Abstract
Free full text

Cleavage Activation of the Human-Adapted Influenza Virus Subtypes by Matriptase Reveals both Subtype and Strain Specificities
Abstract
Cleavage activation of the hemagglutinin (HA) precursor is an essential step in the influenza virus replication cycle that is driven by host cell proteases. HA cleavage activation is required for virus-endosome membrane fusion and the subsequent release of the influenza virus genome into the cytoplasm. Previous studies have determined that HA cleavage is most likely driven by either membrane-bound or extracellular trypsin-like proteases that reside in the respiratory tract. However, there is still uncertainty regarding which proteases are critical for HA cleavage in vivo. Therefore, further investigation of HA cleavage activation is needed in order to gain insight into the critical proteases involved. Matriptase is a member of the type II transmembrane serine protease family that is highly expressed in a membrane-bound form throughout the respiratory tract. One feature of matriptase is that, once activated, the catalytic domain is secreted into the extracellular space and so serves as a functional extracellular protease. In this study, we have determined that the secreted, catalytic domain of matriptase has the ability to cleave and activate HA from the influenza virus H1 subtype but not the H2 and H3 subtypes. Furthermore, matriptase selectively cleaved the HA of particular strains within the H1 subtype, revealing both subtype and H1 strain specificity. Matriptase was also found to activate thrombolytic zymogens that have been shown to cleave and activate the influenza virus HA. Our data demonstrate that matriptase has the ability to cleave HA directly or indirectly by activating HA-cleaving zymogens.
INTRODUCTION
Influenza virus belongs to the Orthomyxoviridae family and is classified into A, B, and C types (26). Influenza A virus is further categorized into 16 hemagglutinin (HA) and 9 neuraminidase (NA) proteins on the basis of their distinct antigenicity. Influenza A virus infects both mammalian and avian species, where each of the influenza A virus subtypes is thought to have originated from waterfowl (23). A few of the influenza A virus subtypes have established themselves in the human population, with current seasonal influenza virus infections being caused by the H1N1 and H3N2 subtypes. The H2N2 influenza virus subtype has also circulated in the human population and caused a pandemic in 1957 but is no longer circulating (22). The more recent 2009 H1N1 pandemic virus caused over 18,000 deaths, many of which were in the young, raising concern for the potential of a high-fatality pandemic (24, 35).
The influenza virus HA is synthesized as a precursor (HA0) that must be cleaved in order to initiate the fusion of the viral envelope with the host cell endosomal membrane, permitting the release of the viral ribonucleoproteins into the cytoplasm (30, 32). HA cleavage is thought to occur by trypsin-like, serine proteases that are found on the plasma membrane or in the extracellular space of the respiratory tract. Extracellular proteases, such as mast cell tryptase, tryptase Clara, and cellular trypsins, have been determined to cleave and activate influenza virus in vitro (8, 16, 17). Moreover, HA cleavage by the extracellular, thrombolytic proteases plasmin, urokinase, plasma kallikrein, and thrombin has also been determined (27). In addition to extracellular proteases, membrane-bound proteases have also been investigated for their ability to cleave HA, namely, the type II transmembrane serine protease (TTSP) family. The TTSP family consists of a number of trypsin-like serine proteases that are expressed in a variety of tissues. These TTSPs are subdivided into the HAT (human airway trypsin-like protease)/DESC (differentially expressed in squamous cell carcinoma gene), hepsin/TMPRSS (transmembrane protease, serine 2), matriptase, and corin families (13). The hepsin/TMPRSS subfamily members TMPRSS2 and TMPRSS4 and the HAT/DESC subfamily member HAT have all been found to cleave and activate influenza virus in vitro (3, 5–7). In addition, TMPRSS2 and HAT have been found to activate entry of other viruses, including the severe acute respiratory syndrome coronavirus (SARS-CoV) (4, 12, 28). To date, only matriptase-3 of the matriptase family has been investigated for the ability to activate HA, where it was determined that the protease was unable to cleave HA (1). Continued studies of this nature are needed in order to gain further insight into the TTSP family members that have the potential to activate influenza virus in vivo (10).
