Abstract
Free full text

Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish
Associated Data
Abstract
Prior studies with transgenic zebrafish confirmed the functionality of the transcription factor Gal4 to drive expression of other genes under the regulation of upstream activator sequences (UAS). However, widespread application of this powerful binary system has been limited, in part, by relatively inefficient techniques for establishing transgenic zebrafish and by the inadequacy of Gal4 to effect high levels of expression from UAS-regulated genes. We have used the Tol2 transposition system to distribute a self-reporting gene/enhancer trap vector efficiently throughout the zebrafish genome. The vector uses the potent, hybrid transcription factor Gal4-VP16 to activate expression from a UAS:eGFP reporter cassette. In a pilot screen, stable transgenic lines were established that express eGFP in reproducible patterns encompassing a wide variety of tissues, including the brain, spinal cord, retina, notochord, cranial skeleton and muscle, and can transactivate other UAS-regulated genes. We demonstrate the utility of this approach to track Gal4-VP16 expressing migratory cells in UAS:Kaede transgenic fish, and to induce tissue-specific cell death using a bacterial nitroreductase gene under UAS control. The Tol2-mediated gene/enhancer trapping system together with UAS transgenic lines provide valuable tools for regulated gene expression and for targeted labeling and ablation of specific cell types and tissues during early zebrafish development.
Introduction
The ability to regulate gene expression along defined temporal and spatial coordinates represents an invaluable tool for investigating gene function in the developing embryo. In Drosophila, spatially restricted transgene expression is frequently accomplished using a bipartite Gal4/UAS system (Duffy, 2002; Fischer et al., 1988). This system relies upon tissue-specific expression of the yeast Gal4 transcriptional activator to drive expression of transgenes placed under the regulation of multimerized Gal4-responsive upstream activator sequences (UAS). Recent refinements have added the capacity for temporal regulation, using either Gal4-steroid receptor fusion proteins or a temperature-sensitive Gal4 antagonist (McGuire et al., 2004). Reflecting the tremendous utility of “Gal4 driver” lines, several large-scale screens have exploited P-elements to distribute Gal4 insertions throughout the Drosophila genome, in the form of gene- or enhancer-traps (Brand and Perrimon, 1993; Lukacsovich et al., 2001). Detailed descriptions of 6,966 separate Gal4 insertions are currently maintained in the Drosophila Gal4 Enhancer Trap Insertion Database, including information on insertion sites, expression patterns and mutant phenotypes (http://flymap.lab.nig.ac.jp/~dclust/getdb.html html). In addition to enabling a wide variety of reverse genetic applications, Gal4 driver lines have also been utilized for genome-wide misexpression screens, in which randomly inserted UAS elements are used to activate endogenous gene expression in an appropriate Gal4 background (Rorth et al., 1998).
In zebrafish, application of the Gal4/UAS system for transgene expression under the control of identified promoters was pioneered by Scheer and Campos-Ortega (Scheer and Campos-Ortega, 1999). Although they were able to show Gal4-mediated activation of 5xUAS reporters in stable transgenics, levels of reporter expression were low. Addressing this concern, Koster and Fraser (Koster and Fraser, 2001) employed the Gal4-VP16 fusion protein comprised of the Gal4 DNA binding domain fused to the transcriptional activator domain of the herpes simplex virus VP16 protein (Sadowski et al., 1988). The Gal4-VP16 fusion protein generated robust expression from a 14xUAS:eGFP reporter in transient activation assays in zebrafish embryos, although recovery of stable transgenic lines was not reported.
In spite of these encouraging early efforts, the use of Gal4/UAS technology has remained relatively limited, exemplified by the fact that only four transgenic Gal4 driver lines are currently registered by the Zebrafish Information Network (www.zfin.org). The paucity of driver lines may reflect the relative inefficiency of conventional methods for zebrafish transgenesis. However, the development of more efficient approaches, utilizing either retroviral vectors (Amsterdam et al., 2004; Ellingsen et al., 2005) or transposable elements (Davidson et al., 2003; Kawakami et al., 1998; Koga and Hori, 2001) facilitates widespread application of Gal4/UAS methodologies in the zebrafish.
The Tol2 transposable element from the Japanese medaka fish, Oryzias latipes, has emerged as a particularly simple and effective system to generate a high rate of stable transgenesis in zebrafish (Kawakami, 2004). Tol2 is a member of the hAT (hobo, Ac, Tam3) family of terminal inverted repeat-type transposable elements (Kempken and Windhofer, 2001), a group of autonomous transposons with internal sequences encoding the transposase protein necessary for mobilization. Derivatives of the Tol2 element lacking the transposase coding sequence can be effectively mobilized by provision of exogenous transposase activity. Transposase-deficient Tol2 vectors have been developed and in combination with transposase mRNA provide highly efficient delivery of transgenes to the zebrafish genome (Kawakami et al., 2000; Urasaki et al., 2006). Rates of germline transmission as high as 30–50% permit Tol2 vectors to be utilized for genome-wide distribution of gene- and/or enhancer-trap elements (Kawakami et al., 2004; Parinov et al., 2004).
As a means to expand available Gal4/UAS capabilities in zebrafish, we have used the Tol2 element to distribute a self-reporting Gal4-VP16;UAS:eGFP gene/enhancer trap construct throughout the zebrafish genome, and conducted a pilot screen for lines displaying Gal4-VP16 dependent activation of the UAS:eGFP reporter. This approach has allowed the recovery of 15 stable transgenic lines displaying distinct temporally and spatially restricted patterns of eGFP expression in a wide variety of cell and tissue types. Most lines are capable of transactivation, driving tissue-specific exogenous genes via UAS regulation. Here, we demonstrate transactivation in UAS:Kaede transgenic fish by photoconversion of Gal4-VP16 expressing cells, a labeling method used to trace migratory cells during development. Transactivation in UAS:nfsB-mCherry transgenic fish leads to production of a fusion protein consisting of nitroreductase (NTR) (Drabek et al., 1997; Pisharath, 2007) and the fluorescent protein mCherry, enabling Gal4-VP16 expressing cells to be monitored and selectively ablated.
Together these results extend the number of currently available zebrafish Gal4-VP16 driver lines, and further support the feasibility of conducting genome-wide screens to produce large panels of zebrafish Gal4 driver lines, analogous to those available in Drosophila.
Materials and Methods
Gene/Enhancer trap vector design
The SAGVG (splice acceptor-Gal4-VP16;UAS:eGFP) vector is a Tol2 transposon-based, bipartite construct consisting of a Gal4-VP16 gene/enhancer trap as well as a cis-linked UAS-regulated eGFP reporter (Fig. 1A). The vector contains a splice acceptor sequence derived from the rabbit β-globin gene (275 base pairs of intron 2 and 45 base pairs of exon 3) which was amplified from the pT2KXIGΔIN vector (Urasaki et al., 2006) and cloned upstream of tandem Gal4-VP16 and UAS:eGFP elements (Koster and Fraser, 2001). The entire unit was subcloned into the pT2KXIGΔIN vector to be flanked by a 535 base pair (bp) right Tol2 arm and a 517 bp left Tol2 arm.
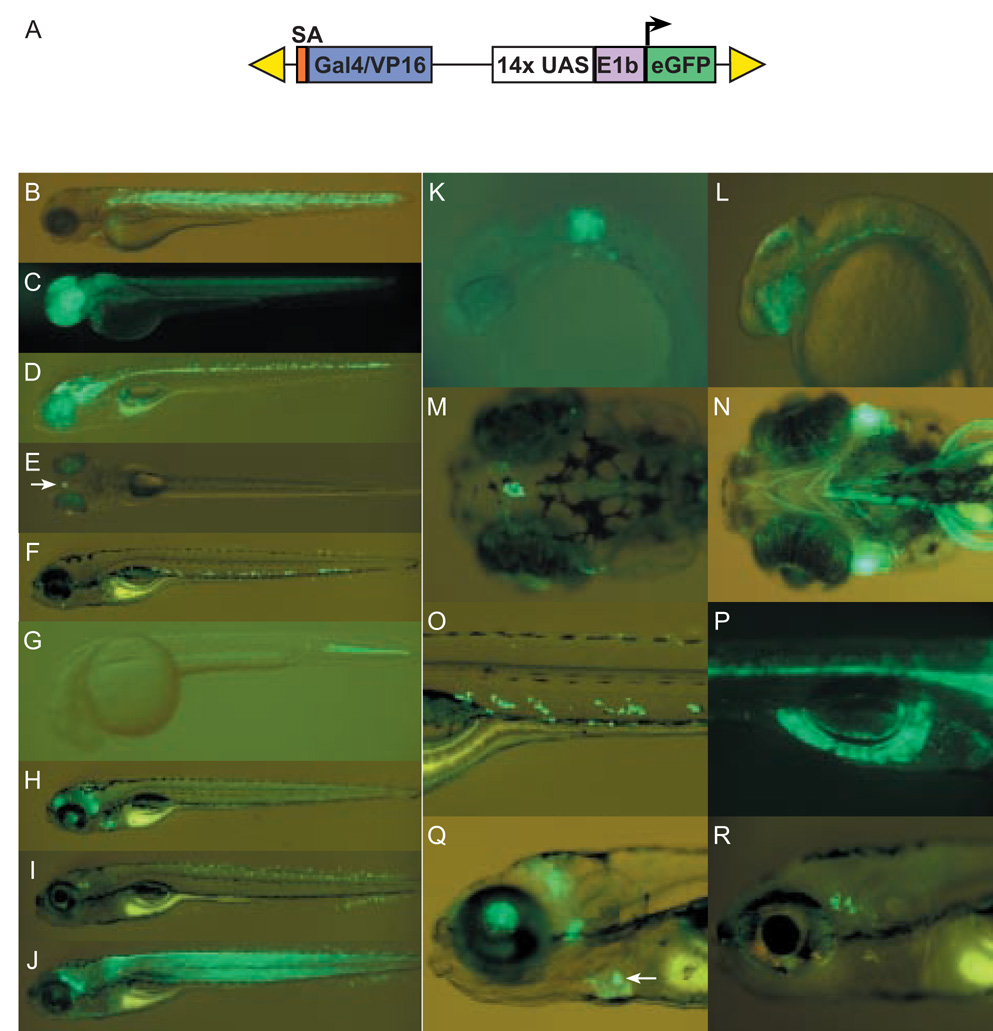
(A) Schematic depiction of SAGVG vector, containing sequence encoding Gal4-VP16 fusion protein, preceded by splice acceptor (orange), and followed by Gal4-VP16 responsive eGFP reporter cassette consisting of Gal4-responsive upstream activating sequence (UAS), adenoviral E1b minimal promoter element (E1b). The entire construct is flanked by right and left arms of Tol2 (yellow triangles). (B) Line c218 with variable expression in somitic muscle fibers at 3 dpf. (C) Line c220 with expression in forebrain and retina at 2 dpf. (D) Line c223 with expression in midbrain, hindbrain and ventral spinal cord at 5 dpf. (E) Line c229 with expression in the pineal (arrow) and early lens expression at 5 dpf. (F) Line c233 with expression in ventral cells dorsal to cardinal vein at 5 dpf. (G) Line c247 with expression in the caudal notochord at 1.5 dpf. (H) Line c236 with expression in subregions of the brain and in the heart at 6 dpf. (I) Line c237 with expression in putative sensory neurons anterior to the otocyst, subsets of spinal cord interneurons, and the ventral tail fin at 6 dpf. (J) Line c240 with expression throughout the skeletal and head musculature at 5 dpf. (K) Line c228 with expression in hindbrain rhombomeres 5 and 6 at 1.5 dpf. (L) Line c223 with expression in retina, dorsal midbrain and hindbrain floor plate at 1.5 dpf. (M) Higher magnification of line c229 demonstrating selective labeling of the pineal at 5 dpf (earlier expression, not shown). (N) Labeling of cranial musculature in c240 at 5 dpf. (O) Higher magnification image of c233 highlighting ventral cells shown in F. (P) c223 also exhibits labeling of the exocrine pancreas at 5 dpf (earlier expression, not shown). (Q) Higher magnification of the heart labeling in line c236 at 6 dpf. High levels of eGFP are observed in heart valves (arrow and refer to supplemental movie). (R) Higher magnification image of putative sensory neurons in line c237. (B–J) lateral views of whole larvae. (K, L, O, Q, and R) lateral views with anterior to the left, (P) is a lateral view with 1 anterior to the right. (M) dorsal and (N) ventral view. All larvae were imaged on a Leica MZFlII stereomicroscope.
