Abstract
Free full text

Role of ATP binding and hydrolysis in assembly of MacAB-TolC macrolide transporter
Summary
MacB is a founding member of the Macrolide Exporter family of transporters belonging to the ATP-Binding Cassette superfamily. These proteins are broadly represented in genomes of both gram-positive and gram-negative bacteria and are implicated in virulence and protection against antibiotics and peptide toxins. MacB transporter functions together with MacA, a periplasmic membrane fusion protein, which stimulates MacB ATPase. In gram-negative bacteria, MacA is believed to couple ATP hydrolysis to transport of substrates across the outer membrane through a TolC-like channel. In this study, we report a real-time analysis of concurrent ATP hydrolysis and assembly of MacAB-TolC complex. MacB binds nucleotides with a low millimolar affinity and fast on- and off-rates. In contrast, MacA-MacB complex is formed with a nanomolar affinity, which further increases in the presence of ATP. Our results strongly suggest that association between MacA and MacB is stimulated by ATP binding to MacB but remains unchanged during ATP hydrolysis cycle. We also found that the large periplasmic loop of MacB plays the major role in coupling reactions separated in two different membranes. This loop is required for MacA-dependent stimulation of MacB ATPase and at the same time, contributes to recruitment of TolC into a trans-envelope complex.
Introduction
The ATP-Binding Cassette (ABC) superfamily of proteins comprises a large number of structurally and functionally diverse transporters that carry out both uptake and export of diverse substrates in all living systems (Davidson et al., 2008). Escherichia coli MacB is the only ABC-type exporter implicated in resistance against antibiotics in this bacterium (Kobayashi et al., 2001). When overproduced in cells lacking the major multidrug efflux pump AcrAB-TolC, this transporter confers resistance against macrolide antibiotics composed of 14- and 15-membered lactones but no or weak resistance against 16-membered ones. This activity of MacB strictly depends on two accessory proteins: MacA from the Membrane Fusion Protein (MFP) family and the outer membrane channel TolC (Kobayashi et al., 2001, Tikhonova et al., 2007). Together MacAB and TolC form an ATP hydrolysis driven efflux pump spanning the inner and outer membranes of E. coli. MacAB homologs were identified in all bacteria with some of them implicated in secretion of toxins and siderophores (Zgurskaya et al., 2009, Dubern et al., 2008, Yamanaka et al., 2008). In contrast, TolC is restricted to Gram-negative bacteria and enables transport of MacAB substrates across the outer membrane.
This study is focused on the mechanism of assembly of MacAB-TolC complex. Previous structural and functional analyses demonstrated that MacB exists as a dimer and contains four domains characteristic for all ABC proteins and a distinctive 200 residues periplasmic loop (Kobayashi et al., 2003, Lin et al., 2009, Tikhonova et al., 2007). The two transmembrane (TM) domains each composed of four transmembrane α-helices, form a substrate binding and translocation pore. The two cytoplasmic nucleotide binding domains (NBDs) hydrolyze ATP and drive conformational changes in TM domains needed to translocate substrate across the membrane. As in other ABC proteins (Davidson et al., 2008), two ATP molecules are bound at the interface formed by NBDs of MacB but the mechanism of ATP hydrolysis is still an open question. In detergents and reconstituted into proteoliposomes, purified MacB possesses a basal ATPase activity with KM for ATP in a low millimolar range (Lin et al., 2009, Tikhonova et al., 2007). This ATPase activity is stimulated by association with MacA, which interacts with MacB on both sides and within the membrane.
Recent studies suggested that MacA and perhaps other MFPs are functional analogs of periplasmic solute binding proteins (PBPs) that function with ABC transporters in uptake of nutrients (Modali & Zgurskaya, 2011). Similar to PBPs, MacA seems to stimulate MacB ATPase by allowing NBDs to close and hydrolyze ATP. However, there is no structural similarity between these two families of proteins and their interactions with transporters appear to be different as well. PBPs are monomeric proteins composed of two lobes connected by one or more polypeptide chains, and the substrate binds between them (Davidson et al., 2008). Each lobe interacts with one of the TM domains of the ABC transporters, so that the substrate-binding cleft of BPs is positioned on the interface between the TM domains. The affinity of PBPs to transporters changes with the availability of substrate and conformational transitions in both PBPs and transporters are linked to hydrolysis of ATP. In contrast, MacA exists as a hexamer, which presumably forms a tunnel-like structure covering the periplasmic exit of MacB (Xu et al., 2011, Yum et al., 2009). The cytoplasmic region of MacA is limited to ten amino acid residues, seven of which are either lysine or arginine. The large periplasmic domain, linked to the cytoplasmic region by a single TM α–helix, is typical for periplasmic MFPs and contains four domains: an α–helical hairpin, a lipoyl, an α–β-barrel and membrane proximal (MP) domains. MacA mutant lacking the cytoplasmic and TM domains binds MacB but fails to stimulate its ATPase activity (Tikhonova et al., 2007). In addition, mutations in the MP domain of MacA inactivate the ATPase activity of MacAB (Modali & Zgurskaya, 2011). These results suggested that MacB ATPase is regulated by protein-protein interactions on both cytoplasmic and periplasmic sides and within the membrane.
