Abstract
Free full text

Feeder-Free Derivation of Human Induced Pluripotent Stem Cells with Messenger RNA
Associated Data
Abstract
The therapeutic promise of induced pluripotent stem cells (iPSCs) has spurred efforts to circumvent genome alteration when reprogramming somatic cells to pluripotency. Approaches based on episomal DNA, Sendai virus, and messenger RNA (mRNA) can generate “footprint-free” iPSCs with efficiencies equaling or surpassing those attained with integrating viral vectors. The mRNA method uniquely affords unprecedented control over reprogramming factor (RF) expression while obviating a cleanup phase to purge residual traces of vector. Currently, mRNA-based reprogramming is relatively laborious due to the need to transfect daily for ~2 weeks to induce pluripotency, and requires the use of feeder cells that add complexity and variability to the procedure while introducing a route for contamination with non-human-derived biological material. We accelerated the mRNA reprogramming process through stepwise optimization of the RF cocktail and leveraged these kinetic gains to establish a feeder-free, xeno-free protocol which slashes the time, cost and effort involved in iPSC derivation.
Shinya Yamanaka's discovery that differentiated cells can be reverted to a pluripotent state by the expression of a select group of transcription factors opened up the prospect that patient-specific cells might be turned into cells of any desired type for use in the study of genetic disease and ultimately for cell-replacement therapy. In Yamanaka's original work the expression of reprogramming factors was achieved using viral vectors which integrate into the genome1, and iPSC derivations are still typically performed with integrating retrovirus or lentivirus. Modification of the genome represents a hurdle to therapeutic application of iPSCs, while the possibility of reactivated expression from integrated viral expression cassettes is a concern even for in vitro studies. Considerable progress has been made in the development of novel RF expression vectors to alleviate this problem. Lentiviral vectors are now available which encode the multiple factors required for iPSC induction within a single polycistronic cassette flanked with recombination sites, allowing almost-seamless excision of the transgene after reprogramming by transient expression of a recombinase2,3,4,5. Transgene insertion with subsequent excision can also be effected by using a transposon vector followed by brief expression of transposase6,7,8. Non-integrating vectors based on adenovirus, plasmids and episomal DNA have been developed to transiently express RFs for a time sufficient to induce pluripotency9,10,11,12,13, and it has proved possible to generate iPSCs by repeated transduction of cells with recombinant RF proteins incorporating cell-penetrating peptides, albeit with low efficiency14,15. Relatively efficient iPSC conversion can now be achieved using Sendai virus16,17,18, which has a completely RNA-based reproductive cycle, and by sustained transfection of synthetic mRNA transcripts encoding the Yamanaka factors19.
The use of synthetic mRNA for cell fate manipulation is appealing because this system allows cellular expression of transcription factors to be modulated on a daily basis simply by adjusting the payload of transcripts added to the culture medium. Once transfection of a particular factor is terminated ectopic expression in the target cells ceases in short order due to the rapid degradation of mRNA in the cytoplasm. In contrast to non-integrating DNA vectors or RNA viruses, no cleanup phase is entailed when reprogramming with mRNA, nor is there any risk of random genomic integration or persistent viral infection. These advantages assume greater significance when we envisage that multiple rounds of ectopic RF expression may ultimately be employed to derive specialized cells of interest from a patient sample via an iPSC intermediate. Nonetheless, there are drawbacks to mRNA-based reprogramming as currently practiced. Expression of RFs is typically robust for about 24 hours after mRNA is transfected, yet it takes about two weeks of factor expression to induce pluripotency in human cells, and the hands-on time involved in performing the repeated transfections needed to generate iPSCs is therefore relatively high. Not all cell types and culture media are equally conducive to transfection, and this is an obstacle to mRNA-based reprogramming of certain cell types, including blood cells. It has also so far been necessary to employ a feeder layer of mitotically-arrested fibroblasts when reprogramming cells using mRNA. The feeder cells buffer the population density of the culture as the target cells grow out from a low starting density over the extended time course required for iPSC induction, evening out the per-cell dosage of RNA and transfection reagent (both of which have associated toxicities) and supporting the viability of the target cells in the face of pro-apoptotic and cytostatic forces engendered by the reprogramming process. This requirement adds complexity and setup time to the procedure and introduces an important source of technical variability, especially given that the feeders are themselves subject to transfection. The presence of a feeder layer also impedes monitoring and analysis of the reprogramming process. Finally, although human feeder cells are currently the standard for mRNA reprogramming, even these cells are a potential source of xeno-biological contamination when non-human animal products are used in their derivation and expansion. We reasoned that the weaknesses inherent in current mRNA-based protocols could be meliorated if reprogramming were accelerated through potentiation of the RF cocktail delivered to the cells. Several recent publications have described novel reprogramming factors or factor combinations which can enhance the kinetics and efficiency of iPSC derivation. Optimization of the established mRNA RF cocktail based on these findings and our own experiments has enabled us to define a revised protocol that compresses and streamlines the mRNA reprogramming process, and which supports the rapid production of footprint-free iPSCs from human fibroblasts without the use of feeder cells or other potentially xeno-contaminated reagents. This work will extend the appeal of the mRNA method and help clear remaining roadblocks to the therapeutic application of iPSC technology.