The TTSP family member matriptase (ST14 [suppression of tumorigenicity 14]) is expressed in a variety of tissues, including the respiratory tract, and plays a key role in maintaining the integrity of epithelial cells (21, 25). Matriptase is thought to mainly reside on the basolateral side of epithelial cells, where it is typically bound to the hepatocyte growth factor activator inhibitor type 1 (HAI-1) (34). The proteolytic activity of matriptase must be tightly regulated and is therefore largely suppressed by HAI-1 on the plasma membrane. In a similar fashion, HAI-1 has recently been shown to be a potent inhibitor and potential regulator of HAT (14). In the case of matriptase, the membrane type 1 matrix metalloproteinase has been found to restore protease activity by cleaving HAI-1 in vitro, which subsequently promotes secretion of the catalytic domain of matriptase (11). A significant proportion of matriptase is thought to be shed from both the basolateral and apical faces of polarized epithelial cells, as demonstrated by the isolation of the protease in human milk (19, 20, 33). On the basis of the similarities to other members of the TTSP family that have been found to activate influenza virus, combined with the known expression in the nasal and bronchial lining, matriptase may also have the potential to activate influenza virus in vivo (15).
In this study, we have investigated whether the secreted catalytic domain of matriptase has the ability to cleave and activate the HA from the human-adapted subtypes. The membrane form of matriptase was not investigated, since it primarily resides on the basolateral face of epithelial cells, whereas influenza virus infects from the apical surface; therefore, the membrane form of matriptase is not likely to be involved in HA cleavage in intact epithelia. In addition, we also investigated whether matriptase has the ability to activate thrombolytic zymogens that have been reported to cleave and activate influenza virus HA, yielding both a direct and an indirect activation by matriptase.
MATERIALS AND METHODS
Cells, viruses, and plasmids.
293T and Vero cells (American Type Culture Collection) were maintained in Dulbecco's modified Eagle medium (DMEM; Cellgro) supplemented with 10% fetal bovine serum (Gibco), 100 units/ml penicillin (Cellgro), and 10 units/ml streptomycin (Cellgro). Plasmids encoding the A/WSN/33 (H1N1), A/PR/8/34 (H1N1), and A/California/04/09 (H1N1) HAs were generated by the methods described by Sun et al. (31). The gene encoding the A/Japan/305/57 (H2N2) HA was subcloned into the pEF4 expression vector (Invitrogen). The plasmid encoding the A/Aichi/2/68 (H3N2) HA was generously donated by David Steinhauer. The plasmids encoding the A/Wyoming/3/03 (H3N2) and A/Wisconsin/67/05 (H3N2) HAs were purchased from Sino Biological Inc. A/PR/8/34 and A/WSN/33 viruses were propagated in eggs and used to produce noncleaved virus.
Proteases.
Tosylsulfonyl phenylalanyl chloromethyl ketone-treated trypsin from bovine pancreas was obtained from Pierce, and bovine thrombin was obtained from Sigma. The catalytic domain of human matriptase was purchased from R&D Systems. Plasminogen, plasmin, prekallikrein, and urokinase were obtained from EMD Biosciences. The mouse prourokinase (pro-UK) gene (ATCC 63256) was obtained from the American Type Culture Collection (2). The pro-UK-specific primers (5′-forward-GGA TCC AGT GTA CTT GGA GCTCCT GAT-3′; 5′-reverse-CTC GAG GAA GGC CAG ACC TTT CTC TTC TCC) were used to amplify the pro-UK gene, which was subsequently cloned into a modified pSUMO vector that was generously donated by Holger Sondermann (Cornell University) and named pro-UK-pSUMO. pro-UK-pSUMO was transformed into Escherichia coli BL21 and grown in 1 liter of Luria broth (LB) containing 35 μg/ml kanamycin. The 1-liter culture was induced with 0.25 mM isopropyl-β-d-thiogalactopyranoside and incubated overnight at 20°C. The cells were then collected by centrifugation at 3,500 rpm for 20 min. The cell pellet was resuspended in buffer A (50 mM Tris, pH 7.8) and lysed by sonication. The bacterial lysate was loaded onto an Ni2+ nitrilotriacetic acid agarose resin column (preequilibrated with buffer A, 3-ml bed volume) and subsequently washed with buffer B (50 mM Tris, 300 mM NaCl, pH 7.8). pro-UK was then eluted with buffer C (50 mM Tris, 300 mM imidazole, pH 7.8), transferred to 3,500-molecular-weight-cutoff dialysis tubing (Spectrapor) along with 1 mg of SUMO protease, and dialyzed overnight in buffer A at 4°C. The protein was then concentrated to 1 mg/ml using a centrifugation vial containing a 10,000-molecular-weight-cutoff membrane (Amicon/Millipore).
