Abstract
Free full text

Human Pumilio Proteins Recruit Multiple Deadenylases to Efficiently Repress Messenger RNAs*
Abstract
PUF proteins are a conserved family of eukaryotic RNA-binding proteins that regulate specific mRNAs: they control many processes including stem cell proliferation, fertility, and memory formation. PUFs repress protein expression from their target mRNAs but the mechanism by which they do so remains unclear, especially for humans. Humans possess two PUF proteins, PUM1 and PUM2, which exhibit similar RNA binding specificities. Here we report new insights into their regulatory activities and mechanisms of action. We developed functional assays to measure sequence-specific repression by PUM1 and PUM2. Both robustly inhibit translation and promote mRNA degradation. Purified PUM complexes were found to contain subunits of the CCR4-NOT (CNOT) complex, which contains multiple enzymes that catalyze mRNA deadenylation. PUMs interact with the CNOT deadenylase subunits in vitro. We used three approaches to determine the importance of deadenylases for PUM repression. First, dominant-negative mutants of CNOT7 and CNOT8 reduced PUM repression. Second, RNA interference depletion of the deadenylases alleviated PUM repression. Third, the poly(A) tail was necessary for maximal PUM repression. These findings demonstrate a conserved mechanism of PUF-mediated repression via direct recruitment of the CCR4-POP2-NOT deadenylase leading to translational inhibition and mRNA degradation. A second, deadenylation independent mechanism was revealed by the finding that PUMs repress an mRNA that lacks a poly(A) tail. Thus, human PUMs are repressors capable of deadenylation-dependent and -independent modes of repression.
Introduction
Messenger RNAs (mRNAs) are subject to extensive regulation throughout their lifespan (1). Synthesis and processing events of precursor mRNAs in the nucleus are regulated to yield mature mRNAs. Once exported to the cytoplasm, translation and stability of mRNAs are controlled to ensure that the appropriate amount of encoded protein is produced at the proper time and cellular location. The discovery of factors and mechanisms responsible for gene regulation is crucial to deepening our understanding of how misregulation contributes to disease.
PUF3 (Pumilio and fem-3 binding factor) proteins are trans-acting factors that regulate mRNAs by binding specific sequences in 3′ untranslated regions (3′ UTR) (2). Members of the PUF family share a conserved RNA binding domain composed of eight α helical repeats (3–8). These PUF repeats adopt a crescent shape, whose concave side binds to specific single-stranded RNA sequences. Each PUF repeat recognizes a single ribonucleotide base, mediated by three RNA recognition amino acids, and these contacts dictate the RNA binding specificity of each individual PUF protein (7).
Humans and other vertebrates possess two canonical PUF proteins, PUM1 and PUM2, collectively referred to as PUMs (9). PUMs share significant sequence similarity: amino acids outside of their RNA binding domains (RBD) share 75% identity, whereas those within are 91% identical (9, 10). Both PUM1 and PUM2 bind with high affinity to the consensus sequence UGUANAUA, hereon referred to as a PUM response element (PRE) (7, 11–13). PUMs are widely expressed in tissues and cell types (9, 14). Given their similar RNA binding specificities and broad expression, it is possible that PUMs compete for many of the same mRNAs, supported by identification of mRNAs that associate with PUMs (13).
Genetics in model organisms demonstrated that PUFs control embryonic development, fertility, stem cell proliferation, and neurological functions, including the formation of memories (4, 5, 15–29). In mice, PUFs are involved in fertility through control of spermatogenesis (30, 31). In cell culture, human PUMs were reported to affect cell proliferation (32). Insight into the functions of human PUMs emerged from identification of mRNAs that co-immunopurified with each PUM (13, 33). Hundreds of mRNAs were enriched in the PUM1 and PUM2 immunoprecipitates, suggesting an extensive regulatory network. The list of putative target mRNAs included genes involved in gene expression, signal transduction, cell cycle and proliferation, among others (13, 33). Importantly, regulation of these mRNAs remains to be demonstrated.
In model organisms, PUFs generally act as repressors, although in several instances they affect mRNA localization and perhaps activation of mRNAs (2, 34–37). In Drosophila and Caenorhabditis elegans, PUF repression correlates with shortening of the poly(A) tail (i.e. deadenylation) (29, 38–41). In yeast, PUFs repress by promoting degradation of target mRNAs, specifically deadenylation and decapping (42–46). Yeast PUFs bind to the Pop2p deadenylase subunit to enhance removal of the poly(A) tail, catalyzed by the Ccr4p deadenylase (43–45). The consequences of deadenylation are known to include translational down-regulation and initiation of mRNA degradation (47). In other cases, PUFs have been reported to directly inhibit translation, with several potential mechanisms having been proposed (5, 48–51).
The mechanism(s) of mRNA regulation by human PUMs remains to be elucidated and a complete understanding of PUM repression will facilitate identification of biologically relevant target mRNAs. A repressive role for human PUMs is supported by several observations. Overexpression of PUM2 reduces expression of reporter genes (52) and overexpression of PUM together with a putative partner NANOS3 was reported to inhibit E2F3 expression (53). Another study reported that reduction of PUM1 by RNA interference stabilized several mRNAs (33). PUMs were reported to repress the mRNA encoding CDKN1B tumor suppressor (32) and, unique to this mRNA, PUM1 was postulated to license microRNA-mediated repression by disrupting basepairing between specific PUM and microRNA binding sites (32).
The role of deadenylases in yeast PUF repression suggested that human deadenylases might serve as PUM co-repressors. Humans possess multiple orthologs of the Pop2p and Ccr4p deadenylase enzymes (47). The human CNOT7 and CNOT8 proteins are related to yeast Pop2p, whereas human CNOT6 and CNOT6L are orthologous to yeast Ccr4p (47, 54–56). All four proteins have been reported to possess deadenylase activity (47, 57–59). Like their yeast counterparts, CNOT7 and CNOT8 form heterodimers with either CNOT6 or CNOT6L, and these pairs assemble with human orthologs of the yeast Not proteins to form large multisubunit complexes referred to as CCR4-NOT (CNOT) complexes (60–62).
In this report, we explore the activities of human PUM1 and PUM2. We show that both PUMs are potent repressors that inhibit protein expression and reduce mRNA levels. We then investigate the mechanism of repression and show that purified PUM complexes contain CNOT deadenylases. Two deadenylase subunits interact directly with the PUMs. In vivo, we find that deadenylases are important PUM co-repressors and the poly(A) tail is necessary for efficient repression. We also present evidence for a poly(A) independent mechanism of PUM repression. This research reveals two modes of PUM repression and thereby enhances our understanding of their regulatory functions to control important biological processes.
EXPERIMENTAL PROCEDURES
Plasmids
Renilla luciferase reporters (RnLUC) are based on psiCheck1 (Promega) with either three wild-type PRE or mutant PRE elements inserted into the XhoI and NotI sites in the 3′ UTR. The PRE sequence is as follows, with the PRE underlined: 5′-TTGTTGTCGAAAATTGTACATAAGCCAA and the PREmt sequence is: 5′-TTGTTGTCGAAAATACAACATAAGCCAA. The altered specificity reporter, RnLUC 3xPRE UGG, was constructed with the following sequence: 5′-TTGTTGTCGAAAATTGGACATAAGCCAA. RnLUC HSL was created by replacing the cleavage/polyadenylation site from the psiCheck1 3′ UTR with a histone stem loop (HSL) sequence from the human H1F3 gene. Two or four PRE sequences were inserted upstream of the HSL to create RnLUC 2xPRE HSL and RnLUC 4xPRE HSL. The firefly luciferase (FfLUC) plasmid pGL4.13 (Promega) was used as a control.
To express proteins as Halotag fusions in human cells, CNOT6, CNOT6L, CNOT7, or CNOT8 were cloned into the vector pFN21A (Promega). Active site mutants, CNOT7 D40A,E42A and CNOT8 D40A,E42A were created by QuikChange mutagenesis (Stratagene). Full-length human PUM1 or PUM2 open reading frames were cloned into pFN21A and site-directed mutagenesis was used to create PUM1 R6as (N1043S Q1047E) and PUM2 R6as (N921S Q925E). Recombinant proteins were expressed as Halotag (HT) fusions from the vector pFN18A (Promega), including HT-CNOT6, HT-PUM1 RBD (aa 828–1176), and HT-PUM2 RBD (aa 705–1050). Renilla and firefly luciferase reporters for Drosophila cells were previously described (63). Drosophila protein expression constructs were made by inserting PUM1 coding sequence into pIZ V5 His6 (Invitrogen).
Cell Culture and Transfections
Human HEK293 cells were cultured at 37 °C under 5% CO2 in DMEM with glucose and 1× penicillin/streptomycin/glutamine and 10% FBS (Invitrogen). Drosophila D.mel-2 cells (Invitrogen) were cultured as previously described (63). Transfections of human cells were carried out with FuGENE HD (Promega) at 3:1 volume of lipid:μg of DNA. For luciferase assays, 2 × 104 cells were plated in white-walled 96-well plates and, after 24 h, were transfected with 100 ng/well of plasmid DNA. For RNA purifications and coimmunoprecipitations, 6 × 105 cells were transfected with 3 μg of plasmid DNA 24 h after seeding. D.mel-2 cells were transfected with Effectene (Qiagen) as previously described (63). For human PUM1 expression and repression assays, 400 ng of PUM1 expression vector was included in the transfection with reporters.