Generation of Tg(Gal4-VP16;UAS:eGFP) lines
One cell stage embryos were injected through the chorion with 1 nl of a solution containing plasmid DNA at a concentration of 12.5 ng/µl, along with Tol2 transposase RNA at 12.5 ng/µl, KCl at 200 mM and phenol red at 0.05% (Kawakami et al., 2004). Injected F0 embryos were raised and the resulting fish interbred to produce F1 progeny. F1 embryos were examined on a fluorescent stereomicrosope at 2 and 5 days post fertilization (dpf). Embryos and larvae exhibiting tissue-restricted eGFP fluorescence were selected and raised. Among 49 pairs of F0 fish, 28 intercrosses generated F1 embryos with tissue-restricted eGFP fluorescence. F1 adults were then outcrossed to wild-type AB fish, and F2 progeny were re-screened. Fifteen stable lines displaying spatially and temporally restricted patterns of eGFP expression were recovered in the F3 generation. Transgenic fish lines are freely available upon request to ude.bmewic@nreplah and will be deposited in ZIRC (http://zfin.org/zirc/home/guide.php). Plasmid requests should be directed to ude.imhj@3nosrapm.
Generation of the Tg(UAS:Kaede)s1999t transgenic line
The Kaede open reading frame (Ando et al., 2002), was kindly provided by Atsushi Miyawaki. Flanking sequences were amplified by PCR and inserted downstream of UAS and the E1b promoter in a 14xUAS:eGFP construct (Koster and Fraser, 2001) using the Xi Clone PCR Cloning System (Gene Therapy Systems, San Diego, CA). I-SceI meganuclease recognition sites were inserted flanking the UAS:Kaede construct to increase rate of transgenesis and ensure single insertions (Thermes et al., 2002). Wild-type TL embryos were injected at the 1–2 cell stage with a solution of 50 ng/µl UAS:Kaede DNA and 5 U/µl I-SceI endonuclease in 0.04% Phenol Red. Injected F0 animals were raised to adulthood and interbred. F1 embryos were pooled and screened by PCR for the presence of the UAS:Kaede transgene. F0 founder animals giving rise to UAS:Kaede-positive offspring were then mated to existing GAL4 driver lines to identify expressors. All transgenic F1 gave the identical expression pattern, regardless of their founder parent. The stable Tg(UAS:Kaede)s1999t line was then propagated by outcrossing a single F0 founder male to wild-type TL fish.
Cloning of insertion alleles linked to eGFP expression
Linker mediated PCR (LM-PCR) was utilized to clone genomic sequences flanking vector insertion sites. Adult F1 or F2 fish were crossed to generate transgenic embryos which were sorted into eGFP-positive and eGFP-negative groups. Genomic DNA was purified from pooled or individual embryos according to previously published protocols (Parsons et al., 2002; Wang et al., 1995). LM-PCR was performed according the protocol described by Dupuy and colleagues (Dupuy et al., 2005), with the following adaptations. Genomic DNA was digested with either NlaIII, BfaI or DpnII. Modified oligonuclotides were annealed to generate linkers and ligated to digested DNA. NlaIII and BfaI Linker oligos were identical to those described by Dupuy; DpnII Linker oligos were as follows: 5’GTAATACGACTCACTATAGGGCTCCGCTTAAGGGAC3’ and (5’ PO4)-GATCGTCCCTTAAGCGGAG-(3’ C3 spacer).
The first round of the nested PCR was performed using linker primer 1 5’GTAATACGACTCACTATAGGGC with either Tol2 Left 1.1 5’TCAAGTAAAGTAAAAATCCCCAAAA3’ or Tol2 Right 1.3 5’TTGAGTAAAATTTTCCCTAAGTAC3’, under the following cycling conditions: 94 deg C (15 sec) – 51 deg C (30 sec) – 68 deg C (1 min), 25–30 cycles. Second round nested PCR was then performed using linker primer 2 5’AGGGCTCCGCTTAAGGGAC3’ with either Tol2 Left 2.1 5’AAAATCCCCAAAAATAATACTTAAGTACAGTAA3’ or Tol2 Right 2.1 5’TGTACTTTCACTTGAGTAAAATTTTTGAG3’ and the following cycling conditions: 94 deg C (15 sec)–57.5 deg C (30 sec) – 68 deg C (1 min), 25–30 cycles. The PCR products were resolved by electrophoresis on a 3% agarose gel. Products selectively amplified in eGFP-positive embryos were cloned and sequenced. Sequences flanking the Tol2 arms were used to search the Ensembl Danio rerio genomic sequence database to position and orient the insert within the zebrafish genome. To confirm the presence of a cloned insertion correlated with eGFP reporter expression, PCR primers were designed based on flanking sequence (see supplementary Table S1).
Gal4-VP16-dependent activation of additional UAS-regulated transgenes
To assess whether stable transgenic lines are capable of activating a second UAS-regulated gene in trans, a UAS:mCherry plasmid construct was synthesized by cloning an mCherry cDNA obtained from R. Tsien (Shaner et al., 2004), downstream from the same 14xUAS, E1b minimal promoter sequence used in the SAGVG vector. Gal4-VP16;UAS:eGFP carriers were crossed to AB fish and resultant embryos were injected with non-linearized UAS:mCherry plasmid DNA at a concentration of 150 ng/µl and examined for transient red fluorescence 24–48 hours later. To confirm that candidate Gal4-VP16 driver lines can activate stably integrated UAS-regulated transgenes, F2 adults were mated with UAS:Kaede transgenic fish. Transgenic embryos were identified on the basis of their tissue-restricted eGFP expression and subjected to green-to-red photoconversion by timed exposure (from 15 to 60 sec) to UV light using a DAPI filter (excitation 365 nM, emission 415 nM) under 10X or 20X objectives on a Zeiss Axioskop microscope.
Enhanced throughput filters (Chroma Technology Corp.) were used to visualize eGFP/Kaede (ET-GFP #49002) or photoconverted Kaede (ET-Cy3 #49004) and produced no overlap in fluorescence. To track the fate of migratory cells in c233, only the most caudal cells of 6 dpf larvae were photoconverted. Control and photoconverted transgenic larvae were allowed to develop in the dark for 2 days and then imaged again at 8 dpf.
Prodrug-dependent cell ablation in Tg(Gal4-VP16;UAS:eGFP) lines
The E.coli gene nfsB, which encodes nitroreductase B, was cloned in frame to mCherry (Pisharath, 2007). The nfsB-mCherry cassette was cloned downstream of the 14xUAS, E1b minimal promoter sequence. The UAS:nfsB-mCherry construct was cloned into the backbone of the T2KXIGΔIN vector to flank the transgene with Tol2 arms. Transgenic fish were generated by Tol2 mediated transgenesis and identified F0 Tg(UAS:nfsB-mCherry) carriers were crossed to c223 and c230 adults. Incubation of compound heterozygous embryos (expressing both mCherry and eGFP) in 10mM of the prodrug metronidazole (Met.; M3761, Sigma®) was carried out for 24–48 hours. Controls included c223 and c230 heterozygous siblings (eGFP positive, mCherry negative) and untreated compound heterozygotes. Individual larvae were examined by fluorescent microscopy before and after prodrug exposure. c223 larvae were fixed in 4% PFA overnight at 4°C, and were sectioned (500 µm, Vibrotome®). Following Hoechst nuclear staining, the pattern of mCherry fluorescence was assessed with confocal microscopy. Widefield and confocal sections were acquired using an Axiovert 200M microscope coupled to the Zeiss LSM 5 Pascal system.