Matriptase HA cleavage analysis.
293T cells were grown in polylysine-coated, 12-well plates and transfected with 1.0 μg of each HA-expressing plasmid using Lipofectamine 2000 (Invitrogen) for 12 h at 37°C. The cells were then washed with phosphate-buffered saline (PBS) and treated with 200 nM matriptase in buffer A (20 mM Tris, 10 mM CaCl2, 150 mM NaCl, pH 7.4) for 1.5 h at 37°C. The HA-expressing cells were also incubated in the absence of protease for 1.5 h at 37°C or treated with 0.7 μM trypsin for 10 min at 37°C as controls. The cells were then washed with PBS and processed by cell surface biotinylation as described by Sun et al. (31). HA cleavage was analyzed by Western blotting using anti-A/PR8 (H1), anti-A/Singapore (H2), and anti-A/Hong Kong/1/68 (H3) antibodies (NIAID Biodefense & Emerging Infections Research Resource Repository). HA cleavage by matriptase was quantified by densitometry.
Cell-cell fusion assay.
Vero cells were grown in 24-well plates that contained glass coverslips and transfected with 0.6 μg of the A/California/04/09, A/South Carolina/1/18, A/Japan/305/57, and A/Aichi/2/68 HA-expressing plasmids using Lipofectamine 2000 for 12 h at 37°C. The HA-expressing cells were washed with PBS and treated with 200 nM matriptase in buffer A for 3 h at 37°C. The cells were then washed with PBS and treated with HBMSS buffer (5 mM HEPES, 5 mM MES [morpholineethanesulfonic acid], 5 mM succinate, 150 mM NaCl, pH 5.0) for 2 min, washed with PBS, and incubated in DMEM for 1 h at 37°C. Cell-cell fusion was analyzed by immunofluorescence staining using the anti-HA antibodies described above coupled with Alexa Fluor 488 anti-goat antibody (Molecular Probes). The nuclei were stained with Hoechst 33258 (Invitrogen).
Virus infection assay.
293T cells were utilized to produce noncleaved virus, since they do not express a protease capable of cleaving HA. In these cells, influenza virus infection undergoes a single replication round, producing only noncleaved virus. Noncleaved A/PR/8/34 and A/WSN/33 were produced by incubation of 1 PFU/cell of egg-derived virus with 293T cells in a 6-cm dish containing RPMI for 1 h at 37°C. The virus solution was subsequently removed, washed with PBS, and incubated in 3 ml of minimal essential medium (MEM) for 3 days at 37°C. The noncleaved virus solution was then treated with 200 nM matriptase for 1 h at 37°C. The virus was also incubated in the absence of proteases or treated with 1 μM trypsin for 15 min at 37°C as controls. The trypsin reaction was stopped by adding 2.5 μg trypsin inhibitor (EMD Chemicals). The resulting viral solutions were incubated for 6 h at 37°C with Vero cells grown on glass coverslips in 24-well plates. Viral infection was analyzed by immunofluorescence using antinucleoprotein coupled with an Alexa Fluor 488 (green) antirabbit antibody. The nuclei were stained with Hoechst 33258 (blue).
Cleavage of peptide mimics of the HA cleavage site by matriptase.
Peptides were designed on the basis of the amino acid sequence of the HA cleavage site region of the H1 (IPSIQSRGL and IPSIQYRGL), H2 (VPQIESRGL), and H3 (VPEKQTRGL) subtypes and with the fluorescence resonance energy transfer pair 7-methoxycoumarin-4-yl acetyl (MCA), added to the N terminus, and N-2,4-dinitrophenyl (DNP) (RS Synthesis), added to the C terminus. Each peptide along with matriptase was diluted to 50 μl in buffer A to produce a final concentration of 100 μM peptide and 40 nM matriptase. The reaction was carried out for 1 h at 37°C and monitored for cleavage by measurement of the change in fluorescence at 390 nm (SpectraMax GeminiXS; Molecular Devices).
Comparison of rate of cleavage of various HA-cleaving proteases.
The cleavage rates of the HA-cleaving proteases urokinase, thrombin, and plasmin were compared to the matriptase rate of cleavage. Each protease along with the H1 cleavage site peptide mimic (IPSIQSRGL) was diluted to 50 μl in buffer A to produce a final concentration of 100 μM peptide and 20 nM protease. The reaction was carried out for 1 h at 37°C and monitored for cleavage by measurement of the change in fluorescence at 390 nm (SpectraMax GeminiXS; Molecular Devices).