Luciferase Assays
Renilla (75 ng) and firefly (25 ng) reporters were transfected into HEK293 cells. Forty-eight hours later, luciferase activity was measured with Dual-Glo reagent using a Glomax Multi+ luminometer (Promega). Relative light unit values were used to calculate a relative response ratio (RRR) by dividing the Renilla value from each well by the corresponding firefly value. Percent repression was then calculated as: (%) = 100 × (1 − RRRvariable/RRRcontrol), where RRRcontrol equals RRR RnLUC 3xPRE mt or RnLUC. A minimum of three replicates were used to calculate mean values and mean ± S.E. All results were verified in multiple independent experiments. Dual luciferase assays from Drosophila cells were performed as previously described (63).
RNA Interference
PUMs were knocked down in HEK293 cells using On-target Plus Smartpool siRNAs for PUM1 (L-014179-00), PUM2 (L-014031-02), GAPDH, or nontargeting control siRNAs (Dharmacon). HEK293 cells (2 × 104 cells per well) were plated into a 96-well plate. After 24 h, cells were transfected with 10 fmol of siRNAs using Dharmafect-1 (Dharmacon). After 48 h, reporters were transfected using FuGENE HD. Twenty-four hours later, cells were analyzed by Dual-Glo assay or whole cell lysates were prepared for Western blot analysis in TNEM (50 mm Tris-HCl, pH 8.0, 0.5% (v/v) Nonidet-P40, 0.5 mm EDTA, 2 mm MgCl2) with 150 mm NaCl and protease inhibitors (1 mm PMSF, 50 μg/ml of aprotinin, 50 μg/ml of pepstatin, 50 μg/ml of leupeptin).
RNAi in D.mel-2 cells was performed as previously described (63) using dsRNAs transcribed from PCR templates generated with the following oligonucleotides: LacZ control, forward primer, 5′-GGATCCTAATACGACTCACTATAGGGTGACGTCTCGTTGCTGCATAAAC, and reverse primer, 5′-GGATCCTAATACGACTCACTATAGGGGGCGTTAAAGTTGTTCTGCTTCATC, Pop2, forward primer, 5′-GGATCCTAATACGACTCACTATAGGGGGACACCGAGTTTCCAGGCG, reverse primer, 5′-GGATCCTAATACGACTCACTATAGGGGAAGAAGGCCATGCCCGTCAGC, Ccr4, forward primer, 5′-GGATCCTAATACGACTCACTATAGGGGGAAGTACGTCGATGGCTGTGC, reverse primer, 5′-GGATCCTAATACGACTCACTATAGGGCGAACGTATAGTTGGTGTGCGGCATT. The T7 promoter sequence is underlined and the gene specific region is in bold.
RNAi of Pop2 and Ccr4 was confirmed by measuring depletion of Halotag-deadenylase fusions. D.mel-2 cells were transfected with 100 ng of pIZ HT-Pop2 or pIZ HT-Ccr4 with 100 ng of control pIZ HT. 1 ml of cell suspension was harvested and lysed for 1 h on ice in TNEM with 150 mm NaCl. HT was labeled with fluorescent Halotag ligand, TMR (Promega), for 30 min. Lysates were analyzed by SDS-PAGE and fluorescence detected with a Typhoon Trio fluorescence imager (GE Healthcare). Depletion was calculated relative to samples treated with LacZ control dsRNA and normalized to HT internal control.
Coimmunoprecipitations
Plasmids expressing FLAG-tagged human PUM1 and PUM2 were transfected into 6 × 105 HEK293 cells with HT fusions of CNOT6, CNOT6L, CNOT7, or CNOT8. Cells were lysed in TNEM with 150 mm NaCl and protease inhibitors. HT fusions were labeled with TMR ligand and treated with 10 units of RNase ONE and 4 μg of RNase A (Promega). Extracts were then bound overnight with end-over-end rotation at 4 °C to pre-equilibrated anti-FLAG beads (Sigma). Beads were washed twice with TNEM with 250 mm NaCl and once with TNEM with 500 mm NaCl. Bound protein complexes were eluted with FLAG peptide (Sigma) at 4 °C and passed over Micro Bio-Spin columns (Bio-Rad) to collect eluates. Eluates were analyzed by SDS-PAGE and fluorescence emission at 580 nm on a Typhoon Trio to detect TMR-labeled Halotag fusions. Western blots were performed and probed with a monoclonal anti-FLAG M2 antibody (Sigma).
Western Blotting
HEK293 cells were lysed in TNEM with 150 mm NaCl and protease inhibitors. D.mel-2 lysates were prepared from 1 ml of cell suspension in 75 μl of TNEM with 150 mm NaCl and protease inhibitors, lysed on ice for 1 h, and centrifuged to remove cell debris. Lysates were analyzed by Western blotting with anti-V5. PUM1 antibodies were from Bethyl Laboratories (A300 201A) and Abcam (80216). PUM2 (K-14) antibody was from Santa Cruz Biotechnology (sc-31535) or Bethyl Laboratories (A300–202A). Antibody to GAPDH was obtained from Applied Biosystems (AM4300). T7 tag antibody was from Novagen (69522-3). V5 antibody was from Invitrogen (37-7500). HRP-conjugated secondary antibodies were obtained from Thermo Scientific (anti-Mouse IgG, 31430) and KPL (anti-goat IgG, 14-13-06, and anti-rabbit IgG, 074-1516).
Purification of Recombinant Proteins
To purify PUM1 (aa 828–1176) and PUM2 (aa 705–1050) RNA binding domains and control CNOT6, pFN18A-based plasmids (Promega) encoding Halotag fusions of each protein were introduced into KRX Escherichia coli strain (Promega) and induced with 0.1% (w/v) rhamnose for 12 h at 20 °C. Proteins were purified using Halolink resin (Promega). Beads were washed extensively with TNEM and 1000 mm NaCl and then equilibrated in TNEM with 250 mm NaCl. To confirm purification of the respective proteins, AcTEV protease (Invitrogen) was used to cleave CNOT6, PUM1 RBD, and PUM2 RBD from an aliquot of the Halolink beads. The eluted proteins were analyzed by Coomassie-stained SDS-PAGE (Fig. 4B). The remaining Halolink bound proteins were used for Halotag pulldown assays. pMAL plasmids (New England Biolabs) encoding maltose-binding protein (MBP)-tagged CNOT6, CNOT7, and CNOT8 were transformed into the BL21 Gold E. coli strain and induced with 0.3 mm isopropyl 1-thio-β-d-galactopyranoside for 16 h at 20 °C. Proteins were purified with the amylose affinity resin (New England Biolabs). Beads were washed three times with TNEM and 1000 mm NaCl and 1 mm DTT and three times with deadenylation buffer (50 mm Tris, pH 8.0, 1 mm MgCl2, 50 mm NaCl, 20% glycerol, and 1 mm DTT). Proteins were eluted with 10 mm maltose in deadenylation buffer.
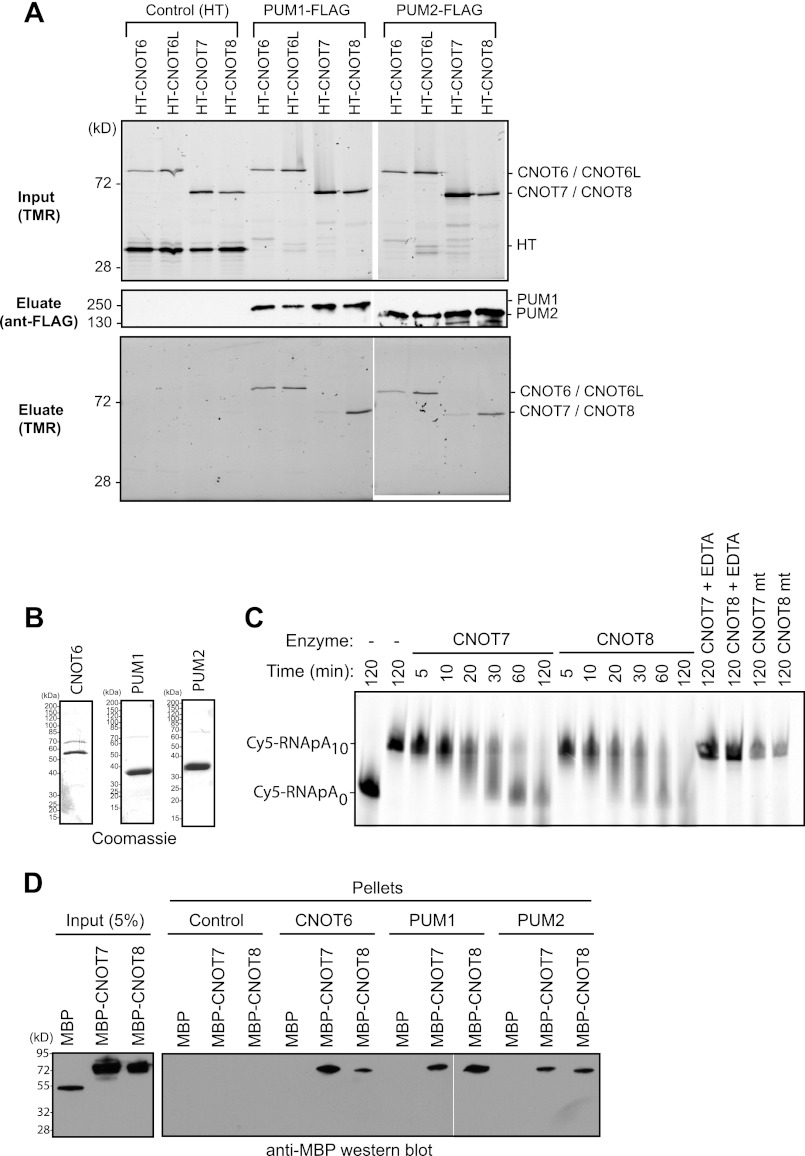
PUM1 and PUM2 interact with deadenylase subunits of the CNOT complex. A, deadenylases, fused to Halotag, coimmunoprecipitate (Eluate) with FLAG-tagged PUM1 and PUM2 from RNase-treated extracts (Input). As a negative control (Control), mock immunoprecipitations were performed with anti-FLAG beads from samples expressing Halotag (HT) protein and Halotag deadenylase fusion proteins. Proteins were detected in input extracts or purified FLAG eluates by fluorescence labeling with the Halotag ligand TMR or by anti-FLAG Western blot. B, Coomassie staining of recombinant, purified bait proteins: CNOT6, PUM1, and PUM2. PUMs were active for RNA binding (Fig. 1B). C, in vitro deadenylation assay using wild-type CNOT7 and CNOT8 or mutant CNOT7 mt and CNOT8 mt with Cy5-labeled RNA substrate with a 10-nucleotide poly(A) tail (Cy5-RNApA10) or, as a marker, substrate lacking a tail (Cy5-RNApA0). EDTA was added as a negative control to chelate Mg2+ and thus inhibit deadenylation. D, Western blot (anti-MBP) of in vitro binding of recombinant, purified PUM1 and PUM2 to CNOT7 and CNOT8. Halolink bound PUM1 and PUM2 were incubated with MBP fusions of CNOT7 or CNOT8. As a positive control, CNOT7 and CNOT8 interacted with CNOT6. Halolink beads alone (Control) and MBP served as negative controls.