Results
Our experiments initially focused on the question of whether mRNA reprogramming could be enhanced using engineered variants of Oct4 incorporating an N-terminal MyoD transactivation domain20,21 or a C-terminal triple repeat of the VP16 transactivation domain22, or by augmenting the cocktail with transcripts for two additional factors, Rarg and Lrh-123. To this end, we carried out a test to compare the performance of six different mRNA cocktails comprising transcripts for the five RFs currently used in mRNA reprogramming (Oct4, Sox2, Klf4, c-Myc-T58A and Lin28), or a 7-factor cocktail including Rarg and Lrh-1, each combination being tried in three variations based on wild-type Oct4 and the MyoD- and VP16-Oct4 fusion constructs (designated M3O and VPx3, respectively). BJ fibroblasts plated at 10K cells per well were transfected in feeder-based cultures for 11 days, by which time advanced morphologies were apparent in several of the wells. Over the next few days colonies with characteristic human embryonic stem cell (hESC) morphology emerged in wells transfected with the cocktails based on wild-type Oct4 and M3O, while the target cells in the VPx3-transfected cultures retained a fibroblastic morphology—albeit while showing accelerated growth and some tendency to aggregate into foci—and no colonies emerged (data not shown). The 5-factor and 7-factor versions of the wild-type Oct4 and M3O-based cocktails demonstrated similar colony productivity, hence no advantage accrued from the inclusion of Rarg and Lrh-1. However, the M3O cocktail gave several times the number of colonies produced with wild-type Oct4. Colonies were picked from the M3O 5-factor well for expansion and further analysis (Fig. 1). Pluripotency of M3O-derived colonies was confirmed by staining for nuclear and cell-surface markers (Fig. 1b), and by in vitro differentiation into the three primary germ layers (Fig. 1c–e and Supplementary Movie 1). Six expanded clones were subjected to karyotype analysis and DNA fingerprinting, karyotypic normality and BJ lineage being confirmed in all cases (data not shown).
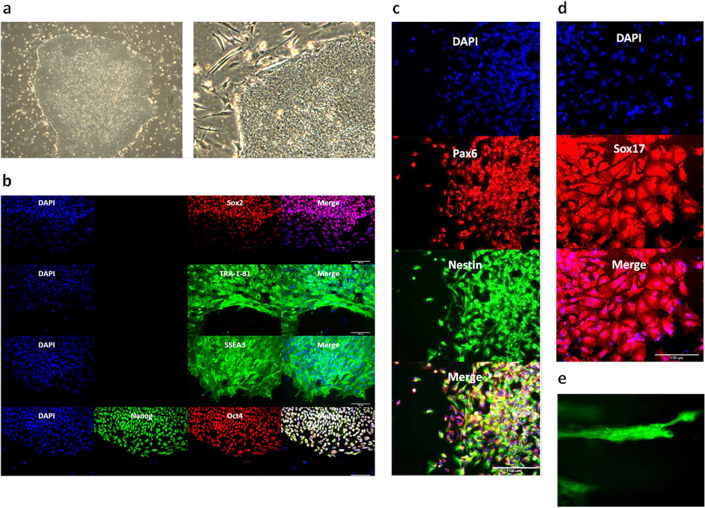
(a) Phase contrast imagery showing morphology of an iPSC colony picked from the first M3O-based BJ reprogramming plate and replated onto feeders. (b) Immunostaining of an expanded clone revealing expression of cell-surface and nuclear pluripotency markers. (c) Immunofluorescence images showing expression of neuro-ectoderm markers after in vitro differentiation of the iPSC clone shown in the previous panel. (d) Immunostaining showing expression of the endodermal marker Sox17 in cells derived from the same clone subjected to in vitro endodermal differentiation. (e) α-Actinin immunostaining of cardiomyocytes derived by mesodermal differentiation of the same iPSC clone (see also Supplementary Movie 1).
In a follow-up experiment we investigated whether the potency of the mRNA cocktail could be further enhanced by the inclusion of Nanog transcripts24. Four wells containing 50K BJ fibroblasts on feeders were transfected with wild-type Oct4 or M3O-based 5-factor or 6-factor cocktails for six days, and each culture was then passaged 1:6 onto fresh feeders to populate a 6-well plate (Fig. 2a). Transfection was continued for up to 5 more days within each plate. Cultures were fixed and stained with TRA-1-60 antibodies on day 18 (where day 0 corresponds to the first transfection) to assess the impact of the different cocktails and transfection time courses on iPSC productivity (Fig. 2b–c). The results showed that adding Nanog to the cocktail was highly beneficial regardless of the Oct4 variant employed, while the greatest conversion efficiency was attained when M3O and Nanog were used together.