Activation of thrombolytic zymogens by matriptase.
Matriptase (0.1 ng/μl) was preincubated with 0.2 μg/μl of plasminogen, prourokinase, and prekallikrein in buffer A for 3 h at 37°C. The same concentrations of matriptase and each zymogen were incubated alone for 3 h at 37°C as controls. Each sample along with the fluorogenic peptide substrate GGGR-7-amino-4-methylcoumarin (GGGR-AMC) (EMD Chemicals) was diluted to a 50-μl total reaction volume, resulting in a 150 μM substrate concentration. The reaction was carried out for 1 h at 37°C, and the rate of cleavage was monitored by measurement of the change in fluorescence at 440 nm.
Matriptase concentration in nasal wash samples.
Nasal wash samples were obtained prior to vaccination from healthy adult subjects participating in an ongoing study of live attenuated influenza vaccine. Serial dilutions of four distinct nasal wash samples were incubated overnight in an Immulon 4HBX plate (Thermo Scientific) and subsequently washed with buffer D (0.05% Triton X-100 in PBS). Serial dilutions of a matriptase standard (R&D Systems) were also incubated overnight and washed with buffer D. The concentration of matriptase was measured by a standard enzyme-linked immunosorbent assay (ELISA) using an anti-matriptase antibody (R&D Systems) coupled with an antimouse secondary antibody (Pierce).
RESULTS
Presence of secreted matriptase in the human respiratory tract.
To determine whether the secreted form of matriptase is present within the respiratory tract, the matriptase concentration in nasal wash samples from four healthy human donors was assessed by ELISA. Analysis of the nasal wash samples resulted in a matriptase concentration of 6.8 ± 1.7 ng/ml. These data confirm the presence of a secreted form of matriptase in the upper respiratory tract, supporting the in vivo relevance of matriptase as a protease capable of activating influenza virus HA.
Cleavage activation of influenza virus HA by matriptase.
To determine whether the catalytic domain of matriptase has the ability to cleave HA, mammalian cells expressing HA from six strains of the H1 subtype, one strain of the H2 subtype, and three strains of the H3 subtype were treated with matriptase and assessed for cleavage by Western blot analysis. In all cases of the H1-subtype strains, cleavage that resembled the migration rate of the HA1 (55-kDa) and HA2 (25-kDa) cleavage products compared to that for the trypsin control was observed (Fig. 1A). Variability lies within the migration rates of HA from each strain due to differences in the abundance of glycosylation, where the HAs from mouse-adapted viruses have a relatively smaller amount of glycosylation. Efficient HA cleavage of the A/PR/8/34, A/New Caledonia/20/99, and A/California/04/09 strains was observed, with the A/California/04/09 strain being cleaved the most efficiently, as determined by quantification of the cleavage products of each strain (Fig. 2). HA cleavage was also observed to a lesser extent for the A/South Carolina/1/18, A/WS/33, and the A/WSN/33 H1 strains with matriptase treatment, giving the appearance of preferential cleavage of particular strains within the H1 subtype. In contrast, trypsin treatment of HA from each H1 strain resulted in a similar cleavage efficiency (Fig. 2). In the case of the H2 subtype, minimal cleavage was observed with matriptase treatment compared to that for the mock control (Fig. 1B). Moreover, in the case of the H3 subtype, minimal cleavage was also observed with matriptase treatment of each strain (Fig. 1C). Thus, on the basis of these results, matriptase has a strong preference for the H1 subtype, with a preference for particular strains within the H1 subtype.

Cleavage of the human-adapted HA subtypes by matriptase. (A) Western blot analysis of HA cleavage from the A/PR/8/34, A/New Caledonia/20/99 (A/NC/99), A/California/04/09 (A/CA/09), A/South Carolina/1/18 (A/SC/18), A/WS/33, and A/WSN/33 H1-subtype strains by matriptase. A/WSN/33 HA in the absence of protease (mock) and in the presence of trypsin was added as a control. (B) Western blot analysis of HA cleavage from the A/Japan/305/57 H2-subtype strain by matriptase. (C) Western blot analysis of HA cleavage from the A/Aichi/2/68, A/Wyoming/3/03 (A/WY/03), and A/Wisconsin/67/05 (A/WI/05) (H3N2) H3-subtype strains by matriptase. A/Aichi/2/68 HA in the absence of protease (mock) and in the presence of trypsin was added as a control.