In Vitro Deadenylation Assays
Deadenylase activity of purified wild-type or mutant CNOT7 and CNOT8 enzymes was confirmed by incubating 1 μm of each enzyme with 200 fmol of a 36-nucleotide RNA substrate with a 5′ Cy5 fluorescent label and, on the 3′ end, a 10-nucleotide poly(A) tail (see PRE RNA sequence below) in 20 μl of deadenylation buffer (64). Control reactions contained 10 mm EDTA to chelate Mg2+. Reactions were incubated at 30 °C for up to 120 min. An equal volume of 98% formamide and 20 mm EDTA was added, the samples were heated to 95 °C for 5 min and then resolved on a 10% polyacrylamide, 7 m urea gel. Products were detected using a Typhoon fluorescence imager.
In Vitro Binding of PUMs and CCR4-NOT Deadenylase Subunits
Recombinant prey proteins included MBP-CNOT7, MBP-CNOT8, or control MBP. For Halotag pulldown assays, 50 nm prey protein was added to 50 μl of TNEM with 250 mm NaCl and 10 μl of Halolink beads bound with HT-CNOT6, HT-CNOT7, or HT-CNOT8 bait proteins (1 μg each). Halolink beads alone served as a negative control. Binding reactions were incubated with rotation for 2 h at 4 °C. Beads were washed 4 times with 1 ml of TNEM containing 500 mm NaCl and 0.5% Tween 20. Beads were collected by centrifugation at 1000 × g for 5 min. Bound proteins were eluted in 20 μl of SDS-PAGE loading dye by heating at 95 °C for 5 min. Fifty percent of eluted proteins were then analyzed by SDS-PAGE and Western blotting using anti-MBP monoclonal antibody conjugated to horseradish peroxidase (New England Biolabs).
Gel Shift Assays
PRE RNA ligand, 5′-TTGTTGTCGAAAATTGTACATAAGCCAAAAAAAAAA, was labeled with Cy5 (Dharmacon). PRE mt RNA ligand, 5′-TTGTTGTCGAAAATACAACATAAGCCAAAAAAAAAA, was labeled with Dylight 650 (Dharmacon). RNA ligands were synthesized, deprotected, and PAGE purified prior to gel shift assays. PUM1 RBD or PUM2 RBD were allowed to bind to 200 fmol (10 nm) of RNA ligand in deadenylation buffer for 30 min at 37 °C. Samples were then analyzed on a 6% polyacrylamide gel with 1× TB running buffer at 300 volts at 4 °C. Gels were imaged with a Typhoon Trio.
Purification of PUMs and Mass Spectrometry
Halotag, HT-PUM1, or HT-PUM2, expressed from plasmid pFN21A, were purified using the Halotag Mammalian Pulldown system (Promega). T150 flasks were transfected with each plasmid and after 48 h, cells were washed with phosphate-buffered saline (PBS), and harvested at 2000 × g at 4 °C. Cells were suspended in 1 ml of Mammalian Lysis Buffer with Protease Inhibitor Mixture (Promega). Cells were passed through a 25-gauge needle 5 times, incubated for 5 min at 4 °C, and then centrifuged for 5 min at 14,000 rpm. Halolink beads were then diluted with TBS (100 mm Tris-HCl, pH 7.5, and 150 mm NaCl) and incubated with the cell extract for 15 min at room temperature with rotation. Beads were washed three times with 10 ml of TNEM with 250 mm NaCl, followed by three washes with the same buffer lacking IGEPAL. Proteins were eluted with 10 units of AcTEV protease (Invitrogen) in 20 mm Tris-HCl, pH 8.0, and 300 mm NaCl. Peptides were prepared from each sample as follows. First, disulfide bonds were reduced with 2 mm DTT at 37 °C for 30 min and blocked with 4 mm iodoacetamide at 23 °C for 30 min in the dark. The blocking reaction was quenched by bringing the final concentration of DTT to 4 mm. Next, sequencing grade trypsin (Promega) at a 1:50 (mass:mass) enzyme to sample ratio was added and incubated overnight at 37 °C. Peptides were then analyzed using nanoflow liquid chromatography (Waters) coupled to an ETD-enabled hybrid linear ion trap-orbitrap mass spectrometer (Thermo Scientific) via electrospray (65). Separation and data-dependent sampling conditions were used as previously described (66, 67). Post-acquisition data processing was performed using a DTA generator and the COMPASS software suite as previously described (68). Protein identifications were assigned by searching the human International Protein Index database with the peptide mass spectra from two independent analyses using the open mass spectrometry search algorithm (OMSSA) (67, 69). A false discovery rate threshold of 1% was applied to filter false positive identifications (67, 70). To eliminate contaminants that bind Halolink resin or Halotag, an identical analysis was performed on control Halotag purifications. All proteins detected in both the control and PUM complexes were excluded.
RNA Purifications and cDNA Preparation
RNA was purified from HEK293 cells harvested 48 h after transfection using the Maxwell 16 simplyRNA LEV cells kit and a Maxwell 16 instrument (Promega). RNA was eluted in 50 μl of nuclease-free water and treated with Turbo DNase (Ambion).
For first strand cDNA synthesis, RNA (1000 ng) was annealed with random hexamers (500 ng) (IDT) at 70 °C for 5 min and cooled on ice. Reverse transcription was performed in reaction buffer with 3 mm MgCl2, 500 mm each dNTP, 0.5 μl of RNasin Plus, and 1 μl of GoScript reverse transcriptase (Promega). RT was omitted in control samples.
Quantitative PCR
Multiplexed quantitative PCR was used to detect Renilla and firefly reporter mRNAs. Reactions were carried out in 25-μl reactions with the Plexor 2-step kit (Promega). 5 μl of cDNA was combined with 2× Plexor Master Mix (Promega) and 100 nm each of the fluorescent primers (Biosearch Technologies). Reactions were performed in triplicate using a CFX96 Real-time PCR instrument (Bio-Rad). The conditions used were: 1) 95 °C for 2 min; 2) 95 °C for 5 s; 3) 60 °C for 35 s. Steps 2 and 3 were repeated a total of 40 cycles. Each reaction was subjected to thermal melting and curves gave single peaks with the expected melting temperature. Amplification efficiencies for each primer set were optimized at 100% efficiency. Cycle thresholds (Ct) were measured using CFX Manager software (Bio-Rad) and imported in Plexor Analysis Software (Promega). Data were analyzed by the comparative Ct method (71, 72). Ct values were measured and normalized to an internal control firefly luciferase mRNA where ΔCt = Ct,Renilla − Ct,firefly. Differences in mRNA levels were calculated using the ΔΔCt method whereby ΔΔCt = ΔCt,target − ΔCt,control. “Control” indicates RnLUC lacking PREs and “target” indicates RnLUC 3xPRE. Changes in mRNA expression are represented as fold-change values, where fold change = 2−ΔΔCt. From fold-change we calculated percent repression, which equals 100 × (1 − fold-change).
To confirm RNAi depletion of deadenylase mRNAs, quantitative PCR was carried out using GoTaq quantitative PCR (Promega). Cycling conditions were as follows: (i) 95 °C for 3 min, (ii) 95 °C for 10 s, (iii) 65 °C for 30 s, and (iv) 72 °C for 40 s. Steps ii–iv were repeated a total of 40 cycles. Negative control reactions were performed in the absence of template or reverse transcriptase. Cycle thresholds (Ct) were measured using the CFX Manager software and analyzed using the ΔΔCt method (71, 72). ΔCt was calculated by normalizing to the Rpl32 gene Ct values. We then calculated ΔΔCt as follows: ΔΔCt = ΔCt(target RNAi) − ΔCt (control RNAi).