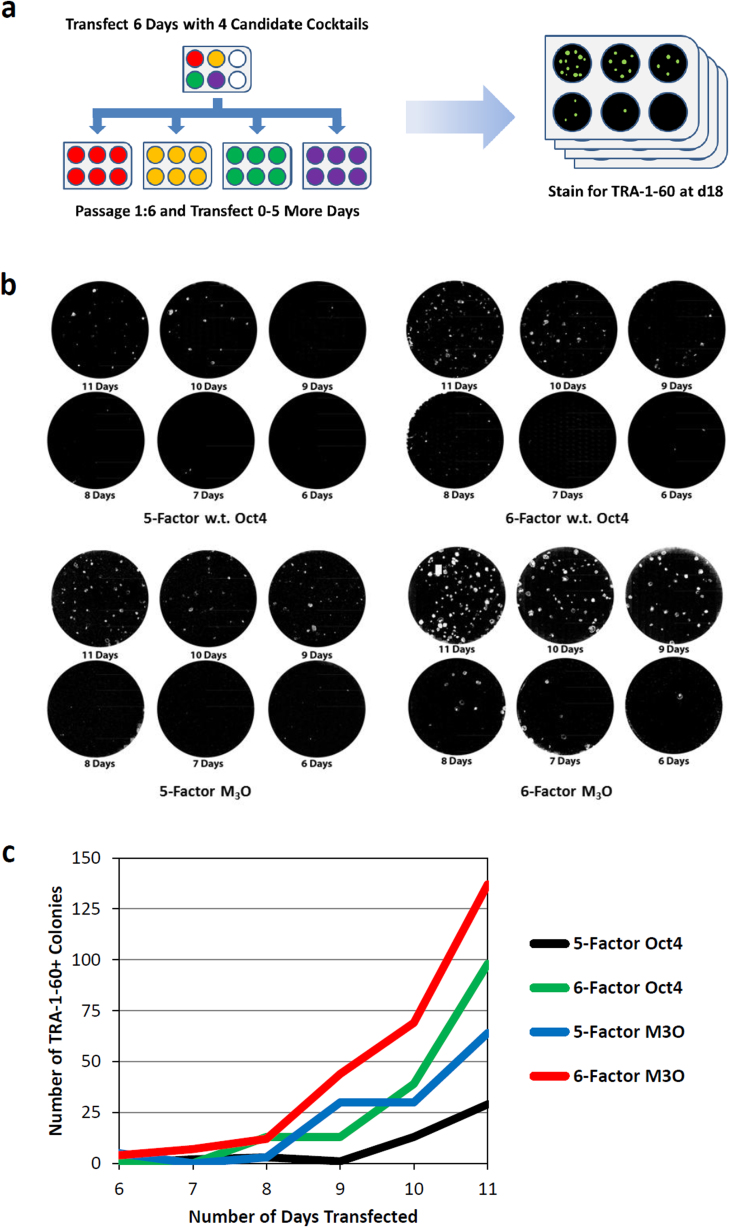
(a) Flowchart summarizing the feeder-based 4-cocktail comparison experiment described in the text. The 5-factor reprogramming cocktails comprised wild-type Oct4 or M3O plus Sox2, Klf4, c-Myc-T58A, and Lin28 mRNA transcripts, while 6-factor cocktails included these RFs plus Nanog. (b) Immunofluorescence imaging showing the yield of TRA-1-60+ colonies produced by the four different cocktails at day 18 of the experiment. The total number of days of transfection is indicated for each culture. (c) Chart showing the yield of TRA-1-60+ colonies at day 18 versus the number of days of transfection using the four cocktails.
We confirmed the efficacy of the M3O-based 5-factor or 6-factor cocktails in additional feeder-based experiments using three additional human fibroblast lines (data not shown). Reprogramming kinetics and efficiency were improved with these low-passage lines (HDF-f, HDF-n and XFF), which displayed faster population doubling times than the relatively late-passage BJ cells in expansion culture. In some cases, hESC-like colonies were obtained following as little as six days of transfection, although the yields were higher when transfection was continued for a few more days. These experiments did reveal a drawback to the M3O-based cocktail in that attrition of the mitotically-arrested feeder cells was markedly accelerated when using the engineered Oct4 (data not shown). The rapid loss of the feeder layer appeared to have an adverse impact on the survival and proliferation of the target cells, suggesting that the performance advantage from using M3O over wild-type Oct4 might be heightened if we could either find a way to sustain the feeder layer or switch to a feeder-independent system. Experiments involving periodic reinforcement of the feeder layer by the addition of fresh cells suggested that while such a strategy might be of some benefit, this would be offset by the added complexity of the protocol (data not shown). We therefore decided to focus on applying the more potent cocktail to establish a streamlined, feeder-free protocol.
Feeder-independent iPSC derivation has generally proved somewhat challenging regardless of the reprogramming technology employed, but raises special difficulties in the context of a sustained transfection regimen. There is a lower limit to the density at which fibroblasts can be plated without compromising cell viability and proliferative activity25. The propensity of cells to undergo mitotic arrest or apoptosis in sparse cultures is exacerbated when they are stressed by transfection and by ectopic expression of reprogramming factors. Moreover, RNA doses which are well tolerated at the high cell densities characteristic of feeder-based reprogramming produce more severe cytotoxic effects when distributed among fewer cells. At the same time, the penetrance of expression achieved with mRNA transfection declines after fibroblasts reach confluence, perhaps due to down-regulation of endocytosis associated with contact inhibition and cell-cycle arrest. Reduced permissiveness to transfection in crowded cultures tends to be alleviated during mRNA reprogramming once cells undergo mesenchymal-to-epithelial transition (MET). However, the ~7 days required for human fibroblasts to reach MET using current mRNA cocktails renders it difficult to avoid fibroblastic overgrowth even when cells are plated at the lowest survivable initial densities. Cultures can be thinned by passaging to postpone this fate, but the plating efficiency of highly stressed reprogramming intermediates is unpredictable, and a split-based protocol would in any case sacrifice convenience and scale poorly to high-throughput applications.