Results
Generation of F1 founders displaying unique eGFP expression patterns
We took a gene trap strategy to generate self reporting Gal4-VP16 driver lines. The SAGVG construct is a Tol2-based vector, designed to express Gal4-VP16 under the control of endogenous regulatory elements in the zebrafish genome (Fig. 1A). The SAGVG construct was injected along with RNA encoding transposase, and the resulting F0 fish were assayed for transmission of eGFP expression patterns in their progeny. Over half of the 49 incrossed F0 pairs (28/49) we examined produced F1 Tg(Gal4-VP16;UAS:eGFP) progeny with restricted eGFP expression. Ultimately, 15 stable lines were generated on the basis of their discrete and reproducible patterns of eGFP expression. Information on these lines and their allele designations is summarized in Table 1. Examples of tissue-specific patterns of expression are depicted in Fig. 1. At the level of a fluorescent stereomicroscope, the recovered lines exhibited robust eGFP fluorescence in a variety of tissues, including somitic muscle, notochord, pancreas, pineal, subregions of the brain, the floor plate, spinal cord neurons, cranial musculature, retina, lens, heart and heart valve (Fig. 1 and supplemental movie 1). In some lines eGFP was detected in only a small subset of cells, such as in the presumptive pineal organ of c229 larvae (Fig. 1E, 1M), putative cranial ganglia (c215, c241; see supplemental Fig. S1) and a previously uncharacterized population of ventral cells (c233; Fig. 1O).
Table 1
Recovered lines and mapping of Gal4-VP16;UAS:eGFP inserts
| Line | Expression Pattern | Transactivation | Linkage Group | Insertion Position |
|---|---|---|---|---|
| c215 | cranial cluster, ear | cranial cluster, ear | Undetermined | N/A |
| c218 | skeletal muscle | skeletal muscle | Zv6, 6:19.451 Mb | Exon of a predicted novel gene with Kelch-BTB/POZ domains; same transcriptional orientation * |
| c220 | retina, forebrain hatching gland | retina, forebrain | Zv6, 23:19.985 Mb | Intron 1 of ikkγ * |
| c223 | CNS, floor plate, pancreas | CNS, floor plate | Zv6, 5:39.948 Mb | A) Intron of a predicted gene; opposite transcriptional orientation |
| Zv6, 5:52.965 Mb | B) Intron of a predicted gene; opposite transcriptional orientation | |||
| Zv6, 5:38.323 Mb | C) Intron of zgc:103557, homologous to CDC42 effector protein 2; same transcriptional orientation * | |||
| c228 | hindbrain (r5, r6) | None | Undetermined | N/A |
| c229 | pineal, lens | pineal, lens | Zv6, 9:10.198 Mb | 0.7 kb 5’ to sequence homologous to FEV and 18 kb 3' to betaA2-2-crystallin * |
| c230 | retina, midbrain, muscle, notochord, hatching gland | muscle, notochord, hatching gland | undetermined | N/A |
| c233 | Uncharacterized ventral cells in trunk | uncharacterized ventral cells in trunk | Undetermined | N/A |
| c235 | forebrain stripe | Some forebrain neurons | Undetermined | N/A |
| c236 | heart, heart valve | None | Undetermined | N/A |
| c237 | cranial ganglia, spinal cord, retina | Cranial ganglia, spinal cord, retina | Zv6, 14:31.580 Mb | 1.8 kb 5' of ngn1 * |
| c239 | yolk | yolk | Zv6, 21:48.028 Mb | 1.5 kb 3' to sara2 * |
| c240 | somitic muscle, head musculature | somitic muscle, head musculature | Undetermined | N/A |
| c241 | cranial cluster | cranial cluster | Undetermined | N/A |
| c247 | Notochord | Notochord | Undetermined | N/A |
Transactivation of UAS-regulated genes by Tg(Gal4-VP16;UAS:eGFP)
The primary goal of this study was to develop methods to generate a diverse collection of zebrafish Gal4-VP16 driver lines for transcriptional activation of target genes of interest. To validate whether reporter eGFP expression is Gal4-VP16 dependent, we assessed transactivation using several UAS-regulated elements, either transiently following injection of plasmid DNA into Tg(Gal4-VP16;UAS:eGFP) embryos or in a stable manner by mating transgenic fish to established UAS lines. Two established driver lines were tested for their ability to transactivate a UAS:mCherry red fluorescent reporter following injection of reporter plasmid DNA into one-cell stage embryos. As demonstrated in Fig. 2, injection of UAS:mCherry plasmid into c218 and c223 embryos resulted mCherry expression in a few cells confined within the domain of eGFP expressing cells. This pattern of expression is produced because of the mosaic inheritance of the plasmid in the cells of the embryo following injection, and the requirement for Gal4-VP16 transactivation for mCherry labeling. For example, in c218 larvae, mCherry fluorescence was only observed within eGFP-positive muscle fibers. The transient assays indicate that Gal4-VP16 insertions can function in trans as tissue-specific transcriptional activators.
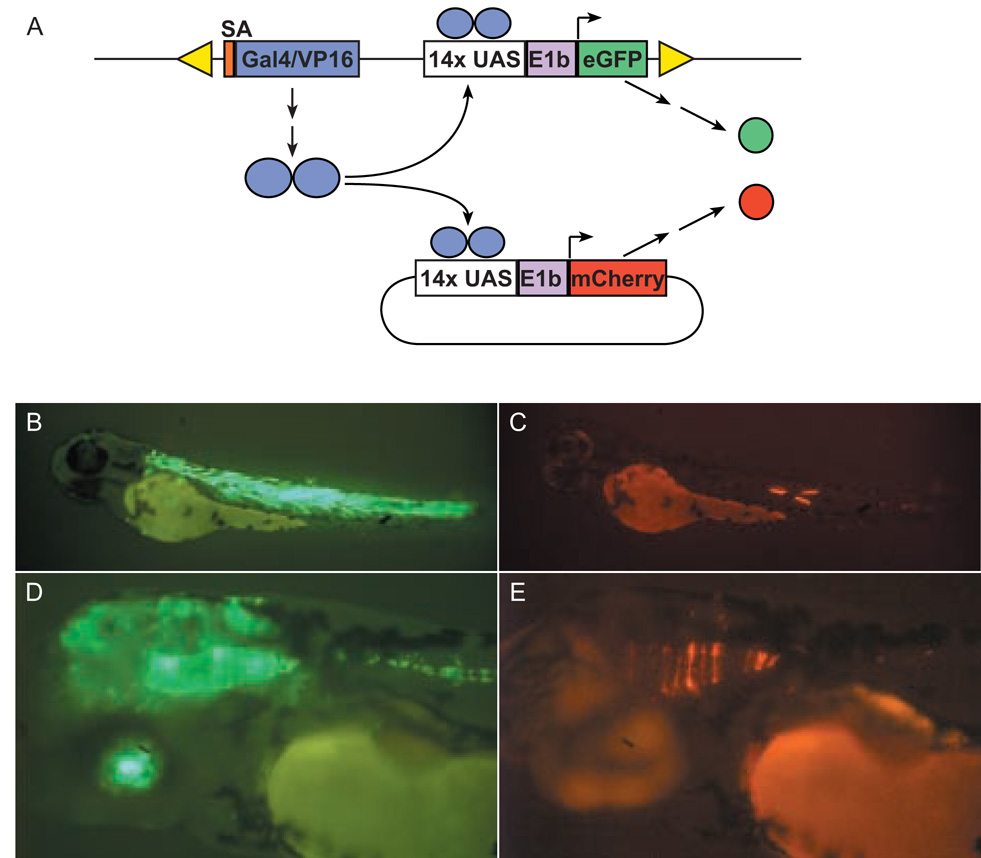
(A) Schematic of transient transactivation experiment. Tg(Gal4-VP16;UAS:eGFP) embryos were injected with non-linearised UAS:mCherry plasmid and then screened for transient expression of mCherry. Because of the mosaic nature of plasmid transmission, activation of UAS:mCherry is observed in a mosaic tissue-specific pattern, entirely restricted to the field of eGFP expression. (B) eGFP expression in skeletal muscle fibers of c218 embryo at 72 hpf. (C) Transient expression of mCherry is confined to eGFP-positive muscle fibers. (D) eGFP expression in retina, midbrain, hindbrain and floorplate of c223 embryo. (E) Transient expression of mCherry in eGFP-positive neuroepithilum.
The ability to transactivate expression in stable UAS lines was evaluated by breeding Tg(Gal4-VP16;UAS:eGFP) fish to Tg(UAS:Kaede) fish. In the Tg(UAS:Kaede) line, a gene encoding the photoconvertible coral fluorescent protein Kaede (Ando et al., 2002), has been placed under the regulation of concatamerized UAS elements. When exposed to UV light, Kaede is photoconverted and fluorescence irreversibly changes from green to red, distinguishing it from cis-activated eGFP. Thirteen out of fifteen putative driver lines mated with Tg(UAS:Kaede), demonstrated robust transactivation (Fig. 3). The two exceptions were c228 and c236, which failed to show transactivation in the hindbrain or heart valve respectively (data not shown), despite highly localized eGFP labeling.