Quantitative PCR primer sequences are as follows: Firefly: forward primer, 5′-dGATCCTCAACGTGCAAAAGAAGC-3′, reverse primer, 5′-d FAM-isoC-TCACGAAGGTGTACATGCTTTGG-3′; Renilla: forward primer, 5′-d CAL Fluor Orange 560-isoC-CGCAACTACAACGCCTACCTTC-3′, reverse primer, 5′-dCCCTCGACAATAGCGTTGGAAAA-3′; Rpl32: forward primer, 5′-dGCCCAAGGGTATCGACAACAG-3′, reverse primer, 5′-dGCACGTTGTGCACCAGGAAC-3′; Dm Pop2: forward primer, 5′-dTGGACAATGCCCTCGGCC-3′, reverse primer, 5′-dGGCCACATAGTGGTACTTCTGCACC-3′; and Dm Ccr4: forward primer, 5′-dCTCGTCATACTCGGCCTCATGG-3′, reverse primer, 5′-dCGTAAAAATGCAGGCTGGTCG-3′.
Poly(A) Selection and Northern Blot Analysis
Total RNA samples from HEK293 cells expressing RnLUC, RnLUC 3xPRE, RnLUC 3xPREmt, and FfLUC control were purified and then polyadenylated mRNA was selected from 20 μg of total RNA by the PolyAtract mRNA Isolation System (Promega). RnLUC HSL and FfLUC RNAs were reverse transcribed and amplified by quantitative PCR as described above.
For Northern blots, mRNA samples were precipitated, suspended in 5 μl of 10 mm EDTA, 10 mm EGTA, and prepared for gel electrophoresis in 5% glycerol, 1 mm EDTA, 0.025% (w/v) bromphenol blue, 0.025% (w/v) xylene cyanol FF with 1× MOPS, 3.7% formaldehyde, and 25% formamide. Samples were boiled for 5 min and separated on a 1% agarose gel with formaldehyde in 1× MOPS. Following transfer to NY+ membrane (Millipore), RNA was UV cross-linked and prehybridized in Ultra-Hyb buffer (Invitrogen) at 68 °C for at least 1 h. Northern probe templates for FfLUC and RnLUC were amplified by PCR using GoTaq (Promega) and the following primers: FF fwd, 5′-CGTGGACGAGGTGCCTAAAG and FF rev, 5′-GGATCCTAATACGACTCACTATAGGTTACACGGCGATCTTGCCGC; Rn fwd, 5′-CAAGCCCGACGTCGTCCAGATT, Rn rev, 5′-GGATCCTAATACGACTCACTATAGGTTACTGCTCGTTCTTCAAGCACGC.
Reverse primers contained the T7 promoter sequence. Riboprobes were transcribed with [α-32P]UTP using T7 MaxiScript kit (Invitrogen) and purified by Sephadex G-25 columns. Blots were hybridized with probes overnight at 68 °C with rotation. Blots were washed twice for 15 min with 2× SSC (300 mm NaCl, 30 mm sodium citrate) + 0.1% SDS and twice for 30 min with 0.1% SSC (15 mm NaCl, 1.5 mm sodium citrate) + 0.1% SDS. Blots were exposed to a phosphorimager screen and visualized with a Typhoon Trio.
RESULTS
Human PUM1 and PUM2 Reduce Protein Expression and mRNA Levels
To study regulation by PUMs, we developed a luciferase reporter assay that recapitulates sequence-specific repression. Three binding sites for PUM1 and PUM2, designated PUM response elements (PRE), were inserted into a minimal 3′ UTR of an mRNA encoding Renilla luciferase (RnLUC 3xPRE, Fig. 1A). This PRE sequence UGUACAUA is a high affinity binding site for PUM1 and PUM2 (7). As a control for specificity, the UGU sequence of the PRE, which is crucial for PUM binding, was mutated to ACA to disrupt PUM binding (Fig. 1A, RnLUC 3xPREmt). Electrophoretic mobility shift assays confirm that PUM1 and PUM2 bind to the PRE with nearly equivalent affinity (Fig. 1B). Importantly, neither PUM bound the PREmt (Fig. 1B). As an additional control, a Renilla luciferase reporter lacking PRE sequences was tested (Fig. 1A, RnLUC).
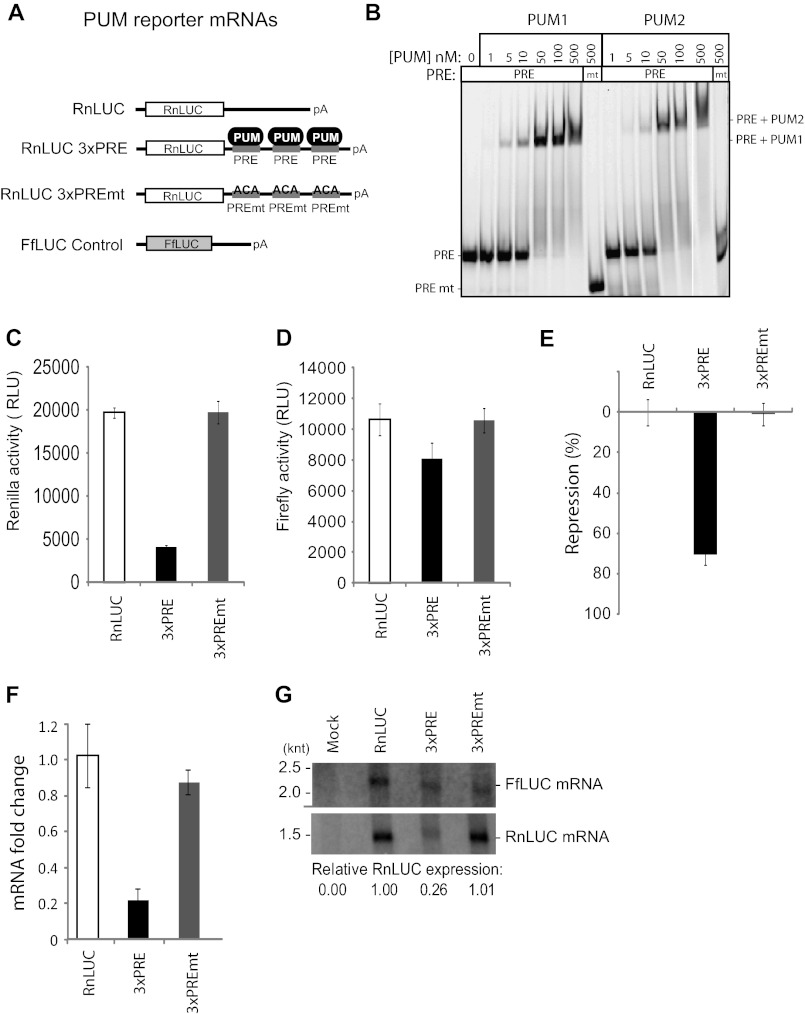
Repression by human PUM1 and PUM2 reduces protein and mRNA levels. A, a luciferase reporter assay was developed to study PUM repression in human cells. Three PRE were inserted into the 3′ UTR of RnLUC to create the reporter RnLUC 3xPRE. As a control, those sites were mutated in RnLUC 3xPREmt to block PUM binding. FfLUC was cotransfected as an internal control. B, gel shift assay showing that PUM1 and PUM2 bind to PRE RNA with nearly equal affinity, but do not bind the mutant PRE (PREmt). PRE RNA was labeled at the 5′ end with Cy5 fluor, whereas PREmt had a Dylight650 fluor, accounting for the difference in mobilities. C, graph of Renilla reporter activity. Relative luminescence units (RLU) are normalized to the activity of the transfection efficiency control, FfLUC, shown in D. E, percent repression of luciferase expression from RnLUC 3xPRE and RnLUC 3xPREmt, calculated relative to RnLUC. F, reporter mRNA levels were measured by multiplexed qRT-PCR and fold-changes, relative to RnLUC and normalized to FfLUC, are plotted. G, Northern blot detection of Renilla reporters and the control, FfLUC, from equal amounts of poly(A) selected mRNA. Relative expression levels were calculated by normalizing Renilla mRNA in each sample to the FfLUC mRNA in that sample. mRNA from mock transfected cells demonstrates specificity of probes. Mean values are graphed and mean ± S.E. is indicated.
Each reporter was transfected into the human HEK293 cell line. As an internal control, a plasmid encoding firefly luciferase was cotransfected (Fig. 1A, FfLUC). Expression of each luciferase was subsequently measured (Fig. 1, C and D). Renilla expression from RnLUC 3xPRE was substantially repressed relative to RnLUC 3xPREmt or RnLUC (Fig. 1C). To normalize variations in transfection efficiency, the Renilla activity for each sample was divided by the corresponding firefly luciferase activity (Fig. 1D). From these values, we calculated a percent repression value, as a measure of PUMs repressive activity (Fig. 1E). The presence of the PRE elements in RnLUC 3xPRE elicited 71% repression relative to control reporters (Fig. 1E), indicating potent, specific repression by endogenous PUM1 and/or PUM2.