Phenotypic indications of MET (involution of fibroblastic processes and the emergence of foci and cobblestone morphologies) were accelerated in experiments with enhanced cocktails, so we attempted feeder-free derivations using the 6-factor M3O cocktail without passaging steps starting with fibroblast cultures seeded at from 50K to 150K cells per well. The fate of the reprogramming cultures proved highly sensitive to the initial seeding density, presumably because the deleterious effects of excessive cytotoxicity and fibroblastic overgrowth compound over the course of days of transfection. In an early experiment employing a uniform RNA dosing regimen, dozens of hESC-like colonies were obtained from neonatal HDF-n and XFF fibroblasts plated at 100K per well, while the population density in 50K wells crashed and 150K wells succumbed to fibroblastic overgrowth, these sub-optimal wells produced only a few colonies each (data not shown). When the same protocol was applied to two other fibroblast lines, BJ and HDF-a (an adult-derived line), even the most promising (150K) cultures became largely quiescent shortly after reaching confluence, subsequently yielding only sporadic colonies with delayed kinetics. In our follow-on experiments we switched from ambient to 5% oxygen culture and ramped to full RNA dosing from a quarter dose over the first four days of transfection, in efforts to minimize the impact of stress-induced cellular senescence26,27,28,29,30,31. In the first trial using these new conditions we tested if substitution of wild-type L-Myc for c-Myc-T58A could further improve reprogramming efficiency32, and in parallel evaluated a simplified transfection scheme by which RNA was added to cells with daily media changes rather than being delivered in a separate step four hours earlier. RNA dosing was scaled down in 24-hour transfection wells to compensate for an expected increase in cytotoxicity, and two different transfection reagents were tested with this regimen (RNAiMAX and Stemfect). XFF cells were plated at 50K per well and transfected for 9 days. Productivity was assessed by counting TRA-1-60+ colonies at the experiment end-point (Fig. 3a–b). The more conventional 4-hour transfection regime yielded about 250 TRA-1-60+ colonies per well by day 15 using the c-Myc-based cocktail, equivalent to a reprogramming efficiency of 0.5% (CV = 21%, n = 3), while the corresponding L-Myc cultures displayed an average reprogramming efficiency of only ~0.02%. Reprogramming kinetics and efficiency were improved across the board with the 24-hour transfection regime, and these cultures were fixed for analysis on day 11 to avoid overgrowth in the most productive wells. Using the c-Myc cocktail, the RNAiMAX condition gave on the order of a thousand colonies per well at all three RNA dosages tested, corresponding to an iPSC conversion efficiency of ~2%. The yield of TRA-1-60+ colonies was still higher using the Stemfect reagent, making it impossible to count colonies in the most productive wells. In the best-performing c-Myc 400 ng/ml condition, the culture was almost completely overrun with hESC-like cells as early as day 9, one day after the last transfection (Fig. 3c). The mechanistic basis for the superior performance of the 24-hour regimen is unknown, but it may be relevant that dose ramping was achieved within these wells by delivering a reduced volume of medium containing a fixed concentration of RNA, and this might have increased the effective density of thinly-plated cultures. When the preferred conditions were applied in a follow-up derivation on HDF-a adult-derived fibroblasts, 50K and 100K cultures gave a handful of colonies each, but wells seeded with 75K cells produced abundant hESC-like colonies with similar kinetics to the highly proliferative XFF cells (Fig 3d). The rapid kinetics of colony formation using these conditions is apparent in time–lapse imaging showing a subsequent XFF-based derivation presented in Supplementary Movie 2.
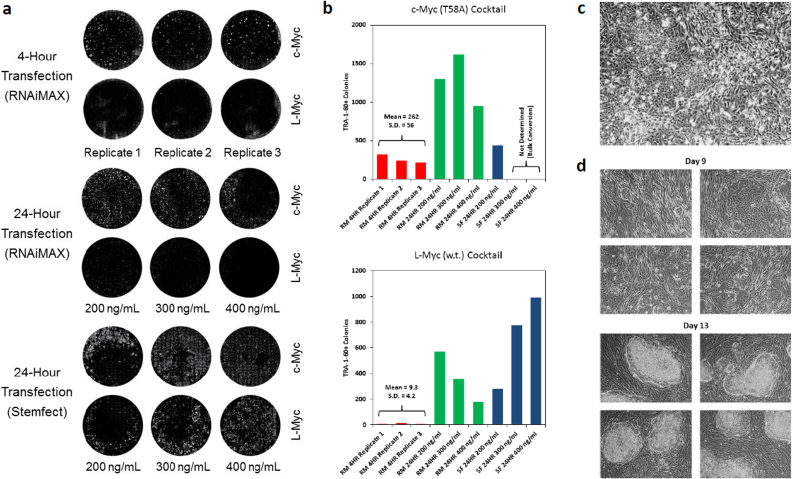
(a) Immunofluorescence imaging showing the TRA-1-60+ colony yield from feeder-free derivations on XFF fibroblasts seeded at 50K per well, comparing c-Myc and L-Myc-based cocktails and 4-hour and 24-hour transfection regimens. All wells were transfected for 9 days. The 4-hour transfection cultures were fixed for staining on day 15 of the experiment, and the 24-hour transfection cultures on day 11. Final media concentration of the transfected RNA is indicated for 24-hour transfection wells. (b) Bar charts showing the TRA-1-60+ colony counts for the wells in the preceding panel. Note that the bulk conversion achieved in the most productive Stemfect wells rendered it impossible to distinguish and count individual iPSC colonies. (c) Phase image of the 400 ng/ml c-Myc Stemfect well from the experiment showing hESC-like colonies overtaking the culture on day 9 of the derivation. (d) Phase imagery showing emergence of colonies from HDF-a adult fibroblast cultures seeded at 75K cells per well and subjected to the same 9-day 400 ng/ml Stemfect transfection regime.