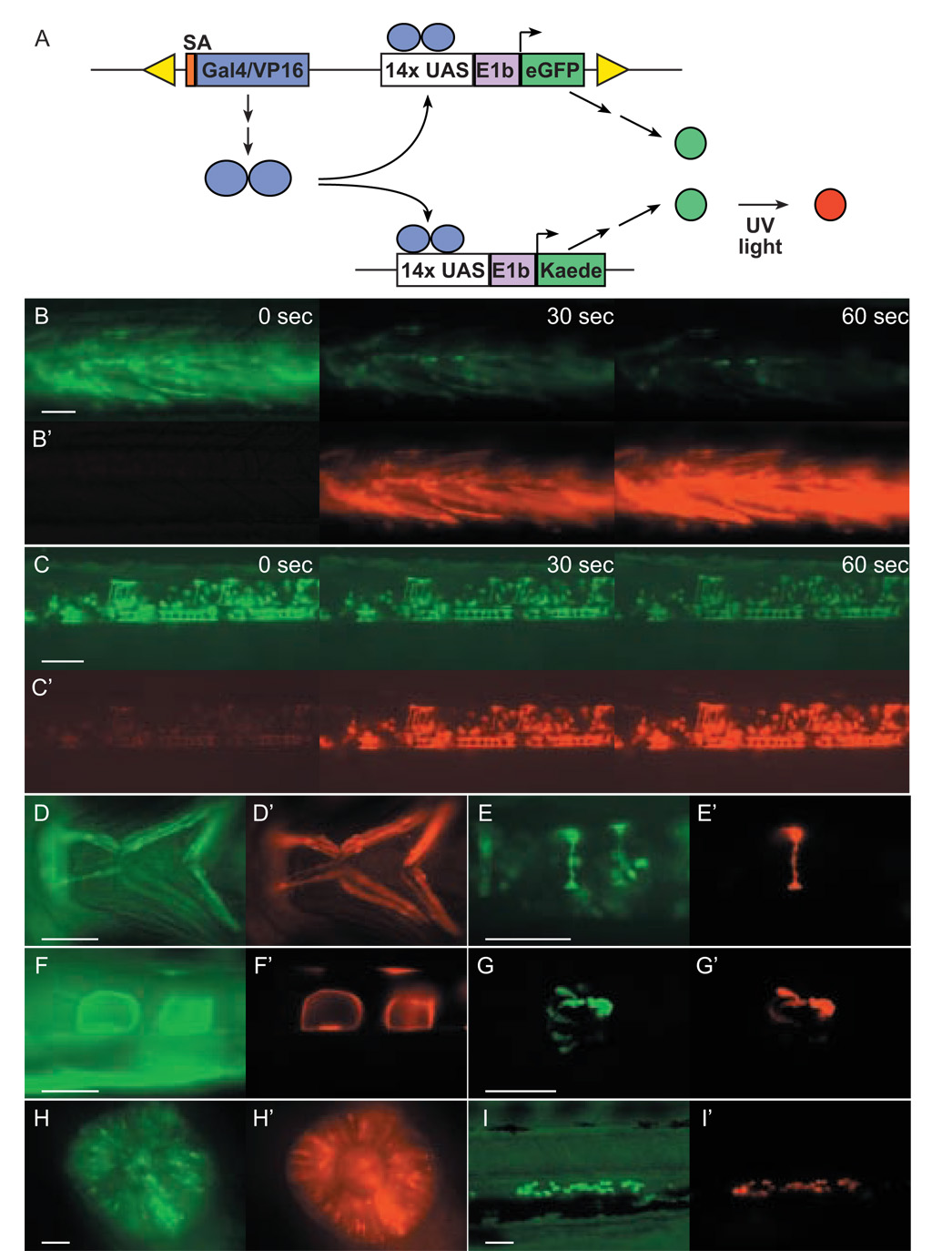
(A) Schematic of UAS:Kaede transactivation experiment. Gal4-VP16 expression from a chromosomal vector insertion site results in vector derived eGFP expression as well as transactivation of Kaede expression. Kaede expression is detected by exposure of embryos to UV light and observing the emergence of red fluorescence in cells in which Gal4-VP16 is expressed. In these experiments Tg(Gal4-VP16;UAS:eGFP) F2 or F3 fish were mated with a stable UAS:Kaede transgenic line. Resultant larvae were imaged for green fluorescent cells potentially resulting from expression of both eGFP and Kaede prior to UV exposure (shown in images B–I) and following exposure to UV light (365 nm) for durations ranging from 30 to 60 sec (shown in images B’–I’). Shown are photoconverted muscle fibers in c218 (B,B’), neurons and floor plate cells in c223 (C,C’), jaw musculature in c240 (D,D’), interneurons in c237 (E,E’), notochord cells in c247 (F,F’), pinealocytes in c229 (G,G’), retinal cells in c220 (H,H’), and ventral migratory cells in c233 (I,I’). All are lateral views except D (ventral) and G (coronal). Images captured before and after UV exposure were digitally processed in an identical manner. Scale bars=50 µm.
Besides demonstrating transactivation in stable transgenic lines, Tg(UAS:Kaede) fish can also be used in cell tracking (Hatta et al., 2006). By photoconverting Kaede in a discrete number of Gal4-VP16 expressing cells in the caudal tail of c233 larvae (6 dpf), we found that many migrated rostrally to reside adjacent to the liver two days later (Fig. 4). Consistent with this finding, fluorescence from eGFP labeling was only detected within a cellular lining on the surface of the liver akin to mesothelial cells in c233 adults (Fig. 4F). The result of cell tracking using Tg(UAS:Kaede) fish indicates that the c233 line expresses Gal4-VP16 in a highly mobile cell type in the embryo, which most likely contribute to the mesothelial layer of the liver.
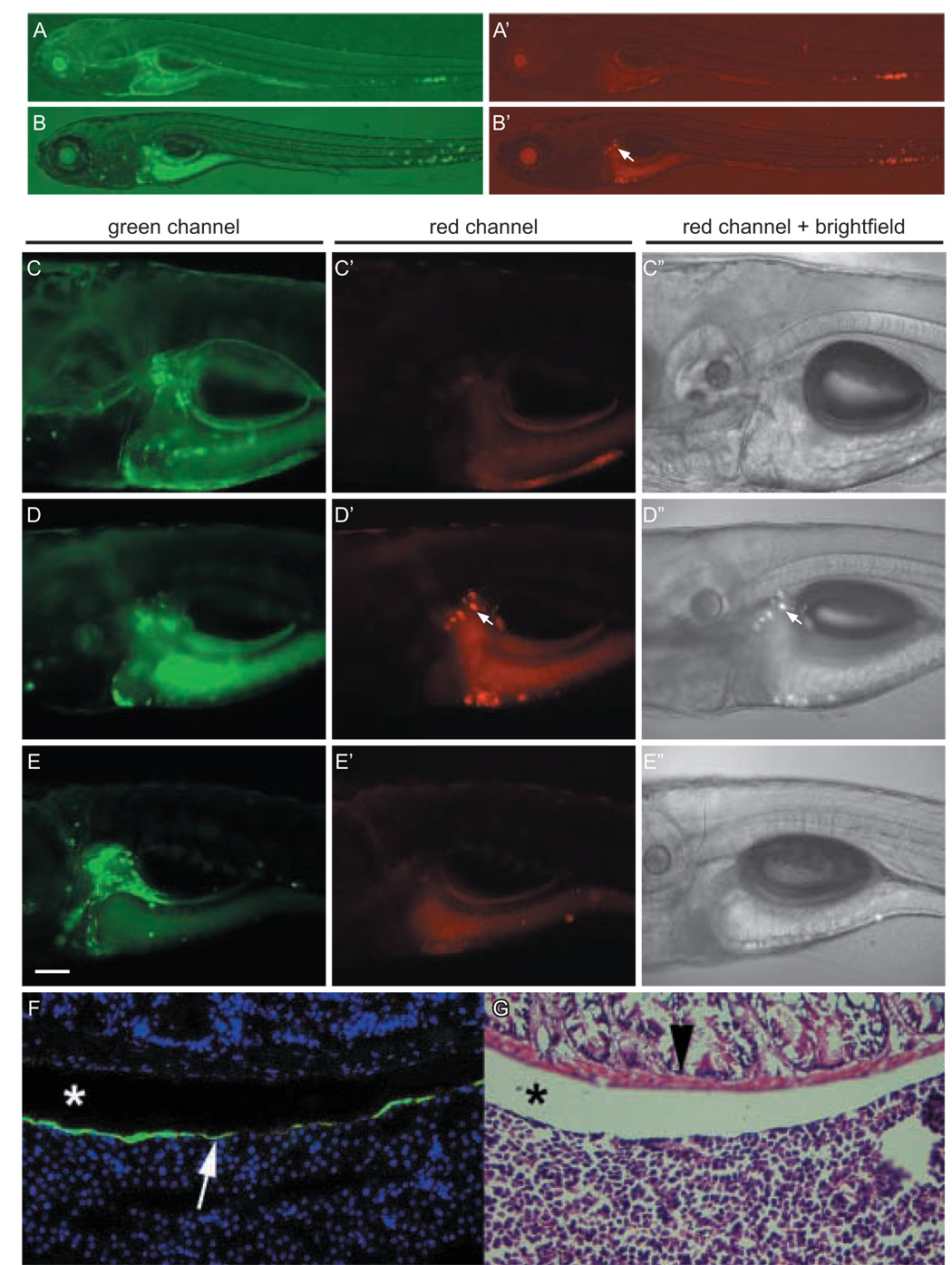
(A, A’) Images of a 6 dpf c233 larva immediately after Kaede photoconversion. Photoconverted red Kaede is restricted to the caudal tail. (B, B’) Same larva at 8 dpf showing the red cells have migrated rostrally (arrow). (C, C’, C”) Higher magnification of same larva as (A) with presumptive liver showing green, but not red cells. (D, D’, D”) At 8 dpf both green and red cells are observed (arrow). (E, E’, E”) Control transgenic larva that was not photoactivated shows only green, around the liver at 8 dpf. All images are lateral views, anterior to the left. Red fluorescent images were taken with the same exposure settings (40 seconds at low magnification, 12 seconds at higher magnification) and digitally processed in an identical manner. (C–E) Scale bar = 100µm. (F) Cryostat section through the viscera of a c233 heterozygous adult showing location of eGFP positive cells on the surface of the liver (arrow). Nuclei are blue due to Hoescht staining. (G) Haemotoxilin and eosin staining of adjacent section to show histology and location of eGFP positive cells. Intestinal smooth muscle (arrowhead) and the coelomic space between the intestines and liver are indicated (*)
Tg(Gal4-VP16;UAS:eGFP) lines for prodrug-dependent cell ablation
The ability to ablate specific cells or a tissue of interest in a temporally-regulated manner allows the investigation of cell-cell interactions and tissue regeneration. To facilitate these experiments, we created a transgenic line (UAS:nfsB-mCherry) in which the E. coli gene nfsB was placed under transcriptional control of UAS sequences. The nfsB gene encodes nitroreductase B (NTR), which can convert prodrugs such as metronidazole (Met) into toxic cellular metabolites (Drabek et al., 1997; Pisharath, 2007). The NTR fusion protein simultaneously renders cells visible due to mCherry fluorescence and susceptible to prodrug treatment (Pisharath, 2007).