Having established that PRE-dependent repression reduces protein output, we wished to determine whether the effect is manifested by changes in mRNA level; therefore, we purified RNA and performed multiplexed quantitative reverse transcription-polymerase chain reaction (qRT-PCR) to measure levels of reporter mRNAs (Fig. 1F). RnLUC Ct values were normalized to the internal control, FfLUC, to yield a ΔCt value (71, 72). From ΔCt values we calculated fold-change for each sample, relative to negative control RnLUC (71, 72). The fold-change of RnLUC 3xPRE mRNA was 0.22, indicating it was reduced by 78% relative to RnLUC mRNA (Fig. 1F, 3xPRE). Consistent with repression by PUMs, mutation of the PREs alleviated regulation (Fig. 1F, 3xPREmt). Northern blotting was then performed using purified mRNA to visualize reporter transcripts (poly(A) affinity purification was necessary for detection). Detection of FfLUC served as an internal control. Quantification of the data revealed that RnLUC 3xPRE mRNA was reduced 74% relative to RnLUC and RnLUC 3xPREmt (Fig. 1G), concordant with qRT-PCR results (Fig. 1F). Together, these findings demonstrated that PUM repression of the PRE bearing reporter substantially reduces protein and mRNA levels, and the reporters provide sensitive sensors for post-transcriptional repression by PUMs.
Both PUM1 and PUM2 are expressed in HEK293 cells (Fig. 2A). To demonstrate that the PRE-dependent repression is caused by endogenous PUM1 and PUM2, each protein was depleted by RNA interference (RNAi). Transfection of nontargeting control siRNAs had no effect on PUM expression (Fig. 2A, Control). Treatment with siRNAs corresponding to PUM1 or PUM2 efficiently depleted the respective proteins (Fig. 2A, PUM1 and PUM2). Treatment of cells with both PUM1 and PUM2 siRNAs substantially depleted both PUM1 and PUM2 (Fig. 2A, PUM1 + PUM2).
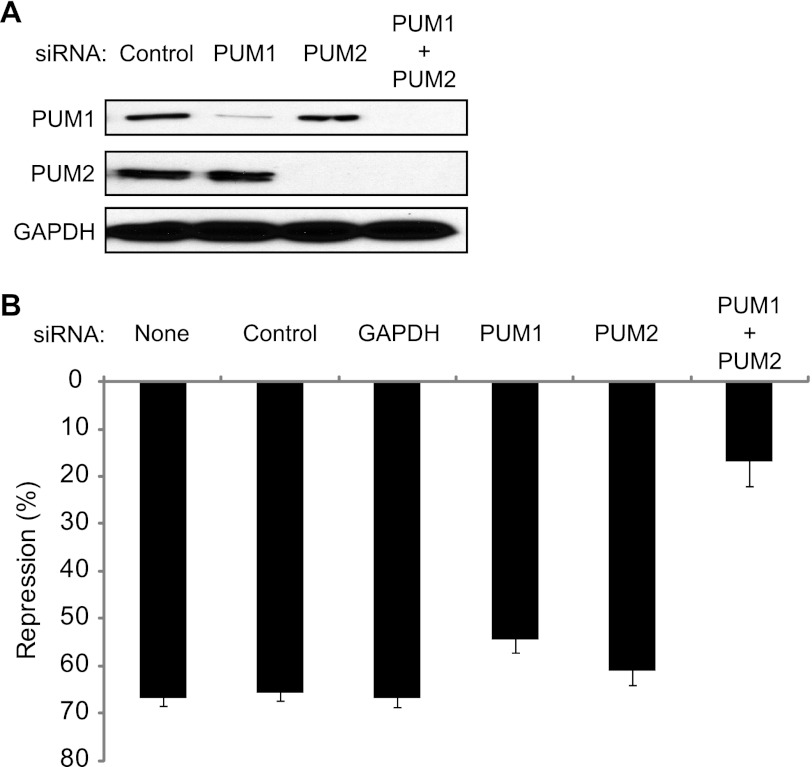
PUM1 and PUM2 repress RnLUC 3xPRE. A, endogenous PUM1 and PUM2 were depleted from HEK293 cells using siRNAs, as indicated at the top. Control indicates nontargeting control siRNAs. Western blot of PUMs using specific antibodies. GAPDH Western blot on the same samples served as a loading control. B, graph of percent repression of RnLUC 3xPRE, calculated relative to RnLUC, in samples treated with the siRNAs indicated at the top. None designates that the samples were mock transfected without siRNAs. Control indicates nontargeting control siRNAs. Mean values are graphed and mean ± S.E. is indicated.
We then measured the effect of depletion of PUM1, PUM2, or both on reporter expression. The control siRNAs had no effect on repression of RnLUC 3xPRE (Fig. 2B, 65% repression) relative to mock transfection without siRNA (Fig. 2B, None). Likewise, transfection of siRNAs to GAPDH had no effect on repression (Fig. 2B, GAPDH). Depletion of each PUM individually caused a modest loss of repression (Fig. 2B). Depletion of both PUM1 and PUM2 together substantially reduced PUM repression to only 15% (Fig. 2B, PUM1 + PUM2). We conclude that both PUMs repress the PRE-bearing reporter. We also tested the impact of overexpression of PUMs but did not observe enhancement of repression (data not shown), indicating that PUM expression is not limiting. Together, these observations indicate that both PUM1 and PUM2 cause PRE-dependent repression, and that they have overlapping regulatory roles. The results in Figs. 1 and and22 validate the specificity and sensitivity of the PUM repression assay.
PUM1 and PUM2 Repress Individually
Having shown that PUMs have overlapping capabilities to repress, we next assessed whether PUM1 and PUM2 individually exhibit repressive activity. To do so, we created a new reporter that responds to exogenously introduced PUM1 or PUM2. First, each PUM was programmed to bind a new PRE sequence (designated PRE UGG) by altering the RNA recognition amino acids of the sixth PUF repeat (R6as)(Fig. 3A) (7, 63). Importantly, wild-type PUMs do not bind UGG efficiently (7, 11). A corresponding reporter, RnLUC 3xPRE UGG, was created by changing the nucleobase at position 3 of the PRE from uracil to guanine (Fig. 3, A and B). The reporters were then transfected into cells and regulation by endogenous PUMs or by PUM1 with altered specificity (PUM1 R6as) was measured. PUM1 R6as, fused to Halotag, was expressed from a transfected plasmid. As a control, a plasmid expressing only Halotag protein was introduced. As observed in Fig. 1, endogenous PUMs repressed the RnLUC 3xPRE but, importantly, did not affect RnLUC 3xPRE UGG, nor the negative controls RnLUC or RnLUC 3xPREmt (Fig. 3C, Halotag). Expression of PUM1 R6as specifically repressed the RnLUC 3xPRE UGG reporter by 64% (Fig. 3C). PUM1 R6as did not change repression of RnLUC 3xPRE by endogenous PUMs, nor did it regulate RnLUC or RnLUC 3xPREmt (Fig. 3C). Next, we compared the repressive activity of PUM1 or PUM2 using the RnLUC 3xPRE UGG. PUM1 R6as repressed the reporter by 75% and PUM2 R6as was repressed by 69% (Fig. 3D), relative to the Halotag control. We conclude that PUM1 and PUM2 can independently repress mRNAs, and that PUMs can be programmed to specifically repress new target mRNAs.
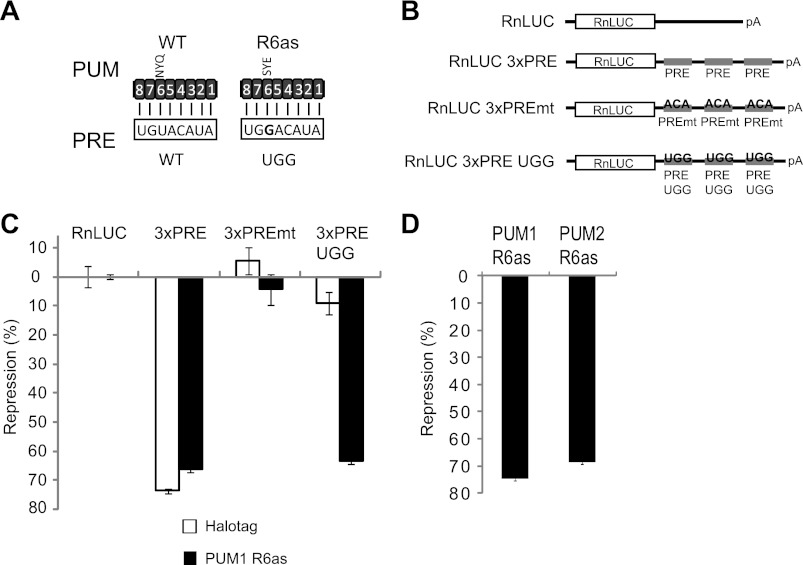
PUM1 and PUM2 repress individually. A, wild-type PUMs bind to the wild-type (WT) PRE sequence. Altered specificity PUMs (R6as) were created by changing RNA recognition amino acids of repeat 6 from NYQ to SYE, thereby binding to the altered PRE with a UGG sequence. Numbers indicate the PUF repeats, aligned to corresponding PRE ribonucleotide. B, diagram of RnLUC reporters with three PREs (RnLUC 3xPRE), with mutant PREs (UGU changed to ACA) that cannot bind PUMs (RnLUC 3xPREmt) or reporter that is bound by altering specificity PUMs (RnLUC 3xPRE UGG). C, regulation of each reporter calculated as percent repression relative to RnLUC reporter. Endogenous PUM1 and PUM2 repress RnLUC 3xPRE but not RnLUC 3xPRE UGG (Halotag samples). Halotag was expressed as a negative control. PUM1 R6as, expressed as a Halotag fusion, specifically represses RnLUC 3xPRE UGG. D, PUM1 R6as and PUM2 R6as repressed the RnLUC 3xPRE UGG reporter. Both PUM1 R6as and PUM2 R6as proteins were expressed as fusions to Halotag. Percent repression was calculated relative to reporter expression in samples transfected with Halotag control. Mean values are graphed and mean ± S.E. is indicated.