Discussion
We have doubtless only scratched the surface in terms of the potential for technical improvements to RNA-based reprogramming. Size-based purification of synthetic mRNA may further reduce activation of cellular antiviral responses and any associated priming of senescence pathways, while possibly obviating the need for an interferon inhibitor33,34,35. Numerous candidate RFs have been identified which might yield additional performance gains if incorporated into mRNA cocktails36,37,38,39,40, while the advantage realized by substituting M3O for wild-type Oct4 offers fresh support for the application of engineered chimeric factors to cell-fate direction. Acceleration of the reprogramming process could likewise come about through optimization of factor stoichiometries41, and we note that the mRNA method opens up the possibility of fine-tuning cocktails to address early and late phases of iPSC induction independently. Finally, the reagents and approach applied here for sustained delivery of mRNA can also be employed to co-transfect other types of RNA including miRNA, which has proven utility for iPSC generation42,43,44. The methodology we describe already represents a major advance over current protocols. It cuts mRNA reprogramming time by as much as a half with even greater reductions in hands-on time and material costs, while removing troublesome steps from the procedure and allowing mRNA to be handled as a media supplement with almost the same facility as growth factors and cytokines. At the same time, the feeder-free protocol meets a key requirement for moving iPSCs from laboratory to clinic by eliminating potential sources of non-human contamination from the derivation process. If this fast, economical footprint-free iPSC origination technique can be combined with advances in high-throughput propagation and quality assurance of pluripotent clones45, the remaining barriers to large-scale, cost-effective production of therapeutically-qualified patient-specific iPSC lines should be few indeed.
Methods
Generation of IVT templates
Plasmid constructs to serve as the basis for generating linear in vitro transcription (IVT) templates were constructed using Ligation Independent Cloning (LIC). We first constructed a parental plasmid (pIVT) incorporating 5′ and 3′ untranslated regions (UTRs) flanking an insertion site designed to accept an open reading frame (ORF) encoding a protein of interest. The ORF-flanking sequences are as described previously19, comprising a low secondary structure 5′ leader and strong Kozak site and the murine α-globin 3′ UTR. A linearized version of the pIVT vector bearing 5′ overhangs was produced by melting and then re-annealing two PCR products amplified from the plasmid using tailed primers. ORF amplicons with complementary overhangs were made by an analogous procedure, pooled with the vector PCR products and transformed into DH5α bacteria to clone gene-specific constructs (pIVT-KLF4, etc.). The transactivation domain-encoding sequences of the M3O and VPx3 inserts were appended to the wild-type Oct4 ORF by tailed PCR and splint-mediated ligation. Purified plasmids were used to template PCRs to generate linear IVT template amplicons incorporating a T7 promoter, a UTR-flanked ORF, and a T120 polynucleotide stretch to drive addition of a polyA tail, as described previously. The T120 sequence was introduced by use of a tailed reverse primer. PCR product stocks were maintained at ~100 ng/uL.
Production of mRNA cocktails
The mRNA synthesis procedure is summarized in Supplementary Fig. S1. Synthetic mRNA was generated in IVT reactions using a 4:1 ratio of ARCA cap analog to GTP to give a high percentage of capped transcripts. Complete substitution of 5m-CTP for CTP and Pseudo-UTP for UTP in the nucleotide triphosphate (NTP) mix was employed to reduce the immunogenicity of the RNA products. Cap analog and modified NTPs were purchased from Trilink Biotechnologies. A 2.5X NTP mix was prepared (ARCA:ATP:5m-CTP:GTP:Pseudo-UTP at 15:15:3.75:3.75:3.75 mM) to replace the standard NTPs provided with the MEGAscript T7 Kit (Ambion) used to perform IVT reactions. Each 40 uL IVT reaction comprised 16 uL NTP mix, 4 uL 10X T7 Buffer, 16 uL DNA template and 4 uL T7 polymerase. Reactions were incubated 4–6 hours at 37°C and treated with 2 uL TURBO DNase for a further 15 minutes at 37°C before being purified on MEGAclear (Ambion) spin columns, the RNA products being eluted in a volume of 100 uL. To remove immunogenic 5′ triphosphate moieties from uncapped transcripts, 10 uL of Antarctic Phosphatase reaction buffer and 3 uL of Antarctic Phosphatase (NEB) was added to each prep. Phosphatase reactions were incubated for 30 minutes at 37°C and the IVT products were repurified. RNA yield was quantitated by Nanodrop (Thermo Scientific), and the preps were subsequently adjusted to a standardized working concentration of 100 ng/uL by addition of TE pH 7.0 (Ambion). IVT product quality was checked by running 500 ng of RNA per lane on 2% agarose SYBR Safe E-gels (Invitrogen). Cocktails were assembled by pooling preps representing the various RFs in the desired stoichiometric ratios. The fraction of each RF used took into account the predicted molecular weight of the respective transcript, all RFs being equimolar except for Oct4 and its derivatives, which were included at 3X molar concentration. A 10% spike of mRNA encoding a nuclearized monomeric LanYFP fluorescent protein was added to cocktails to facilitate monitoring of transfection efficiency during reprogramming trials.