Comparison of the H1 HA cleavage efficiency of trypsin and matriptase. Bar graph depicting the cleavage efficiency of A/PR/8/34, A/New Caledonia/20/99 (A/NC/99), A/California/04/09 (A/CA/09), A/South Carolina/1/18 (A/SC/18), A/WS/33, and A/WSN/33 H1-subtype strains by trypsin and matriptase. The percent HA cleavage was determined by densitometry of the Western blot shown in Fig. 1.
To determine whether matriptase cleavage produces a fusogenic HA, we conducted a cell-cell fusion assay on the A/California/04/09 (H1N1), A/South Carolina/1/18 (H1N1), A/Japan/305/57 (H2N2), and A/Aichi/2/68 (H3N2) HAs, and syncytium formation was assessed. A high degree of syncytium formation was observed with matriptase treatment of the H1 influenza virus A/California/04/09, with the efficiency being similar to that for the trypsin control (Fig. 3). A lower abundance of syncytia was observed for A/South Carolina/1/18 HA than for A/California/04/09 (Fig. 3), which corresponds to the cleavage efficiency observed by Western blot analysis of HA cleavage (Fig. 1). However, no syncytium formation was observed for either the H2 or H3 HA, which is consistent with the results of Western blot cleavage analysis (Fig. 3). These results, taken together with the results of Western blot analysis, further demonstrate the H1 subtype specificity of matriptase.

Cleavage by matriptase produces a fusogenic HA. Immunofluorescence staining of cells expressing A/California/04/09 (A/CA/09), A/South Carolina/1/18 (A/SC/18), A/Japan/305/57, and A/Aichi/2/68 after treatment with either trypsin, no protease treatment (mock), or matriptase. The HA from each subtype was labeled with anti-influenza HA antibody and Alexa Fluor 488 secondary antibody (green), and the cell nuclei were stained with Hoechst 33258 (blue).
As an additional means of determining whether matriptase cleaves HA in a functionally active manner, noncleaved A/PR/8/34 and A/WSN/33 viruses were produced by a single replication round in 293T cells, in order to determine whether matriptase cleaves HA in the virion. Only the H1-subtype viruses were assessed due to the lack of cleavage and syncytium formation described above for the H2 and H3 subtypes. Matriptase was incubated with the noncleaved virus and subsequently incubated with mammalian cells and assessed for infection by immunofluorescence. Successful infection was observed with the matriptase-treated A/PR/8/34 virus and was well above the background infection observed in the mock control and comparable to that of the trypsin control (Fig. 4). However, infection was not observed with the matriptase-treated A/WSN/33 virus (Fig. 4), which is most likely due to the limited HA cleavage by the protease (Fig. 1). Thus, matriptase has the ability to activate influenza virus, a process that is dependent on the HA cleavage efficiency of the protease.
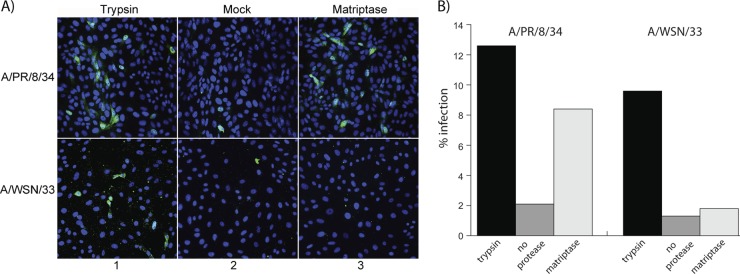
Cleavage activation of A/PR/8/34 and A/WSN/33 H1N1 influenza viruses by matriptase. (A) Immunofluorescence staining of the viral nucleoprotein of infected cells after treatment with trypsin, no protease treatment (mock), and matriptase. The viral nucleoprotein was stained with anti-influenza nucleoprotein antibody and Alexa Fluor 488 secondary antibody (green), and the cell nuclei were stained with Hoechst 33258 (blue). (B) Quantitation of the percent infection from panel A.
Cleavage of peptide mimics of the HA cleavage site by matriptase.