PUM1 and PUM2 Interact with CCR4-NOT Deadenylase Complex Subunits
We hypothesized that PUM1 and PUM2 may recruit co-repressor proteins to mediate repression. PUM complexes had not been previously biochemically analyzed. To identify co-repressors, we purified PUM1 and PUM2 complexes and identified associated proteins. First, PUMs were expressed in HEK293 cells as fusions to Halotag and affinity purified. Purified complexes were eluted and tryptic digests were then analyzed by nanoflow reversed-phase liquid chromatography and electrospray ionization using a hybrid linear ion trap-orbitrap mass spectrometer. Peptide sequences and protein identifications were assigned by use of high accuracy mass spectral data (<10 ppm mass measurement) with a 1% false discovery rate cut off (67, 70). To eliminate false-positives, an identical analysis was performed on control Halotag purifications; proteins detected in both the control and PUM complexes were excluded as contaminants. As a result of this analysis, multiple subunits of the CCR4-NOT (CNOT) deadenylase complex (47, 61) were detected in purified PUM complexes including CNOT1, CNOT2, CNOT4, and CNOT10 (data not shown).
Association of CNOT subunits with PUMs prompted us to investigate interaction of deadenylase enzyme subunits with PUMs. The CNOT complex interacts with heterodimers formed by pairing CNOT6 or CNOT6L with CNOT7 or CNOT8 deadenylases (47, 60–62). To analyze association of PUMs with these enzymes, FLAG-tagged PUM1 and PUM2 were expressed in cells that co-expressed Halotag fusion proteins of CNOT7, CNOT8, CNOT6, or CNOT6L. Cell extracts were prepared and treated with RNase One and RNase A to destroy RNA. Halotag fusions were fluorescently labeled with TMR fluor and detected in the cell lysates (Fig. 4A, Input). Next, PUM complexes were immunopurified using anti-FLAG monoclonal antibody and specifically eluted with FLAG peptide. Purification of PUM1 and PUM2 was confirmed by Western blot of the eluates (Fig. 4A). CNOT6, CNOT6L, and CNOT8 were strongly detected in both PUM1 and PUM2 eluates, whereas CNOT7 was weakly detected (Fig. 4A). The interactions were specific, because none of the deadenylases or the Halotag control protein associated with the anti-FLAG resin (Fig. 4A, Control). This data demonstrates that PUMs associate with CNOT deadenylase complexes. Because the PUM-deadenylase association was detected in RNase-treated extracts, protein interactions likely mediate the contacts and not RNA.
PUM1 and PUM2 Bind the CNOT7 and CNOT8 Deadenylases in Vitro
To further investigate the interaction of PUMs with deadenylase enzymes, we performed in vitro protein interaction assays. As bait proteins, recombinant Halotag fusions of PUM1 and PUM2 were purified and immobilized to halolink beads (Fig. 4B). These proteins were active in RNA binding assays (Fig. 1B). As a positive control, a Halotag fusion of CNOT6 was also purified. Recombinant CNOT7 and CNOT8, fused to MBP were then purified and used as prey proteins. First, the enzymatic activity of each deadenylase was demonstrated by deadenylating a 5′ Cy5 fluorescently labeled RNA substrate with a 10-nucleotide poly(A) tail (Fig. 4C). CNOT7 and CNOT8 progressively deadenylated the substrate over time. As a control, chelation of Mg2+ with EDTA inactivated CNOT7 and CNOT8 (Fig. 4C, EDTA). Furthermore, mutation of the magnesium coordinating residues (Asp-40 and Glu-42) within the active site of each enzyme to alanine blocked deadenylation (Fig. 4C).
Having demonstrated that CNOT7 and CNOT8 were active, we then measured binding to PUMs. Each prey was added to beads bound with CNOT6, PUM1, PUM2, or negative control beads. None of the prey proteins bound to control beads (Fig. 4D). The positive control, CNOT6, bound both CNOT7 and CNOT8, as expected (47), but not MBP (Fig. 4D). PUM1 and PUM2 bound to both CNOT7 and CNOT8, but not the control MBP (Fig. 4D). Therefore, human PUMs specifically interact with POP2 orthologs in vitro. Together with the results from co-immunoprecipitation studies (Fig. 4A), we conclude that PUMs bind either CNOT7 or CNOT8. We speculate that the preference for CNOT8 observed in Fig. 4A could result from additional factors in vivo that might modulate the interaction or differences in relative affinity. Because CNOT6 and CNOT6L bind CNOT7 or CNOT8, their co-purification with PUMs is likely the result of heterodimerization.
Deadenylation Inhibitors Alleviate PUM Repression
The observation that PUMs bind deadenylases suggests that deadenylation may be required for PUM-mediated repression. To address this hypothesis, we used the observation that mutations in the catalytic residues of deadenylases render them inactive (Fig. 4C), and when overexpressed in cells, these mutants block deadenylation in a dominant-negative manner (74–77). Therefore, we expressed mutant CNOT8 (CNOT8 mt) in which magnesium ion coordinating residues Asp-40 and Glu-42 were changed to alanine. The impact of these mutant deadenylases on PUM repression of RnLUC 3xPRE reporter was then measured. CNOT8 mt expression plasmid was transfected over a range from 0 to 85 ng (Fig. 5A). The CNOT8 mt protein was fused to Halotag to facilitate detection (Fig. 5B). A reciprocal gradient of the plasmid expressing only Halotag was used to balance transfections and Halotag alone served as a negative control. When Halotag alone was expressed (Fig. 5A, 0 ng CNOT8 mt), PUMs repressed the RnLUC 3xPRE by 77% relative to RnLUC, consistent with earlier observations (Fig. 1). Transfection of 20, 50, and 85 ng of the CNOT8 mt plasmid reduced PUM repression in a dose-dependent manner to 58, 51, and 40%, respectively (Fig. 5A). The effect of CNOT8 mt was specific to PUM repression; neither RnLUC nor RnLUC 3xPREmt reporter was affected (Fig. 5A). Dose-dependent expression of HT and HT-CNOT8 mt in these samples was confirmed by fluorescence detection (Fig. 5B). We conclude that CNOT8 mt has a dominant-negative effect that inhibits repression by PUMs.
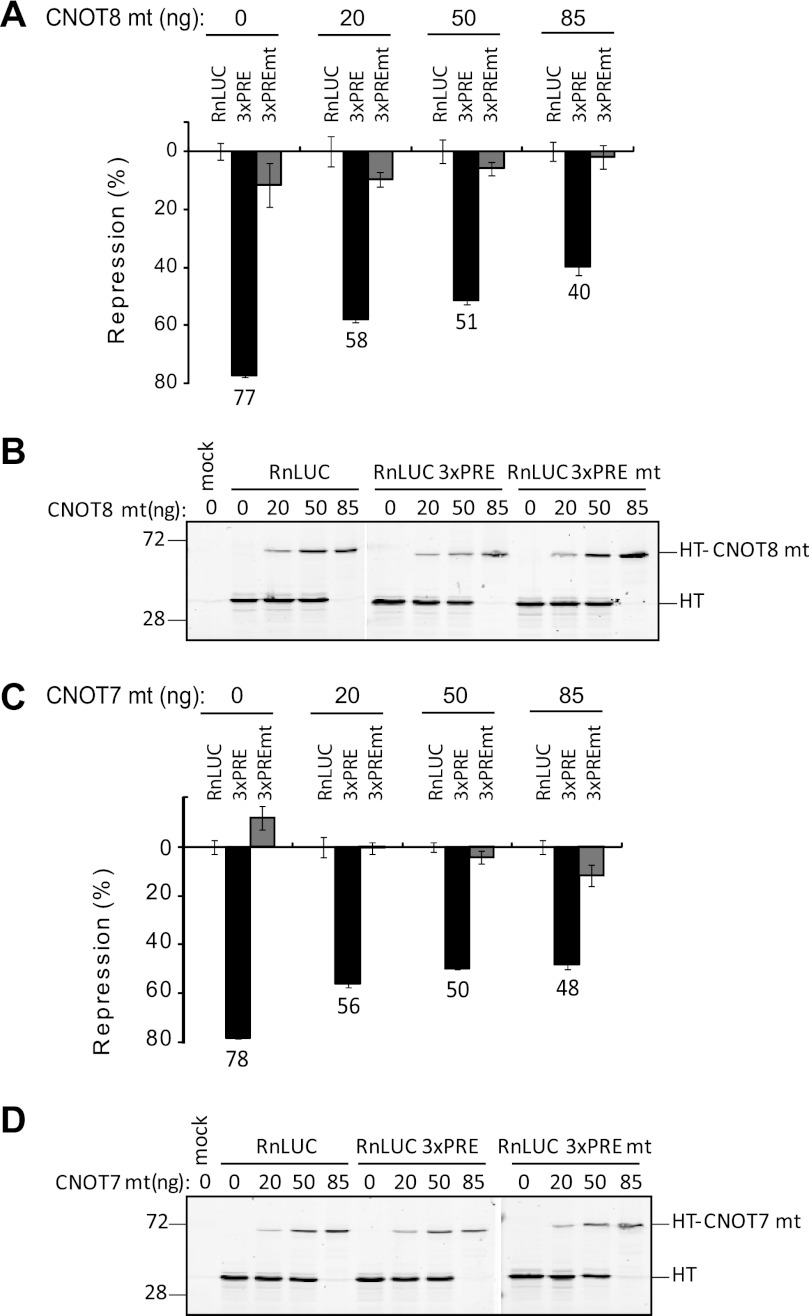
Dominant-negative deadenylases alleviate PUM repression. A, expression of a dominant-negative CNOT8 mt inhibits PUM repression in HEK293 cells. Graph of percent repression, relative to the RnLUC, of the indicated reporters in cells transfected with increasing amounts of plasmid expressing CNOT8 mt protein is shown. B, expression of the Halotag-CNOT8 mt fusion protein was confirmed by fluorescent TMR labeling and detection on a SDS-PAGE gel from the samples in panel A. Halotag alone was used to balance the transfected mass of DNA and is therefore also detected. C, dominant-negative CNOT7 mt inhibits PUM repression. Graph of percent repression relative to RnLUC control of the indicated reporters in cells transfected with increasing amounts of plasmid expressing CNOT7 mt protein. D, expression of Halotag-CNOT7 mt fusions was confirmed by TMR detection on SDS-PAGE from the samples in panel C. Halotag alone was used to balance the transfected mass of DNA and is therefore also detected. In all graphs, mean values are graphed with mean ± S.E.