Cells and growth media
Cells targeted for reprogramming included BJ neonatal fibroblasts (ATCC), HDF-f fetal fibroblasts, HDF-n neonatal fibroblasts and HDF-a adult fibroblasts (ScienCell), and XFF xeno-free neonatal fibroblasts (Millipore). Expansion culture was carried out in BJ medium (DMEM + 10% FBS), ScienCell Fibroblast Medium, and FibroGRO Xeno-Free Human Fibroblast Expansion Medium (Millipore) for BJ, HDF and XFF cells respectively. The feeder cells used were 3001G irradiated neonatal human foreskin fibroblasts (GlobalStem) and FibroGRO mitomycin C-inactivated xeno-free human neonatal fibroblasts (Millipore). Cell passaging steps pertaining to xeno-free feeder-based and feeder-free reprogramming trials were conducted using TrypLE Select (Gibco), an animal product-free cell dissociation reagent.
Reprogramming of human fibroblasts
All reprogramming experiments described were performed in 6-well tissue culture plates coated with CELLstart (Gibco) xeno-free substrate in accordance with the manufacturer's directions. GlobalStem feeders were plated at 250K per well in the first BJ reprogramming experiment using FBS-containing BJ medium. In some of the later feeder-based trials the seeding density was increased and feeders were supplemented ad hoc during medium changes in an effort to sustain near-confluent feeder layers in response to the higher attrition rates encountered using novel RF cocktails. Xeno-free feeders were plated in Pluriton xeno-free medium (Stemgent) without serum. Target cells were seeded in Pluriton supplemented with 200 ng/ml B18R interferon inhibitor (eBioscience). Media was replaced daily during and after reprogramming, B18R supplementation being discontinued the day after the final transfection. In experiments in which cells were split during reprogramming, 10 uM Y27632 (Stemgent) was added to the medium used for replating. Transfections commenced the day after seeding of target cells, and were repeated every 24 hours for the durations indicated. An RNA dose of 1200 ng was delivered to each well using RNAiMAX (Invitrogen) 4 hours prior to daily medium change, except as otherwise noted. For dose ramping, the amount of RNA delivered was reduced to 25%, 50% and 75% of the maintenance dose on the first three transfections. For 24-hour transfections, dose ramping was achieved by delivering 0.5 mL, 1 mL, 1.5 mL or 2 mL of medium supplemented with RNA at the concentrations specified in Fig. 3a. RNAiMAX transfection cocktails were assembled by diluting 100 ng/uL RNA 5X and 5 uL of RNAiMAX per microgram of RNA 10X separately in DPBS without calcium or magnesium, then pooling to give a 10 ng/uL RNA/vehicle suspension and dispensing to culture media after a 15-minute incubation. For transfections with Stemfect (Stemgent), RNA and Stemfect (4 uL per microgram of RNA) were prepared for 3 days of transfection in the provided buffer at an RNA concentration of 10 ng/uL. The mixture was incubated for 15 minutes and added to culture media directly or refrigerated for use within 48 hours.
Characterization of iPSC colonies
To assess iPSC colony productivity in reprogramming plates, cells were fixed with 4% paraformaldehyde (PFA) in DPBS with calcium and magnesium and immunostained with StainAlive TRA-1-60 Alexa 488 antibody (Stemgent) diluted 1:100. Colony picking, expansion, and subsequent immunostaining and trilineage differentiation assays for molecular and functional validation of pluripotency were carried out by the Sanford Burnham Medical Research Institute. For pluripotency immunostaining panels, clones were cultured on Matrigel (BD Biosciences) in mouse embryonic fibroblast (MEF)-conditioned medium, fixed with 4% PFA and permeabilized with 0.3% Triton X-100 (for nuclear markers), then stained with primary antibodies against SSEA3 (Millipore), TRA-1-81 (Cell Signaling), Oct3/4 (Santa Cruz) and Sox2 (Cell Signaling). Ectodermal differentiation was performed by breaking up Dispase-detached colonies into chunks and culturing them in neural progenitor cell (NPC) media within low-attachment plates for 6 days, after which the resulting NPCs were transferred to a tissue-culture plate coated with Fibronectin (Sigma). The plated cells were stained after 2–3 days using primary antibodies against Nestin and Pax6 (Abcam) diluted 1:200. For endodermal differentiation, iPSCs were cultured in MEF-conditioned media on Matrigel-coated plates and switched to RPMI + 0.5% FBS + 100 ng/mL activin-A for 5 days, then stained with Sox17 primary antibody (Santa Cruz) diluted 1:200. Cardiomyocyte differentiation was done by detaching confluent colonies using Dispase, breaking them up using manual technique and culturing the suspended clumps in Embryoid Body (EB) media (DMEM/F12 + 20% FBS) for 7-10 days in low-attachment plates, then plating the EBs to tissue culture plates coated with 0.1% gelatin in EB media. Cardiomyocytes were stained with 1:200 diluted mouse anti-human α-Actinin primary antibody (Sigma).
Author Contributions
LW and JW planned the project. YN and XG participated in discussion. XG provided funding and support. LW conducted the majority of the experiments. JW and YN performed certain plasmid construction work. LW wrote the manuscript; YN helped with the videos; JW finalized the paper.
Supplementary Material
Supplementary Information
Supplementary Movie 1.
Supplementary Movie 2.
Acknowledgments
We thank Drs. Brandon Nelson and Yang Liu of the Sanford Burnham Medical Research Institute and Dr. Kim Thien Ly for their assistance with the isolation, propagation and analysis of iPSC clones, and WiCell for DNA fingerprinting and karyotype analyses. This work was supported in part by grants from the National Natural Science Foundation of China (81100618, 30973231) and the Jiangsu medical research program (LJ201108) to XG.