To demonstrate the utility of this approach, c223 carriers were mated to UAS:nfsBmCherry transgenic fish. Compound heterozygous larvae displayed colocalization of eGFP and mCherry fluorescence in the expected tissue specific pattern. Larvae were subjected to treatment with 10mM Met for 24 hours (from 56 to 80 hours post fertilization - hpf) and then imaged by confocal microscopy to detect the presence of NTR-mCherry-expressing cells (Fig. 5). When compared to untreated siblings, c223 larvae incubated in prodrug for 24 hrs displayed a dramatic reduction in the number of mCherry-positive cells in the floor plate (Fig. 5B). The few remaining mCherry positive cells displayed nuclear fragmentation, an indicator of apoptosis (inset, Fig. 5C), suggesting that ablation of the floor plate was complete. When c223 larvae lacking the UAS:nfsB-mCherry transgene were treated with Met, eGFP expression was unaffected (5D).
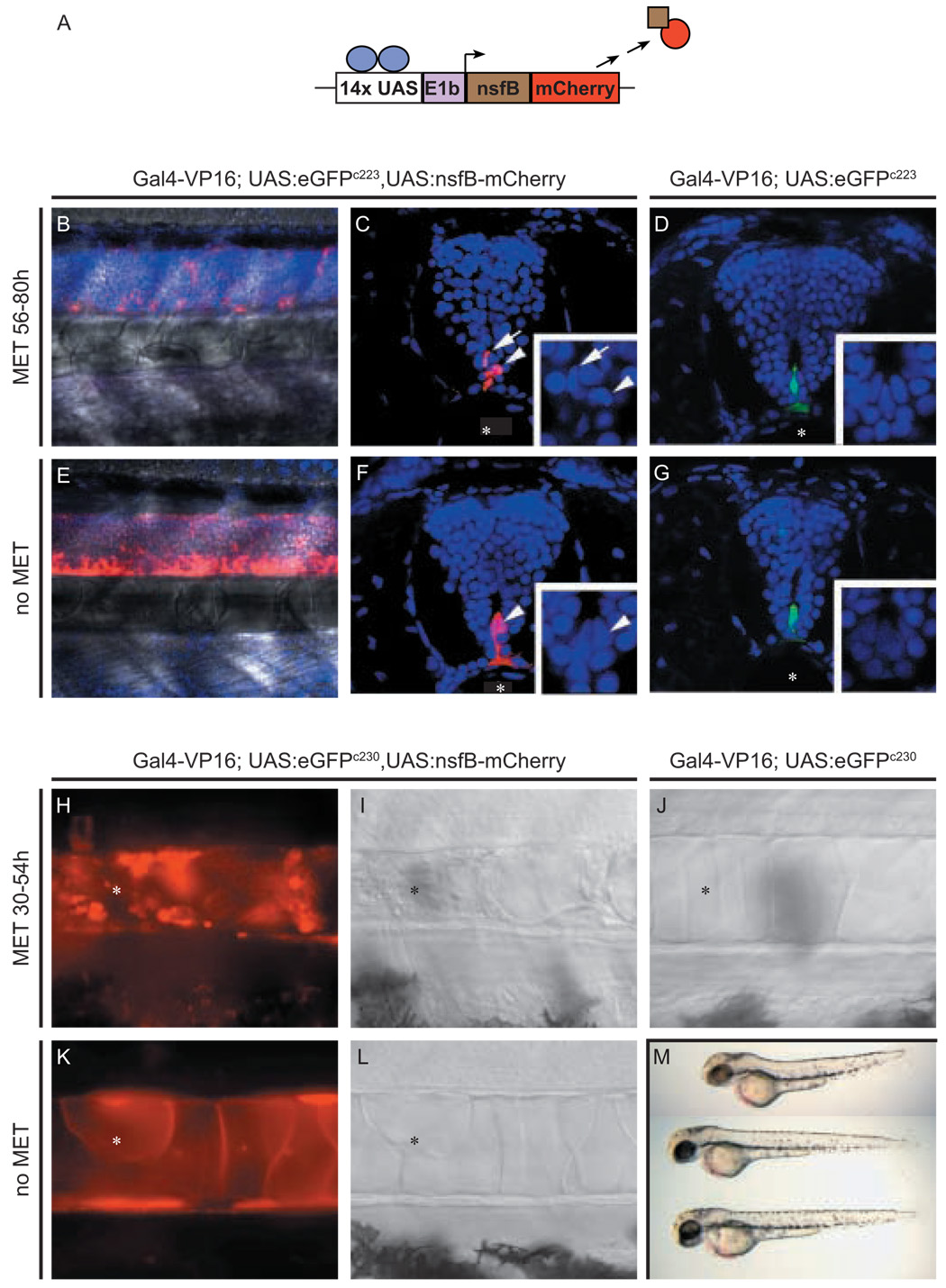
(A) Schematic of NTR-mCherry fusion protein generated by Gal4-VP16 transactivation of UAS:nfsB-mCherry transgene. Transgenic c223 larvae at 80 hpf (B–G) and c230 larvae at 54 hpf (H-M) were treated with Met from 56 to 80 hpf (B, C, D) and 30 to 54 hpf (H, I, J), respectively. Negative, age-matched controls had either one (E, F, K, L) or no copies (D, G, J) of the UAS:nfsB-mCherry. Transactivation of the NTR-mCherry fusion protein in c223 in Z-stack projections of the trunk neural tube superimposed over bright field images (B and E) or in transverse vibratome sections of the neural tube (C and F), and in the c230 (H, K) notochord (asterisks). Larvae expressing NTR-mCherry show a prodrug dependent loss of fluorescent cells in the developing CNS (B, C) and notochord (H, I corresponding DIC image) when compared to untreated controls (E, F and K, L corresponding DIC image, respectively). In B to G, nuclei are counterstained with Hoechst dye. Inset panels show high magnification of the nuclei in cells of the ventral neural tube overlaying the notochord (asterisks)
Arrows and arrowheads (C, D and F, G) mark individual fluorescent floor plate cells corresponding to identical cells in high magnification inserts. Following prodrug treatment, the few remaining NTR-mCherry expressing floor plate cells possess fragmented nuclei indicative of being apoptotic (inset in F). (M) Morphology of c230 larvae at 54 hpf either treated with Met (upper, as in H,I) or untreated (middle as in J, or lower as in K,L) demonstrating that ablating notochord cells leads to reduced body length.
UAS:nfsB-mCherry transgenic fish were also mated to c230 carriers and resulting larvae were treated with Met from 30 to 54 hpf ( Fig. 5H, I). Live imaging of the prodrug treated compound heterozygotes, revealed abnormal morphology and apoptotic mCherry positive cells in the notochord, which eventually lead to a shortening of the body axis (Fig. 5M). These defects were not observed in either untreated compound heterozygotes (Fig. 5K, L), or Met-treated c230 siblings that lacked the UAS:nfsB-mCherry transgene (Fig. 5J). Together the analyses of c223 and c230 larvae demonstrates that tissue restricted cell death is both prodrug and UAS:nfsB-mCherry transgene dependent.
These results indicate that Gal4-VP16 driver lines can be used to achieve prodrug-dependent, tissue-specific cell ablation in UAS:nfsB-mCherry transgenic zebrafish.
Identifying and mapping insertions linked to eGFP expression
To gain insight into the regulation of Gal4-VP16 transcription in recovered lines, we used LM-PCR to amplify and clone the genomic sequences flanking vector insertions. Six lines, shown to transactivate UAS-regulated gene expression, were selected for evaluation. LM-PCR indicated multiple vector insertions in all lines tested (from 3 to 10, data not shown). In all but one line (c223), a single insertion was isolated that segregated with eGFP expression. In c223, 3 insertions linked on chromosome 5, segregated with eGFP expression. Table 1 lists the data from the insertions in the 6 lines examined; including the chromosomal location and the identity of the nearest known gene. Flanking genomic sequences are listed in Supplemental Table 1.
As shown in Table 1, 8 insertions that segregated with eGFP expression could be mapped to chromosomal loci, and three mapped insertions possess the necessary transcriptional orientation within a gene to be potential gene traps. The c218 insertion occurs in the terminal coding exon of a predicted gene that encodes a protein with Kelch and BTB/POZ domains. The c223 insertion C maps to the first intron of a gene encoding a protein with homology to Cdc42 effector protein 2. The c220 insertion also maps to the first intron of a gene called inhibitor of kappa light polypeptide gene enhancer in B-cells kinase gamma (ikkγ) (Rothwarf et al., 1998), also known as NF-κB essential modulator (NEMO) (Yamaoka et al., 1998). The c220 insertion occurs in an intron upstream of the first coding exon of ikkγ and is linked to a late onset phenotype causing abnormal body curvature and lethality (supplemental Fig. S2). Consistent with a potentially mutagenic gene trapping event, a single transcript was detected using RT-PCR with primers to Gal4 and ikkγ, and following sequencing shown to be due to a fusion of the upstream non-coding ikkγ exon spliced onto Gal4-VP16. This result shows that the SAGVG construct can act as a genuine gene trap construct.