We next tested the ability of a catalytically inactivated mutant CNOT7 to affect PUM repression by the same strategy. Transfection of 20, 50, and 85 ng of CNOT7 mt expression plasmid reduced PUM repression from 78 to 56, 50, and 48%, respectively (Fig. 5, C and D). Again, the effect was specific to the 3xPRE bearing reporter; RnLUC and RnLUC 3xPREmt reporters were not significantly affected. Together these results demonstrate that dominant-negative mutant deadenylases block PUM repression, indicating that deadenylation plays an important role in PUM repression.
Depletion of Deadenylases Inhibits PUM Repression
To corroborate the results above, we attempted to measure the impact of depletion of human deadenylases on PUM repression. Although we tested multiple siRNAs for each deadenylase, we were unable to substantially deplete CNOT7/8 and CNOT6/6L. Instead, we employed Drosophila D.mel-2 cells, which offer three advantages: 1) RNA interference elicited by dsRNA is highly efficient in these cells; 2) Drosophila possess one copy each of POP2 (i.e. CAF1) and CCR4 (i.e. TWIN), thus circumventing the potential redundancy of deadenylases in human cells (47, 78, 79); and 3) human PUMs actively repress in D.mel-2 cells (see below).
We first confirmed the efficacy of RNAi-mediated knockdown of deadenylases. To measure depletion of each protein, Halotag fusions of POP2 or CCR4 were co-expressed with a Halotag internal control. Cells were then treated with dsRNAs corresponding to either POP2 or CCR4 and, after 48 h, levels of the Halotag fusion proteins were measured. POP2 and CCR4 were depleted by 99 and 94%, respectively (Fig. 6A), demonstrating efficient RNAi knockdown.
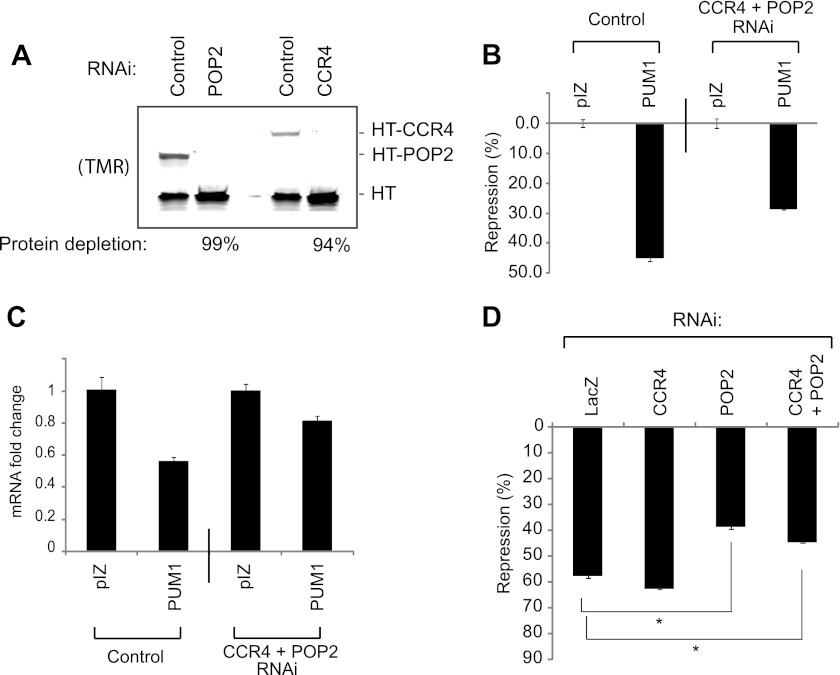
Depletion of deadenylases reduces PUM repression. A, RNAi depletion of Halotag fusions of Drosophila CCR4 (HT-CCR4) and POP2 (HT-POP2) deadenylases in D.mel-2 cells, assayed by SDS-PAGE and TMR fluorescence detection. Halotag (HT) alone served as an internal control. Percent knockdown of each protein is indicated below the figure. B, human PUM1 represses RnLUC 3xPRE reporter by 45% in D.mel-2 cells, relative to empty expression vector, pIZ. RNAi depletion of endogenous CCR4 and POP2 reduced repression to 28%. C, RnLUC 3xPRE mRNA levels were measured from samples in panel B using multiplexed qRT-PCR to determine the fold-change in mRNA levels relative to empty expression vector, pIZ. PUM1 reduced mRNA levels 44% on the control sample versus only 19% when deadenylases were depleted by RNAi. D, RNAi depletion of endogenous POP2 inhibits PUM1 repression, whereas knockdown of endogenous CCR4 does not. Nontargeting double-stranded RNA corresponding to the bacterial LacZ gene served as a negative control. Statistical significance is indicated with *, representing p < 0.0001 by a two-tailed, unpaired t test.
We then tested the ability of human PUM1 to repress RnLUC 3xPRE in D.mel-2 cells. PUM1 repressed reporter protein expression by 45% relative to the empty expression vector (Fig. 6B). Simultaneous depletion of CCR4 and POP2 reduced repression to 28% (Fig. 6B). This effect was reflected at the mRNA level: PUM1 reduced RnLUC 3xPRE mRNA by 44% (Fig. 6C) and depletion of CCR4 and POP2 alleviated PUM1-mediated reduction of the mRNA to 19% (Fig. 6C). Pop2 and Ccr4 mRNAs were depleted from these samples by 82 and 94%, respectively, as ascertained by qRT-PCR. Therefore, the POP2 and CCR4 deadenylases are necessary for efficient repression by PUM1.
We sought to determine whether the reduction in PUM1 repression was due to depletion of CCR4, POP2, or both. RNAi knockdown of CCR4 did not reduce PUM1 repression (Fig. 6D, CCR4, 58% repression), whereas PUM1 repression was significantly abrogated by knockdown of POP2 (Fig. 6D, POP2, 39% repression). This result may reflect the fact that POP2 is the predominant deadenylase in Drosophila (78, 79). This finding supports the conclusion that deadenylases are important for PUM repression, and that PUM1 can repress by recruiting the deadenylase complex via a conserved interaction with Pop2p orthologs.
PUMs Also Repress by a Poly(A) Independent Mechanism
We next asked is the poly(A) tail, and therefore deadenylation, absolutely necessary for repression by PUMs? Replication-dependent histone mRNAs lack a poly(A) tail; rather, their 3′ ends are formed by cleavage after a HSL structure (80). Translation of histone mRNAs is promoted by the 5′ cap and HSL, and degradation occurs via the 5′ decapping pathway (80). Consequently, the HSL provides a means of examining PUM repression in the absence of a poly(A) tail. We removed the cleavage/polyadenylation elements from the Renilla luciferase reporter and, in its place, inserted sequences encoding the HSL to drive 3′ end formation of the RnLUC HSL reporter (Fig. 7A). To verify that the RnLUC HSL lacked a poly(A) tail, this mRNA was expressed in cells. As a positive control, the polyadenylated RnLUC reporter was separately expressed. As an internal control, both samples also expressed the polyadenylated FfLUC mRNA. Total RNA was purified from each sample and the mRNAs were then purified using oligo(dT) magnetic beads to enrich poly(A) mRNA. Using qRT-PCR, each mRNA was detected in the poly(A)-selected fraction and normalized to the total amount. The poly(A)-selected RNA contained less than 6% of RnLUC HSL mRNA, whereas 100% of the control RnLUC mRNA was poly(A) selected (Fig. 7B). As expected, the FfLUC internal control was highly enriched in the poly(A) fraction (80–100%). These results confirm that at least 94% of the RnLUC HSL mRNA is not polyadenylated.
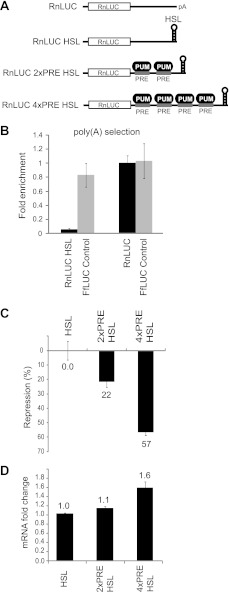
Poly(A) independent repression by PUMs. A, RnLUC reporters that lack a 3′ poly(A) tail were created by replacing the cleavage/polyadenylation sites with a HSL processing signal. Two or four PREs were inserted into the 3′ UTR to created RnLUC 2xPRE HSL and RnLUC 4xPRE HSL, respectively. B, graph of fold-enrichment of RnLUC HSL, RnLUC, and FfLUC internal control mRNAs in poly(A) selected fraction isolated using oligo(dT) affinity purification. Fold-enrichment was measured by qRT-PCR analysis of poly(A) selected mRNA, normalized to total, and calculated relative to polyadenylated RnLUC. C, graph of percent repression relative to RnLUC HSL for the indicated reporters showing that endogenous PUMs repress 2x and 4xPRE HSL reporters. D, graph of fold-change in reporter mRNA levels measured by multiplexed qRT-PCR and calculated relative to RnLUC HSL control.