Footnotes
LW is a consulting scientist and YN and JW are employees of Allele Biotechnology & Pharmaceuticals, Inc., San Diego, CA. A patent has been filed on the reported technologies.
References
- Takahashi K. & Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–76 (2006). [Abstract] [Google Scholar]
- Carey B. W. et al. Reprogramming of murine and human somatic cells using a single polycistronic vector. Proc Natl Acad Sci U S A 106, 157–62 (2009). [Europe PMC free article] [Abstract] [Google Scholar]
- Chang C. W. et al. Polycistronic lentiviral vector for "hit and run" reprogramming of adult skin fibroblasts to induced pluripotent stem cells. Stem Cells 27, 1042–9 (2009). [Abstract] [Google Scholar]
- Sommer C. A. et al. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells 27, 543–9 (2009). [Abstract] [Google Scholar]
- Somers A. et al. Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem Cells 28, 1728–40 (2010). [Europe PMC free article] [Abstract] [Google Scholar]
- Woltjen K. et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature 458, 766–70 (2009). [Europe PMC free article] [Abstract] [Google Scholar]
- Kaji K. et al. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature 458, 771–5 (2009). [Europe PMC free article] [Abstract] [Google Scholar]
- Yusa K., Rad R., Takeda J. & Bradley A. Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat Methods 6, 363–9 (2009). [Europe PMC free article] [Abstract] [Google Scholar]
- Okita K., Nakagawa M., Hyenjong H., Ichisaka T. & Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science 322, 949–53 (2008). [Abstract] [Google Scholar]
- Zhou W. & Freed C. R. Adenoviral gene delivery can reprogram human fibroblasts to induced pluripotent stem cells. Stem Cells 27, 2667–74 (2009). [Abstract] [Google Scholar]
- Yu J. et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science 324, 797–801 (2009). [Europe PMC free article] [Abstract] [Google Scholar]
- Jia F. et al. A nonviral minicircle vector for deriving human iPS cells. Nat Methods 7, 197–9 (2010). [Europe PMC free article] [Abstract] [Google Scholar]
- Yu J., Chau K. F., Vodyanik M. A., Jiang J. & Jiang Y. Efficient feeder-free episomal reprogramming with small molecules. PLoS One 6, e17557 (2011). [Europe PMC free article] [Abstract] [Google Scholar]
- Zhou H. et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell 4, 381–4 (2009). [Abstract] [Google Scholar]
- Kim D. et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell 4, 472–6 (2009). [Europe PMC free article] [Abstract] [Google Scholar]
- Fusaki N., Ban H., Nishiyama A., Saeki K. & Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci 85, 348–62 (2009). [Europe PMC free article] [Abstract] [Google Scholar]
- Ban H. et al. Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. Proc Natl Acad Sci U S A 108, 14234–9 (2011). [Europe PMC free article] [Abstract] [Google Scholar]
- MacArthur C. C. et al. Generation of human-induced pluripotent stem cells by a nonintegrating RNA Sendai virus vector in feeder-free or xeno-free conditions. Stem Cells Int. 2012, 564612 (2012). [Europe PMC free article] [Abstract] [Google Scholar]
- Warren L. et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 7, 618–30 (2010). [Europe PMC free article] [Abstract] [Google Scholar]
- Hirai H. et al. Radical acceleration of nuclear reprogramming by chromatin remodeling with the transactivation domain of MyoD. Stem Cells 29, 1349–61 (2011). [Europe PMC free article] [Abstract] [Google Scholar]
- Hirai H., Katoku-Kikyo N., Karian P., Firpo M. & Kikyo N. Efficient iPS Cell Production with the MyoD Transactivation Domain in Serum-Free Culture. PLoS One 7, e34149 (2012). [Europe PMC free article] [Abstract] [Google Scholar]
- Wang Y. et al. Reprogramming of mouse and human somatic cells by high-performance engineered factors. EMBO Rep. 12, 373–8 (2011). [Europe PMC free article] [Abstract] [Google Scholar]
- Wang W. et al. Rapid and efficient reprogramming of somatic cells to induced pluripotent stem cells by retinoic acid receptor gamma and liver receptor homolog 1. Proc Natl Acad Sci U S A 108, 18283–8 (2011). [Europe PMC free article] [Abstract] [Google Scholar]
- Yu J. et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–20 (2007). [Abstract] [Google Scholar]
- Freshney R. I. Culture of Animal Cells: A Manual of Basic Technique and Specialized Applications, (Wiley-Blackwell, 2010). [Google Scholar]
- Marion R. M. et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 460, 1149–53 (2009). [Europe PMC free article] [Abstract] [Google Scholar]
- Kawamura T. et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460, 1140–4 (2009). [Europe PMC free article] [Abstract] [Google Scholar]
- Li H. et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 460, 1136–9 (2009). [Europe PMC free article] [Abstract] [Google Scholar]
- Utikal J. et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature 460, 1145–8 (2009). [Europe PMC free article] [Abstract] [Google Scholar]
- Hong H. et al. Suppression of induced pluripotent stem cell generation by the p53–p21 pathway. Nature 460, 1132–5 (2009). [Europe PMC free article] [Abstract] [Google Scholar]