In order to give more biochemical insight into the cleavage patterns observed among the different HA strains and subtypes, fluorogenic peptides of the HA cleavage site region of A/California/04/09 (H1), A/WSN/33 (H2), A/Japan/305/57 (H2), and A/Aichi/2/68 (H3) were designed with the goal of determining whether the differences in the amino acid sequence among each subtype affect cleavage by matriptase. Matriptase efficiently cleaved the peptide mimic of the A/California/04/09 cleavage site (IPSIQSRGL), which represents the H1 consensus sequence (Fig. 5). However, a marked decrease in the rate of cleavage with the A/WSN/33 cleavage site peptide mimic (IPSIQYRGL) compared to that with the H1 consensus peptide was observed (Fig. 5). Matriptase cleavage of the A/Japan/305/57 (H2) peptide mimic (VPQIESRGL) also resulted in a low rate of cleavage compared to that for the H1 consensus (Fig. 5). In the case of the H3 HA cleavage site peptide mimic (VPEKQTRGL), efficient cleavage that was comparable to that for the H1 consensus peptide and well above the rate of cleavage of the A/WSN/33 and H2 peptide mimics was observed with matriptase treatment.
The amino acid residues neighboring the HA cleavage site Arg affect cleavage by matriptase. The cleavage rate of the fluorogenic peptides IPSIQSRGL (H1 consensus), IPSIQYRGL (A/WSN/33 cleavage site), VPQIESRGL (H2 consensus), and VPEKQTRGL (H3 consensus) after treatment with matriptase. The rate of cleavage was determined by monitoring the increase in fluorescence at 390 nm. RFU, relative fluorescence units.
Along with matriptase, we also assessed the cleavage rate of other known soluble HA-cleaving proteases to determine their relative enzymatic activities, using the H1 consensus cleavage site peptide mimic. Thrombin, urokinase, and plasmin all displayed relatively low activity toward the H1 peptide (Table 1). In the case of plasmin and thrombin, the low relative activity is most likely due to their known preference for particular residues directly N terminal to the cleavage site arginine (27, 31). In contrast, matriptase displayed a high activity toward the H1 peptide, suggesting that it is a highly efficient enzyme for HA cleavage (Table 1).
Table 1
Comparison of cleavage rates of various HA-cleaving proteases with matriptase
| Protease | Vmax (RFUa/min) | Vmax protease/Vmax matriptase |
|---|---|---|
| Matriptase | 834.42 ± 80.0 | |
| Plasmin | 90.34 ± 9.3 | 0.11 |
| Urokinase | 48.4 ± 7.3 | 0.06 |
| Thrombin | 15.26 ± 9.8 | 0.02 |
Indirect HA cleavage activation by matriptase.
Plasmin, urokinase, and plasma kallikrein are thrombolytic blood proteases that have all been found to cleave and activate various influenza virus subtypes (27). The majority of these proteases are circulating in their inactive form in blood serum but can localize to the respiratory tract when tissue damage is caused by influenza virus infection (27). To determine whether matriptase has the ability to cleave and activate each protease, matriptase was incubated with each zymogen and monitored for the conversion to the active form by the change in fluorescence upon cleavage of a fluorogenic peptide (GGGR-AMC). In all cases, matriptase efficiently activated the zymogens (Fig. 6). Prourokinase is a known substrate of matriptase, and our results are consistent with this (18). Thus, matriptase has the ability to both directly and indirectly activate the influenza virus HA.

Cleavage activation of thrombolytic zymogens by matriptase. Cleavage of the fluorogenic peptide, GGGR-AMC by plasmin (1,611.7 relative fluorescence units [RFU]/min) (A), urokinase (2,855.1 RFU/min) (B), and plasma kallikrein (3,012.3 RFU/min) (C) after treatment with matriptase. Each graph includes the change in fluorescence at 440 nm of matriptase alone (squares), the zymogen alone (circles), and matriptase incubated with the zymogen (diamonds).
DISCUSSION
The influenza virus HA is synthesized as an inactive precursor that must be cleaved by host proteases to enable fusion with the host endosome. While certain proteases have been identified, uncertainty surrounding which host proteases are responsible for HA cleavage in the human respiratory tract remains. The membrane-bound proteases TMPRSS2, TMPRSS4, and HAT are some of the more recent examples of respiratory tract-localized proteases that have been shown to be capable of cleaving HA in vitro or in cell culture models (3, 5–7). To date, the extracellular proteases known to cleave HA, such as plasmin, urokinase, and mast cell tryptase, generally rely on in vitro experiments (8, 27).