The remaining mapped lines contain insertions that do not predict conventional gene traps. Two of these lines had insertions that mapped less than 20kb from genes whose endogenous expression is similar to the pattern of eGFP expression associated with that particular line. The c237 insertion segregates with eGFP expression in presumptive cranial ganglia, CNS neurons and in the retina. This insertion resides 1.8 kb 5’ of neurogenin1, which is also expressed in these tissues (Nechiporuk et al., 2005). The c229 allele, which segregates with eGFP expression in pineal gland and lens, maps to a position 18 kb 3’ of betaA2-2-crystallin (zgc:92722), a gene which is highly represented in a cDNA library constructed from zebrafish lens tissue (http://www.ncbi.nlm.nih.gov/UniGene/clust.cgi?ORG=Dr&CID=31370 31370).
Discussion
The results described here confirm that Tol2 integrated Gal4-VP16 transgenes not only self report via an UAS:eGFP cassette, but can function as driver lines to activate UAS responder lines in trans. The pilot screen has efficiently generated lines displaying restricted eGFP expression in a wide variety of tissues. The SAGVG vector is designed to take advantage of the amplification inherent in the Gal4-VP16/UAS system (Koster and Fraser, 2001) and is facilitated by recent advances in transgenic methods (Kawakami et al., 2000). With Gal4-VP16 activation, detectable eGFP expression is expected even if transcription levels from endogenous regulatory elements are low. Ideally, this maximizes the number of driver lines that can be obtained in a screen.
Most Gal4-VP16 driver lines maintained robust expression of eGFP over 3 generations. In some cases, such as c218, we have observed a decrease in the frequency of embryos/larvae expressing eGFP, in each new generation. This occurrence could be due to transgene silencing, but as SAGVG trapping events are self reporting, green fluorescence can be used to easily identify the progeny that have escaped this phenomenon. Alternatively, as each line contains several copies of SAGVG distributed throughout the genome, random assortment could be reducing the copy number of UAS:eGFP insertions available for Gal4 activation, thereby affecting the amount of eGFP expressed. For some lines, special efforts may be needed to maintain adequate templates for transactivation.
To identify candidate genes associated with particular expression patterns, and to gain insight into the mechanisms underlying Gal4-VP16 expression, we determined the chromosomal localization of insertions responsible for eGFP expression patterns of interest. We examined 6 lines and found a total of 8 insertions co-segregating with eGFP expression (c223 had three linked insertions). Genuine gene trap events are potentially mutagenic (Friedrich and Soriano, 1991). Although the SAGVG construct was designed to function as a gene-trap, we found that only three insertions (c218, c220, c223-C) were consistent with a gene trap mechanism (depicted in Fig. 6A and B), on the basis of the identity and orientation of flanking genomic sequences.
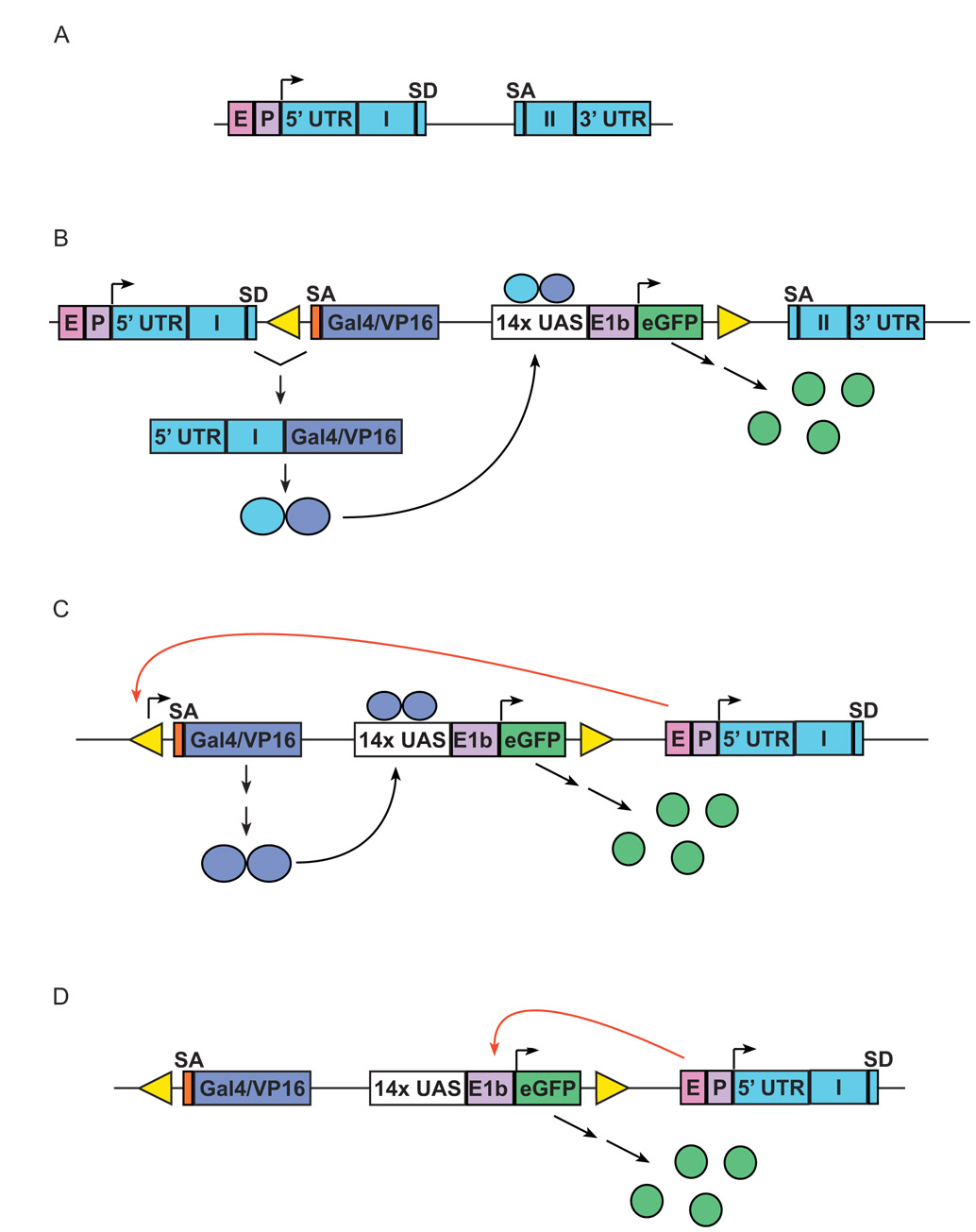
(A) Schematic representation of a target endogenous gene, with two exons (Roman numerals); E, endogenous enhancer; P, endogenous promoter. (B) Insertion of SAGVG into the intron of gene, leading to activation of eGFP expression through traditional gene trap event; SA, splice acceptor; UAS, Gal4-responsive upstream activating sequence; E1b, adenoviral E1b minimal promoter element. After splicing of Gal4-VP16 to upstream endogenous coding sequence, the resulting fusion protein provides in cis activation of UAS:eGFP expression. (C and D) two methods by which enhancer trapping can lead to expression of eGFP expression (nb in reality both the orientation and distance from endogenous enhancer can be varied due to the nature of enhancer activity). (C) Gal4-VP16-dependent activation of eGFP expression by enhancer trapping. Endogenous enhancer “E” drives Gal4-VP16 expression through interaction with cryptic promoter elements in either Tol2 vector or in genomic DNA adjacent to insertion. (D) Gal4-VP16-independent activation of eGFP expression by enhancer trapping. Endogenous enhancer “E” drives eGFP expression directly through interaction with E1b minimal promoter in UAS:eGFP cassette.
The c220 insertion is in the first intron of a known gene and does indeed create a mutant fusion transcript, as demonstrated by sequencing the RT-PCR product. When bred to homozygosity, c220 fish display abnormal morphology and late lethality (supplemental Fig.S2). IKKγ mutations in humans lead to a range of defects that include ectodermal dysplasia and immunodeficiency (Smahi et al., 2000). The Ikkγ gene is X-linked in mammals, and male mice hemizygous for a null Ikkγ mutation die during mid-gestation. Although heterozygous females are viable, likely due to random X inactivation of the one wild type copy of the gene they display ectodermal dysplasia (Schmidt-Supprian et al., 2000). It is clear that SAGVG can function as a bona fide gene trap vector with the potential to be mutagenic, and work is underway to demonstrate whether the linked late phenotype seen in c220 fish is solely due to the disruption in zebrafish ikkγ.
Two other lines (c229 and c237) appear to represent Gal4-VP16 dependent enhancer traps (mechanism depicted in Fig. 6C), because the SAGVG insertions are located outside of known transcribed regions. Activation of eGFP expression is most likely mediated by Gal4-VP16, which in these lines is capable of activating other UAS reporters in trans (see below). Specific enhancer trap studies in the zebrafish have shown that active insertions are often in close proximity to genes with similar patterns of expression (Balciunas et al., 2004; Ellingsen et al., 2005). In the case of c229 and c237, the SAGVG construct has inserted within 20kb of genes with tissue-specific expression predicted to be similar to the observed eGFP labeling patterns. This suggests that nearby endogenous genes and Gal4-VP16 are under the control of a common regulatory enhancer(s). In these two cases, the site of transcription initiation is unknown: it may be that there are cryptic promoter sequences in upstream genomic DNA or within the vector itself.
Lines c228 and c236 are most likely examples of Gal4-VP16 independent eGFP expression (mechanism depicted in Fig. 6D). Both insertions generate highly restricted eGFP expression, in hindbrain rhombomeres (c228) or in the heart (c236), respectively. However, when fish bearing either insertion were bred to Tg(UAS:Kaede) fish, the resultant eGFP+ larvae failed to show any evidence of Kaede photoconversion (see below). This finding is consistent with an SAGVG insertion in which transcription from the minimal promoter of eGFP is under the direct influence of a local enhancer (Fig. 6D) rather than mediated via Gal4-VP16 activation.