- Yoshida Y., Takahashi K., Okita K., Ichisaka T. & Yamanaka S. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell 5, 237–41 (2009). [Abstract] [Google Scholar]
- Nakagawa M., Takizawa N., Narita M., Ichisaka T. & Yamanaka S. Promotion of direct reprogramming by transformation-deficient Myc. Proc Natl Acad Sci U S A 107, 14152–7 (2010). [Europe PMC free article] [Abstract] [Google Scholar]
- Kariko K., Muramatsu H., Ludwig J. & Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 39, e142 (2011). [Europe PMC free article] [Abstract] [Google Scholar]
- Mah N. et al. Molecular insights into reprogramming-initiation events mediated by the OSKM gene regulatory network. PLoS One 6, e24351 (2011). [Europe PMC free article] [Abstract] [Google Scholar]
- Moiseeva O., Mallette F. A., Mukhopadhyay U. K., Moores A. & Ferbeyre G. DNA damage signaling and p53-dependent senescence after prolonged beta-interferon stimulation. Mol. Biol. Cell 17, 1583–92 (2006). [Europe PMC free article] [Abstract] [Google Scholar]
- Chang R., Shoemaker R. & Wang W. Systematic search for recipes to generate induced pluripotent stem cells. PLoS Computational Biology 7, e1002300 (2011). [Europe PMC free article] [Abstract] [Google Scholar]
- Fischedick G. et al. Zfp296 is a novel, pluripotent-specific reprogramming factor. PLoS One 7, e34645 (2012). [Europe PMC free article] [Abstract] [Google Scholar]
- Singhal N. et al. Chromatin-Remodeling Components of the BAF Complex Facilitate Reprogramming. Cell 141, 943–55 (2010). [Abstract] [Google Scholar]
- Zhao Y. et al. Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell 3, 475–9 (2008). [Abstract] [Google Scholar]
- Wang T. et al. The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell 9, 575–87 (2011). [Abstract] [Google Scholar]
- Papapetrou E. P. et al. Stoichiometric and temporal requirements of Oct4, Sox2, Klf4, and c-Myc expression for efficient human iPSC induction and differentiation. Proc Natl Acad Sci U S A 106, 12759–64 (2009). [Europe PMC free article] [Abstract] [Google Scholar]
- Anokye-Danso F. et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 8, 376–88 (2011). [Europe PMC free article] [Abstract] [Google Scholar]
- Liao B. et al. MicroRNA cluster 302-367 enhances somatic cell reprogramming by accelerating a mesenchymal-to-epithelial transition. J. Biol. Chem. 286, 17359–64 (2011). [Europe PMC free article] [Abstract] [Google Scholar]
- Miyoshi N. et al. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell 8, 633-8 (2011). [Abstract] [Google Scholar]
- Valamehr B. et al. A novel platform to enable the high-throughput derivation and characterization of feeder-free human iPSCs. Scientific Reports 2, 213 (2012). [Europe PMC free article] [Abstract] [Google Scholar]
Articles from Scientific Reports are provided here courtesy of Nature Publishing Group
Full text links
Read article at publisher's site: https://doi.org/10.1038/srep00657
Read article for free, from open access legal sources, via Unpaywall:
https://www.nature.com/articles/srep00657.pdf
Citations & impact
Impact metrics
Article citations
Rapid iPSC inclusionopathy models shed light on formation, consequence, and molecular subtype of α-synuclein inclusions.
Neuron, 112(17):2886-2909.e16, 29 Jul 2024
Cited by: 3 articles | PMID: 39079530 | PMCID: PMC11377155
The occurrence and development of induced pluripotent stem cells.
Front Genet, 15:1389558, 18 Apr 2024
Cited by: 0 articles | PMID: 38699229 | PMCID: PMC11063328
Review Free full text in Europe PMC
Engineered T cells from induced pluripotent stem cells: from research towards clinical implementation.
Front Immunol, 14:1325209, 12 Jan 2024
Cited by: 1 article | PMID: 38283344 | PMCID: PMC10811463
Review Free full text in Europe PMC
Cell Reprogramming and Differentiation Utilizing Messenger RNA for Regenerative Medicine.
J Dev Biol, 12(1):1, 20 Dec 2023
Cited by: 0 articles | PMID: 38535481 | PMCID: PMC10971469
Review Free full text in Europe PMC
Learning Towards Maturation of Defined Feeder-free Pluripotency Culture Systems: Lessons from Conventional Feeder-based Systems.
Stem Cell Rev Rep, 20(2):484-494, 11 Dec 2023
Cited by: 0 articles | PMID: 38079087
Review
Go to all (91) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Non-integrating episomal plasmid-based reprogramming of human amniotic fluid stem cells into induced pluripotent stem cells in chemically defined conditions.
Cell Cycle, 15(2):234-249, 01 Jan 2016
Cited by: 19 articles | PMID: 26654216 | PMCID: PMC4825845
Reprogramming of human fibroblasts to induced pluripotent stem cells under xeno-free conditions.
Stem Cells, 28(1):36-44, 01 Jan 2010
Cited by: 51 articles | PMID: 19890879
Establishment and optimal culture conditions of microrna-induced pluripotent stem cells generated from HEK293 cells via transfection of microrna-302s expression vector.
Nagoya J Med Sci, 74(1-2):157-165, 01 Feb 2012
Cited by: 8 articles | PMID: 22515122 | PMCID: PMC4831261
An Insight into DNA-free Reprogramming Approaches to Generate Integration-free Induced Pluripotent Stem Cells for Prospective Biomedical Applications.
Stem Cell Rev Rep, 15(2):286-313, 01 Apr 2019
Cited by: 38 articles | PMID: 30417242
Review