To measure PUM repression, PREs were inserted into the 3′ UTR to create RnLUC 2xPRE HSL and 4xPRE HSL (Fig. 7A). The two and four PREs conferred 22 and 57% repression, respectively (Fig. 7C). To determine whether repression of HSL reporters by PUMs affected their mRNA level, we measured the levels of each mRNA by qRT-PCR. PUM repression did not reduce the RnLUC 2xPRE HSL reporter mRNA and, in fact, the 4xPRE HSL mRNAs was more abundant than RnLUC HSL mRNA (Fig. 7D). This indicates that PUM repression of the HSL reporters may occur at the translational level, rather than by direct activation of mRNA degradation pathways. From these data we conclude that PUMs can repress mRNAs lacking poly(A) tails and, therefore, can also repress by a deadenylation independent mechanism.
DISCUSSION
Our results demonstrate that both human PUM1 and PUM2 are potent repressors that reduce levels of target mRNAs and cause a corresponding decrease in protein expression (Fig. 1). Endogenous PUMs have overlapping function and act redundantly to repress protein expression (Fig. 2). We show that human PUM1 and PUM2 repress autonomously and can be programmed to regulate new mRNAs, which offers potential therapeutic value for developing designer PUMs to reduce expression of deleterious genes (Fig. 3) (81, 82). Furthermore, our results identify two modes of repression: deadenylation-mediated repression and a deadenylation-independent mechanism.
Our data provide the first evidence that human PUMs use deadenylase enzymes as co-repressors. PUMs physically associate with CNOT deadenylase subunits, including the four known deadenylase enzymes, CNOT6, -6L, -7, and -8 (Fig. 4), mediated by direct binding to human Pop2 orthologs, CNOT7 and CNOT8 (Fig. 4). The association of CNOT6 and -6L with PUMs is likely bridged via CNOT7 and CNOT8. Thus, we propose that PUMs recruit multiple deadenylase complexes to efficiently repress target mRNAs. The regulatory role of deadenylases in PUM repression is supported by the ability of dominant-negative CNOT7 and CNOT8 mutants to inhibit PUM repression (Fig. 5). Importantly, these dominant-negative mutants were previously shown to inhibit deadenylation when expressed in vivo (74–77, 79). Further support is provided by data showing that depletion of deadenylase enzymes reduces the magnitude of PUM repression (Fig. 6). Therefore, we conclude that deadenylation is necessary to achieve robust repression.
Removal of the poly(A) tail through concerted action of PUMs and deadenylases is anticipated to reduce translation efficiency and, at the same time, initiate degradation of the mRNA by either 5′ decapping mediated decay, 3′ decay by the exosome, or both pathways (83). This model is supported by our observation that protein and mRNA levels are concomitantly reduced by PUM repression. In accordance with this model, a previous study concluded that PUM1 promoted degradation of target mRNAs (33). It is noteworthy that we were unable to detect partially or fully deadenylated mRNAs, likely because these intermediates are unstable and low abundance.
Our results, combined with past data, indicate that PUF repression via deadenylation is a conserved mechanism. Correlations between deadenylation and PUF regulation were documented by studies in model organisms (29, 35, 38–41). Compelling evidence came from yeast, where PUFs were shown to accelerate mRNA deadenylation and degradation (42–45, 84, 85). The Ccr4p-Pop2p deadenylase is required for repression by yeast PUFs (43–45). In all cases, including human PUMs (Fig. 4), the highly conserved PUF RNA binding domain was sufficient for interaction with Pop2p orthologs (35, 43, 45, 86); therefore, the PUF repeat domain is likely responsible for deadenylation-mediated repression.
Although deadenylases are important for PUM repression, several observations provide evidence for a second poly(A)-independent repression mechanism. First, dominant-negative CNOT7/8 mutants do not completely block repression in vivo (Fig. 5). Second, depletion of deadenylases does not fully alleviate Pum repression (Fig. 6). The third, more telling finding is that PUMs repress target mRNAs with a 3′ HSL, indicating that a poly(A) tail, and consequently deadenylation, is not absolutely essential. Taken together, these data support an additional deadenylation-independent repression mechanism.
Deadenylation-dependent and -independent mechanisms may function together to achieve maximal regulation. Indeed, the magnitude of repression of HSL mRNAs was less than that observed with polyadenylated reporter. Our results are reminiscent of a study that analyzed repression by the Drosophila PUF protein, PUMILIO, wherein embryos were injected with reporter mRNAs either bearing a poly(A) tail or lacking a tail. PUMILIO repressed the poly(A) mRNA most efficiently and, to a lesser degree, the tail-less RNA (40). Other mRNA regulators have also been reported to repress by deadenylation-independent mechanisms. For instance, artificial tethering of the miRNA effector protein GW182 or the CNOT complex can inhibit HSL reporters (87, 88), suggesting that PUM recruitment of CNOT might cause translation repression independent of deadenylation.
How do PUMs cause deadenylation-independent PUM repression? In addition to deadenylation, PUMs could activate another mRNA decay step, such as decapping; although the observation that the PRE containing HSL target mRNA were not degraded argues against this hypothesis (Fig. 7). Alternatively, PUMs might interfere with translation, supported by work in model organisms indicating that PUFs can inhibit translation (48, 51, 73). Germane to this idea, PUFs were recently reported to bind to a translation elongation factor (51). Furthermore, we recently characterized conserved repression domains in the N terminus of Drosophila and human PUMs that may elicit deadenylation independent repression (63). Future investigations will evaluate these possible mechanisms.
Acknowledgments
We thank Dr. David Turner for comments on this manuscript, Nathan Blewett and Nathan Raynard for helpful suggestions, and May Tsoi for assisting with a multitude of lab tasks.
*This work was supported in part by a grant from the University of Michigan, Center for Genetics in Health and Medicine, Genomics and Genetics, and a grant from Promega. T. L. S, spouse of A. C. G., is an employee of Promega Corporation.
3The abbreviations used are:
- PUF
- Pumilio and Fem-3 binding factor
- RBD
- RNA binding domain
- PRE
- PUM response element
- HT
- Halotag
- MBP
- maltose-binding protein
- HSL
- histone stem loop
- RnLUC
- Renilla luciferase
- FfLUC
- firefly luciferase.
REFERENCES
Articles from The Journal of Biological Chemistry are provided here courtesy of American Society for Biochemistry and Molecular Biology
Full text links
Read article at publisher's site: https://doi.org/10.1074/jbc.m112.373522
Read article for free, from open access legal sources, via Unpaywall:
http://www.jbc.org/content/287/43/36370.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Diversity and structural-functional insights of alpha-solenoid proteins.
Protein Sci, 33(11):e5189, 01 Nov 2024
Cited by: 0 articles | PMID: 39465903 | PMCID: PMC11514114
Review Free full text in Europe PMC
Single-cell total-RNA profiling unveils regulatory hubs of transcription factors.
Nat Commun, 15(1):5941, 15 Jul 2024
Cited by: 1 article | PMID: 39009595 | PMCID: PMC11251146
Regulation of the Drosophila transcriptome by Pumilio and the CCR4-NOT deadenylase complex.
RNA, 30(7):866-890, 17 Jun 2024
Cited by: 0 articles | PMID: 38627019
Chemi-Northern: a versatile chemiluminescent northern blot method for analysis and quantitation of RNA molecules.
RNA, 30(4):448-462, 18 Mar 2024
Cited by: 1 article | PMID: 38282416
PUF partner interactions at a conserved interface shape the RNA-binding landscape and cell fate in Caenorhabditis elegans.
Dev Cell, 59(5):661-675.e7, 29 Jan 2024
Cited by: 1 article | PMID: 38290520 | PMCID: PMC11253550
Go to all (125) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The RNA binding domain of Pumilio antagonizes poly-adenosine binding protein and accelerates deadenylation.
RNA, 20(8):1298-1319, 18 Jun 2014
Cited by: 54 articles | PMID: 24942623 | PMCID: PMC4105754
Human Pumilio proteins directly bind the CCR4-NOT deadenylase complex to regulate the transcriptome.
RNA, 27(4):445-464, 04 Jan 2021
Cited by: 32 articles | PMID: 33397688 | PMCID: PMC7962487
The enzyme activities of Caf1 and Ccr4 are both required for deadenylation by the human Ccr4-Not nuclease module.
Biochem J, 469(1):169-176, 06 May 2015
Cited by: 22 articles | PMID: 25944446 | PMCID: PMC4613498
Roles of Puf proteins in mRNA degradation and translation.
Wiley Interdiscip Rev RNA, 2(4):471-492, 16 Dec 2010
Cited by: 96 articles | PMID: 21957038
Review
Funding
Funders who supported this work.
NIGMS NIH HHS (2)
Grant ID: T32 GM007544
Grant ID: 5T32GM007544-33