Although a somewhat unexpected finding, the multiple mechanisms by which eGFP expression can be achieved from the SAGVG construct likely accounts for the very high frequency with which distinct expression patterns were observed during the screening phase of this study. Fifty-seven percent (28/49) paired crosses of F0 fish resulted in restricted patterns of eGFP expression. We also demonstrated in this report the utility of the SAGVG insertions to function as Gal4-VP16 driver lines by transactivating 3 different UAS-regulated genes to produce mCherry, Kaede and NTR-mCherry. Of the 15 lines assayed for transactivation, only 2 (c228 and c236) failed to show labeling of other fluorescent proteins within eGFP-expressing tissues. Significant resources will be required to maintain a large panel of transactivating fish lines with restricted patterns of eGFP expression generated from a more extensive screen for Gal4-Vp16 insertions. Hence, it is satisfying that Tol2 insertional events failing to lead to transactivation are uncommon.
Spatially restricted photoconversion of Kaede can be used to label individual cells, such as neurons (e.g., Figs. 3E, 3E’) and to trace their axonal projections (Sato et al., 2006). This system is also well suited to cell tracking (Hatta et al., 2006). Using this technique we tracked an unknown population of photoconverted cells over time and demonstrated that they migrated rostrally and came to lie in close proximity to the liver. Cell position at 8 dpf strongly correlated with the tissue location of eGFP positive cells in the mesothelium of the adult liver. Thus, transactivation of Kaede by SAGVG transgenic lines and subsequent photoconversion is a powerful method to monitor the behavior of cells and their ultimate tissue specific fate.
In several experiments where Tg(UAS:Kaede) fish were bred to Gal4-VP16 lines, we noted a mosaic pattern of transactivation. For instance larvae carrying the c237 allele possess interneurons that fluoresce green. In some interneurons photoconversion of Kaede was not observed (Fig. 3E’), even though adjacent cells with similar morphology converted to red fluorescence following exposure to UV light. This presumed variegation in transactivation may represent differences in chromatin state between otherwise similar cells. Such epigenetic effects are known to cause variegation in transcription levels and to be influenced by Gal4 activity (Ahmad and Henikoff, 2001). Tissue variability in expression has also been previously documented in Drosophila using the Gal4-VP16/UAS system (Fischer et al., 1988) and may also influence transactivation in zebrafish. Such potentially complicating phenomena must be taken into account when interpreting experiments where Gal4-VP16 drivers are used to misexpress genes in given cell types and may necessitate fluorescently tagging activated proteins of interest.
Two Gal4-VP16 lines were bred to fish transgenic for UAS:nfsB-mCherry. The production of the NTR-mCherry fusion protein led to prodrug dependent cell ablation. Self-reporting Gal4-VP16 drivers can be used for selective cell ablation in a tissue-specific manner and for monitoring cell loss over time. From our work and the work of others it is clear that NTR activity can be used to ablate a wide range of cell types throughout the embryo/larva (Pisharath, 2007) and D. Stainier (personal communication). As more Gal4-VP16 driver lines become available, the UAS:nfsB-mCherry transgenic line will constitute an important resource in studying cellular regeneration and cell-cell interactions.
Gal4-VP16 technology revolutionized experimental design in Drosophila (Brand and Perrimon, 1993; Duffy, 2002) and we envision the same advantages can be readily applied to zebrafish. Future applications of in the zebrafish could include the rescue of mutations by expression of wild-type genes in specific tissues (Zars et al., 2000a; Zars et al., 2000b), the generation of cancer models by expression of oncogenes in tissues of interest (Folberg-Blum et al., 2002), dissection of signal transduction pathways in specific cells by expression of dominant negative (Elefant and Palter, 1999) or constitutively active pathway components, and probing the neuronal basis of behavior (Brand and Dormand, 1995). The binary nature of the Gal4-VP16 activation system will permit the production of many genetic tools for zebrafish that cannot be generated as simple transgenic lines due to lethality.
In conclusion, the current results demonstrate that distribution of SAGVG throughout the zebrafish genome by Tol2 transposition can generate a collection of effective Gal4-VP16 driver lines. Self-reporting transgenic lines are useful for labeling cell types, and for transactivating other UAS regulated genes. Mapping has shown that there are apparently multiple mechanisms by which transcription of Gal-VP16 can be induced and may explain the efficient recovery of eGFP-expressing lines. We anticipate that the widespread application of this technology will provide an important new resource for genetic manipulation in zebrafish.
Supplementary Material
01
Supplemental Fig. S1:Tissue-specific eGFP expression and Kaede photoconversion in putative cranial ganglia. (A) c215 larvae express eGFP in clusters of cells posterior to the eye (arrowhead) and in the ear (out of focal plane) at 5 dpf. (B, C) Higher magnification of Kaede photoconversion in a cranial cluster after 90s exposure to UV light. Kaede is also activated in trans in the ear (not shown). (D) c241 larvae express eGFP in a small subset of cells between the eye and ear at 5 dpf. (E, F) Higher magnification after 90s UV photoconversion of Kaede. All images are lateral views, anterior to the left. (A and D) images captured on a Leica MZFLIII stereomicroscope. (B, C, E, and F) images using a 20X objective on a Zeiss Axioskop microscope. Scale bars = 50µM.
02
Supplemental Fig. S2:The c220 insertion resides in the inhibitor of kappa light polypeptide gene enhancer in B-cells kinase gamma (ikkγ) locus. (A) Intercrosses of c220 transgenic heterozygotes produced some progeny that exhibited variable body curvature by three months and were not viable beyond 4 months of age (n=30). (B) Multiplex PCR of genomic DNA derived from fin clips of three month old fish demonstrates that the mutant phenotype correlates with homozygosty for the c220 insertion.
Primers to amplify mutant allele (lower band): Forward primer (Tol2) TTACTCAAGTACTTTACACCTC;
Reverse primer (ikkγ downstream of DNA element) CTTATGAGTCATTCAATGACAC.
Primers to amplify wild-type allele (Upper band): Forward primer (ikkγ exon1) GGAAGACCGACAGACAGTGA;
Reverse primer (ikkγ downstream of DNA element) CTTATGAGTCATTCAATGACAC
(C) The Tol2 insertion causes the first exon of ikkγ to splice to the β-globin exon 3 splice acceptor site in SAGVG and produces an ikkγ exon 1 and Gal4 fusion transcript. The sequence across the splice junction, as determined from the analysis of RT-PCR products from RNA derived from c220 heterozygous larvae, is color coded as in the schematic.
RT used oligodT priming. For PCR Forward (ikkγ exon1) GGAAGACCGACAGACAGT GA; Reverse (Gal4) TCATGTCAAGGTCTTCTCGAG
03
04
Supplementary Movie 1:In c236 larvae, specific GFP labeling reveals functioning of heart valves during normal blood flow (anterior to the left). A live 6 dpf larva was anesthetized, mounted in 2% low-melting agarose, and viewed ventrally under a Plan-Apochromat 20x/0.8 objective on a Zeiss LSM 5 Live DuoScan. Digital images were captured at 60 frames per second, false colored and exported at 30 frames per second with Zeiss LSM Image Examiner v.4.0.0.91. The final movie was adjusted to real-time with Apple QuickTime v.7.1.3
Acknowledgements
The authors wish to acknowledge Koichi Kawakami and Scott Fraser for reagents, Harshan Pisharath, Shannon Fisher and Andrew McCallion for useful advice, Michelle Swanson, Yangseon Park, Melinda Campbell, Nicole Gabriel and Kate Lewis for expert technical support and Bryan Bowman (Zeiss), Ruth Chalmers-Redman (Zeiss) and Chroma Technology Corp. for assistance with microscope filters. HB was supported by a Sandler Opportunity Award, a Byers Award, and a grant from the March of Dimes Foundation. NJG was funded by an NRSA Predoctoral Fellowship. This work was supported in part by NIH grants DK61215 and DK56211 (to SDL) and HD042215 (to MEH). SDL is also supported by the Paul K. Neumann Professorship at Johns Hopkins University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmad K, Henikoff S. Modulation of a transcription factor counteracts heterochromatic gene silencing in Drosophila. Cell. 2001;104:839–847. [Abstract] [Google Scholar]
- Amsterdam A, Nissen RM, Sun Z, Swindell EC, Farrington S, Hopkins N. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci U S A. 2004;101:12792–12797. [Europe PMC free article] [Abstract] [Google Scholar]
- Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc Natl Acad Sci U S A. 2002;99:12651–12656. [Europe PMC free article] [Abstract] [Google Scholar]
- Balciunas D, Davidson AE, Sivasubbu S, Hermanson SB, Welle Z, Ekker SC. Enhancer trapping in zebrafish using the Sleeping Beauty transposon. BMC Genomics. 2004;5:62. [Europe PMC free article] [Abstract] [Google Scholar]