Here we identify matriptase as a human respiratory tract-expressed protease involved in HA cleavage activation. Matriptase has characteristics that are common to each of the HA-cleaving proteases currently identified, being a trypsin-like serine protease that is specific for cleavage after an arginine residue and is highly expressed in the respiratory tract. An interesting feature of matriptase is that the protease is found both on the plasma membrane and in the extracellular space of the respiratory tract. In the case of TMPRSS2 and HAT, however, the majority of the protein was found to exclusively reside on the plasma membrane in vitro, and furthermore, the TMPRSS2 and HAT that were secreted were shown to be unable to cleave HA (6). We have determined that secreted matriptase is present in the nasal wash of human subjects; however, it is difficult to compare the abundance of this secreted protease to that of the other proteases relevant for influenza virus cleavage, which are transmembrane proteases and so are not present in a nasal wash. Although it is active for only a short time in vivo (9), matriptase does display high catalytic activity compared to other proteases examined by our laboratory (Table 1). However, determining if the matriptase identified in the nasal wash samples is in an active form poses a significant challenge (29).
It has been a long-standing belief that influenza virus activation is mediated in a paracrine fashion by a secreted, extracellular protease. However, the identification of such a secreted protease in the human respiratory tract has proved elusive. Instead, attention has recently focused on transmembrane proteases known to be expressed in the human respiratory tract (i.e., TMPRSS2, TMPRSS4, and HAT). Here, we provide the first identification of a secreted protease normally present in the human respiratory tract that can cleave and activate influenza virus. Overall, it is likely that both secreted and transmembrane proteases have the ability to activate influenza virus in humans and must be considered candidates for therapeutic development. Within this spectrum of proteases, it should also be considered which proteases are most important in a given situation, such as the localization within the respiratory tract, between individuals, or with individuals with underlying conditions that might affect protease expression, activity, or distribution.
One notable feature of our studies is that matriptase cleavage was limited to the H1 influenza virus subtype, whereas no cleavage was observed with the H2 and H3 subtypes. This finding is distinct from that for other proteases identified in humans, where it is assumed that all human-adapted subtypes are cleaved in an equivalent manner. Our finding that cleavage by matriptase showed a preference for particular strains within the H1 subtype raises the question of whether each human-adapted virus has evolved to be activated by a particular set of host proteases that is distinct from the set used to activate the other subtypes and, furthermore, whether each strain from a given subtype has a preference for particular proteases within that set. One virus of particular interest in this regard is A/WSN/33, which is unusual among H1 viruses in having a bulky aromatic residue (tyrosine) directly N terminal to the cleavage site arginine in the P-2 cleavage site position in place of the highly conserved serine. This change at the P-2 position (S-Y) appears to not be suitable for binding to the matriptase active site and could offer an explanation for the limited cleavage observed for matriptase treatment of the A/WSN/33 HA. Recent data from our laboratory have demonstrated that cleavage of the A/WSN/33 HA by plasmin was enhanced by the P-2 tyrosine substitution, supporting that idea that residues flanking the cleavage site P-1 arginine in the HA cleavage site region can affect proteolytic cleavage (31). In agreement with HA cleavage data, matriptase efficiently cleaved the peptide mimic of the H1 cleavage site consensus, and a marked decrease in the rate of cleavage was observed with the A/WSN/33 cleavage site peptide mimic compared to that for the H1 consensus peptide (Fig. 5). However, matriptase cleavage was also low for other H1 strains that retain the consensus P-2 serine residue, i.e., A/SC/18 and A/WS/33, and so features of the HA in addition to the P-2 residue cleavage site flanking region must also be involved. Matriptase cleavage of the H2 peptide mimic also resulted in an expected low rate of cleavage compared to that for the H1 consensus (Fig. 5). However, in the case of the H3 HA cleavage site peptide mimic, with matriptase treatment, efficient cleavage that was comparable to that for the H1 consensus peptide was observed. This result is intriguing, since virtually no cleavage was observed by Western blot analysis of each H3 HA examined. In this case, other factors, such as HA glycosylation or secondary structure, must also contribute to the inability of matriptase to cleave the H3 subtype. Overall, the features that allow matriptase cleavage activation of selected H1 viruses remain to be explored.
