11 Oct 2024
Cited by: 0 articles | PMID: 39394526
Review
Frequency and spectrum of mutations in human sperm measured using duplex sequencing correlate with trio-based de novo mutation analyses.
Sci Rep, 14(1):23134, 08 Oct 2024
Cited by: 0 articles | PMID: 39379474 | PMCID: PMC11461794
Effect of sequencing platforms on the sensitivity of chemical mutation detection using Hawk-Seq™.
Genes Environ, 46(1):20, 09 Oct 2024
Cited by: 0 articles | PMID: 39385252 | PMCID: PMC11462924
Go to all (548) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Targeted sequencing of both DNA strands barcoded and captured individually by RNA probes to identify genome-wide ultra-rare mutations.
Sci Rep, 7(1):3356, 13 Jun 2017
Cited by: 7 articles | PMID: 28611392 | PMCID: PMC5469810
Detection of Low-Frequency Mutations and Identification of Heat-Induced Artifactual Mutations Using Duplex Sequencing.
Int J Mol Sci, 20(1):E199, 08 Jan 2019
Cited by: 9 articles | PMID: 30625989 | PMCID: PMC6337315
Sequence error storms and the landscape of mutations in cancer.
Proc Natl Acad Sci U S A, 109(36):14289-14290, 21 Aug 2012
Cited by: 5 articles | PMID: 22912407 | PMCID: PMC3437884
Application of next-generation sequencing in the detection of low-abundance mutations.
Yi Chuan, 46(2):126-139, 01 Feb 2024
Cited by: 0 articles | PMID: 38340003
Review
Funding
Funders who supported this work.
Intramural NIH HHS (1)
Grant ID: Z01 AG000751
NCI NIH HHS (2)
Grant ID: R01 CA102029
Grant ID: R01 CA115802
NIA NIH HHS (5)
Grant ID: T32 AG000057
Grant ID: P01 AG0751
Grant ID: F30 AG039173
Grant ID: F30 AG033485
Grant ID: P01 AG001751
NIGMS NIH HHS (1)
Grant ID: T32 GM007266