- Brand AH, Dormand EL. The GAL4 system as a tool for unravelling the mysteries of the Drosophila nervous system. Curr Opin Neurobiol. 1995;5:572–578. [Abstract] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. [Abstract] [Google Scholar]
- Davidson AE, Balciunas D, Mohn D, Shaffer J, Hermanson S, Sivasubbu S, Cliff MP, Hackett PB, Ekker SC. Efficient gene delivery and gene expression in zebrafish using the Sleeping Beauty transposon. Dev Biol. 2003;263:191–202. [Abstract] [Google Scholar]
- Drabek D, Guy J, Craig R, Grosveld F. The expression of bacterial nitroreductase in transgenic mice results in specific cell killing by the prodrug CB1954. Gene Ther. 1997;4:93–100. [Abstract] [Google Scholar]
- Duffy JB. GAL4 system in Drosophila: a fly geneticist's Swiss army knife. Genesis. 2002;34:1–15. [Abstract] [Google Scholar]
- Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–226. [Abstract] [Google Scholar]
- Elefant F, Palter KB. Tissue-specific expression of dominant negative mutant Drosophila HSC70 causes developmental defects and lethality. Mol Biol Cell. 1999;10:2101–2117. [Europe PMC free article] [Abstract] [Google Scholar]
- Ellingsen S, Laplante MA, Konig M, Kikuta H, Furmanek T, Hoivik EA, Becker TS. Large-scale enhancer detection in the zebrafish genome. Development. 2005;132:3799–3811. [Abstract] [Google Scholar]
- Fischer JA, Giniger E, Maniatis T, Ptashne M. GAL4 activates transcription in Drosophila. Nature. 1988;332:853–856. [Abstract] [Google Scholar]
- Folberg-Blum A, Sapir A, Shilo BZ, Oren M. Overexpression of mouse Mdm2 induces developmental phenotypes in Drosophila. Oncogene. 2002;21:2413–2417. [Abstract] [Google Scholar]
- Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. [Abstract] [Google Scholar]
- Hatta K, Tsujii H, Omura T. Cell tracking using a photoconvertible fluorescent protein. Nature Protocols. 2006;1:1–8. [Abstract] [Google Scholar]
- Kawakami K. Transgenesis and gene trap methods in zebrafish by using the Tol2 transposable element. Methods Cell Biol. 2004;77:201–222. [Abstract] [Google Scholar]
- Kawakami K, Koga A, Hori H, Shima A. Excision of the tol2 transposable element of the medaka fish, Oryzias latipes, in zebrafish, Danio rerio. Gene. 1998;225:17–22. [Abstract] [Google Scholar]
- Kawakami K, Shima A, Kawakami N. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc Natl Acad Sci U S A. 2000;97:11403–11408. [Europe PMC free article] [Abstract] [Google Scholar]
- Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7:133–144. [Abstract] [Google Scholar]
- Kempken F, Windhofer F. The hAT family: a versatile transposon group common to plants, fungi, animals, and man. Chromosoma. 2001;110:1–9. [Abstract] [Google Scholar]
- Koga A, Hori H. The Tol2 transposable element of the medaka fish: an active DNA-based element naturally occurring in a vertebrate genome. Genes Genet Syst. 2001;76:1–8. [Abstract] [Google Scholar]
- Koster RW, Fraser SE. Tracing transgene expression in living zebrafish embryos. Dev Biol. 2001;233:329–346. [Abstract] [Google Scholar]
- Lukacsovich T, Asztalos Z, Awano W, Baba K, Kondo S, Niwa S, Yamamoto D. Dual-tagging gene trap of novel genes in Drosophila melanogaster. Genetics. 2001;157:727–742. [Europe PMC free article] [Abstract] [Google Scholar]
- McGuire SE, Roman G, Davis RL. Gene expression systems in Drosophila: a synthesis of time and space. Trends Genet. 2004;20:384–391. [Abstract] [Google Scholar]
- Nechiporuk A, Linbo T, Raible DW. Endoderm-derived Fgf3 is necessary and sufficient for inducing neurogenesis in the epibranchial placodes in zebrafish. Development. 2005;132:3717–3730. [Abstract] [Google Scholar]
- Parinov S, Kondrichin I, Korzh V, Emelyanov A. Tol2 transposon-mediated enhancer trap to identify developmentally regulated zebrafish genes in vivo. Dev Dyn. 2004;231:449–459. [Abstract] [Google Scholar]
- Parsons MJ, Pollard SM, Saude L, Feldman B, Coutinho P, Hirst EM, Stemple DL. Zebrafish mutants identify an essential role for laminins in notochord formation. Development. 2002;129:3137–3146. [Abstract] [Google Scholar]
- Pisharath H, Rhee JM, Swanson MA, Leach SD, Parsons MJ. Targeted ablation of beta cells in the embryonic zebrafish pancreas using E.coli nitroreductase. Mech Dev. 2007 In press. [Europe PMC free article] [Abstract] [Google Scholar]
- Rorth P, Szabo K, Bailey A, Laverty T, Rehm J, Rubin GM, Weigmann K, Milan M, Benes V, Ansorge W, Cohen SM. Systematic gain-of-function genetics in Drosophila. Development. 1998;125:1049–1057. [Abstract] [Google Scholar]
- Rothwarf DM, Zandi E, Natoli G, Karin M. IKK-gamma is an essential regulatory subunit of the IkappaB kinase complex. Nature. 1998;395:297–300. [Abstract] [Google Scholar]
- Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. [Abstract] [Google Scholar]
- Sato T, Takahoko M, Okamoto H. HuC:Kaede, a useful tool to label neural morphologies in networks in vivo. Genesis. 2006;44:136–142. [Abstract] [Google Scholar]
- Scheer N, Campos-Ortega JA. Use of the Gal4-UAS technique for targeted gene expression in the zebrafish. Mech Dev. 1999;80:153–158. [Abstract] [Google Scholar]
- Schmidt-Supprian M, Bloch W, Courtois G, Addicks K, Israel A, Rajewsky K, Pasparakis M. NEMO/IKK gamma-deficient mice model incontinentia pigmenti. Mol Cell. 2000;5:981–992. [Abstract] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. [Abstract] [Google Scholar]
- Smahi A, Courtois G, Vabres P, Yamaoka S, Heuertz S, Munnich A, Israel A, Heiss NS, Klauck SM, Kioschis P, Wiemann S, Poustka A, Esposito T, Bardaro T, Gianfrancesco F, Ciccodicola A, D'Urso M, Woffendin H, Jakins T, Donnai D, Stewart H, Kenwrick SJ, Aradhya S, Yamagata T, Levy M, Lewis RA, Nelson DL. Genomic rearrangement in NEMO impairs NF-kappaB activation and is a cause of incontinentia pigmenti. The International Incontinentia Pigmenti (IP) Consortium. Nature. 2000;405:466–472. [Abstract] [Google Scholar]
- Thermes V, Grabher C, Ristoratore F, Bourrat F, Choulika A, Wittbrodt J, Joly JS. I-SceI meganuclease mediates highly efficient transgenesis in fish. Mech Dev. 2002;118:91–98. [Abstract] [Google Scholar]
- Urasaki A, Morvan G, Kawakami K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics. 2006 [Europe PMC free article] [Abstract] [Google Scholar]
- Wang K, Gan L, Boysen C, Hood L. A microtiter plate-based high-throughput DNA purification method. Anal Biochem. 1995;226:85–90. [Abstract] [Google Scholar]
- Yamaoka S, Courtois G, Bessia C, Whiteside ST, Weil R, Agou F, Kirk HE, Kay RJ, Israel A. Complementation cloning of NEMO, a component of the IkappaB kinase complex essential for NF-kappaB activation. Cell. 1998;93:1231–1240. [Abstract] [Google Scholar]
- Zars T, Fischer M, Schulz R, Heisenberg M. Localization of a short-term memory in Drosophila. Science. 2000a;288:672–675. [Abstract] [Google Scholar]
- Zars T, Wolf R, Davis R, Heisenberg M. Tissue-specific expression of a type I adenylyl cyclase rescues the rutabaga mutant memory defect: in search of the engram. Learn Mem. 2000b;7:18–31. [Europe PMC free article] [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.ydbio.2007.01.033
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3470427?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Peds1 deficiency in zebrafish results in myeloid cell apoptosis and exacerbated inflammation.
Cell Death Discov, 10(1):388, 29 Aug 2024
Cited by: 0 articles | PMID: 39209813 | PMCID: PMC11362147
Neural progenitor-derived Apelin controls tip cell behavior and vascular patterning.
Sci Adv, 10(27):eadk1174, 05 Jul 2024
Cited by: 0 articles | PMID: 38968355 | PMCID: PMC11225789
Satellite glial cell manipulation prior to axotomy enhances developing dorsal root ganglion central branch regrowth into the spinal cord.
Glia, 72(10):1766-1784, 22 Jun 2024
Cited by: 0 articles | PMID: 39141572
A nox2/cybb zebrafish mutant with defective myeloid cell reactive oxygen species production displays normal initial neutrophil recruitment to sterile tail injuries.
G3 (Bethesda), 14(6):jkae079, 01 Jun 2024
Cited by: 0 articles | PMID: 38696730 | PMCID: PMC11152067
Genetically defined nucleus incertus neurons differ in connectivity and function.
Elife, 12:RP89516, 31 May 2024
Cited by: 1 article | PMID: 38819436 | PMCID: PMC11142643
Go to all (243) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Zebrafish transgenic lines co-expressing a hybrid Gal4 activator and eGFP in tissue-restricted patterns.
Gene Expr Patterns, 11(8):517-524, 10 Sep 2011
Cited by: 2 articles | PMID: 21930245
Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish.
Proc Natl Acad Sci U S A, 105(4):1255-1260, 17 Jan 2008
Cited by: 328 articles | PMID: 18202183 | PMCID: PMC2234125
zTrap: zebrafish gene trap and enhancer trap database.
BMC Dev Biol, 10:105, 18 Oct 2010
Cited by: 84 articles | PMID: 20950494 | PMCID: PMC2970601
Gal4 Driver Transgenic Zebrafish: Powerful Tools to Study Developmental Biology, Organogenesis, and Neuroscience.
Adv Genet, 95:65-87, 13 Jun 2016
Cited by: 35 articles | PMID: 27503354
Review
Funding
Funders who supported this work.
NICHD NIH HHS (2)
Grant ID: R01 HD042215
Grant ID: HD042215
NIDDK NIH HHS (9)
Grant ID: DK61215
Grant ID: R01 DK061215
Grant ID: R01 DK061215-03
Grant ID: R01 DK061215-04
Grant ID: R01 DK056211-07
Grant ID: P30 DK079637
Grant ID: R01 DK056211-08
Grant ID: DK56211
Grant ID: R01 DK056211
NINDS NIH HHS (1)
Grant ID: 1F31NS05097