To conclude, with uncertainty over the host proteases involved in HA cleavage in vivo still remaining, it is important to investigate the diversity of host proteases for their ability to cleave HA, in attempts to determine the critical host protease(s) involved in influenza virus activation in the respiratory tract. Here we show that matriptase cleaved and activated HA from human-adapted, influenza virus subtypes. Importantly, our studies reveal both a subtype and strain specificity for cleavage and fusion activation. Our studies highlight the role of matriptase as a secreted, extracellular protease important for influenza virus infection, alongside transmembrane proteases such as TMPRSS2 and HAT. To our knowledge, matriptase (ST14) is the first example of a member within the matriptase TTSP subfamily having the ability to cleave HA. With renewed effort to develop alternative influenza therapeutics, an improved understanding of the spectrum of proteases critical for activation of the virus in vivo will be essential for the development of effective, protease-based therapeutics. Our finding that matriptase may have both direct and indirect effects via activation of other serum and tissue proteases argues in favor of the development of broad-spectrum protease inhibitors.
ACKNOWLEDGMENTS
We thank Theresa Fitzgerald for providing nasal wash samples, Nadia Chapman for technical assistance, Jean Millet for critical reading of the manuscript, and all members of the G. R. Whittaker lab for helpful discussions.
These studies were funded by United States Department of Health and Human Services contract HHSN266200700008C (NIAID Centers of Excellence for Influenza Research and Surveillance). Work in the GRW laboratory is also supported by a research grant from the National Institutes of Health (R01 AI48678).
REFERENCES
Articles from Journal of Virology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jvi.00306-12
Read article for free, from open access legal sources, via Unpaywall:
https://jvi.asm.org/content/jvi/86/19/10579.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/jvi.00306-12
Article citations
Identification of Embryonic Chicken Proteases Activating Newcastle Disease Virus and Their Roles in the Pathogenicity of Virus Used as In Ovo Vaccine.
J Virol, 97(5):e0032423, 12 Apr 2023
Cited by: 0 articles | PMID: 37042750 | PMCID: PMC10231145
Aprotinin-Drug against Respiratory Diseases.
Int J Mol Sci, 24(13):11173, 06 Jul 2023
Cited by: 3 articles | PMID: 37446350 | PMCID: PMC10342444
Review Free full text in Europe PMC
Advances in deciphering the interactions between viral proteins of influenza A virus and host cellular proteins.
Cell Insight, 2(2):100079, 31 Jan 2023
Cited by: 8 articles | PMID: 37193064 | PMCID: PMC10134199
Review Free full text in Europe PMC
In Vitro Pharmacokinetic Behavior of Antiviral 3-Amidinophenylalanine Derivatives in Rat, Dog and Monkey Hepatocytes.
Biomedicines, 11(3):682, 23 Feb 2023
Cited by: 1 article | PMID: 36979660 | PMCID: PMC10045298
Hemagglutinin Subtype Specificity and Mechanisms of Highly Pathogenic Avian Influenza Virus Genesis.
Viruses, 14(7):1566, 19 Jul 2022
Cited by: 19 articles | PMID: 35891546 | PMCID: PMC9321182
Review Free full text in Europe PMC
Go to all (50) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Cleavage activation of human-adapted influenza virus subtypes by kallikrein-related peptidases 5 and 12.
J Biol Chem, 288(24):17399-17407, 23 Apr 2013
Cited by: 32 articles | PMID: 23612974 | PMCID: PMC3682540
Human matriptase/ST 14 proteolytically cleaves H7N9 hemagglutinin and facilitates the activation of influenza A/Shanghai/2/2013 virus in cell culture.
Influenza Other Respir Viruses, 14(2):189-195, 09 Dec 2019
Cited by: 6 articles | PMID: 31820577 | PMCID: PMC7040964
Hemagglutinins of Avian Influenza Viruses Are Proteolytically Activated by TMPRSS2 in Human and Murine Airway Cells.
J Virol, 95(20):e0090621, 28 Jul 2021
Cited by: 15 articles | PMID: 34319155 | PMCID: PMC8475512
Aprotinin and similar protease inhibitors as drugs against influenza.
Antiviral Res, 92(1):27-36, 23 Jul 2011
Cited by: 72 articles | PMID: 21802447
Review
Funding
Funders who supported this work.
NIAID NIH HHS (3)
Grant ID: R01 AI048678
Grant ID: HHSN266200700008C
Grant ID: R01 AI48678