Also in the periplasm, MacA and TolC form a dynamic complex with a high nanomolar affinity (Tikhonova et al., 2009). The periplasmic coiled coil domain of TolC protrudes halfway across the periplasm and is believed to be a docking site for the α-helical hairpin of MacA and possibly for the large periplasmic loop (LPL) of MacB. Interactions with TolC in vivo bring notable conformational changes into the MP domain of MacA (Modali & Zgurskaya, 2011), which could lead to activation of MacB ATPase. However, how ATP hydrolysis by MacAB is integrated into assembly and disassembly of the complex with TolC remains unknown. In this study, we investigated the kinetics of MacAB-TolC assembly and the role of ATP in this process.
Results
MacA binds MacB with nanomolar affinity
To characterize protein-protein interactions in MacAB-TolC complex, we used Surface Plasmon Resonance (SPR) approach, which relies on immobilization of functionally active proteins onto surfaces. For immobilization purposes, a functionally silent D643C substitution (Kobayashi et al., 2003) was introduced into the C-terminus of MacB tagged with six histidine residues (6His-tag), MacBwt, or its derivative, in which the intrinsic C56 located in the NBD of MacB was mutated into alanine. Analysis of ATPase activities of purified proteins showed that the D643C substitution had no effect on MacB activity (Fig. 1A). In contrast, MacBC56A and its double C56A/D643C derivative lost ~60% of MacB activity, indicating that C56A substitution is detrimental for MacB ATPase. The C56 amino acid residue is located eight positions down-stream from the Walker A motif of MacB, and could contribute to binding and/or hydrolysis of ATP.
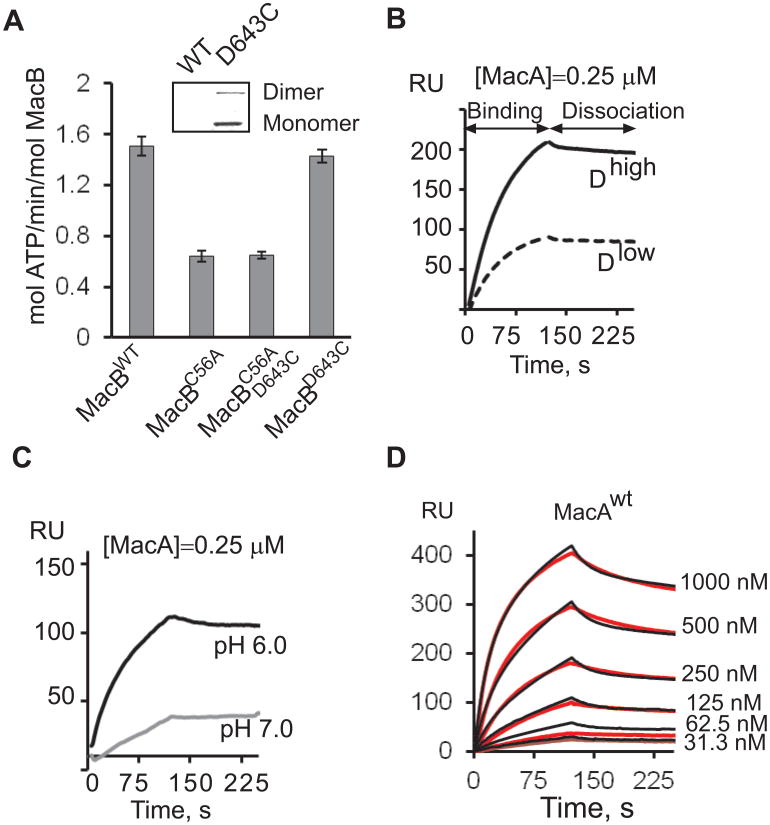
A. Specific ATPase activity of MacBwt and its derivatives measured in HEPES reaction buffer (pH 7.0). All reactions contained 0.42 μM MacB and 1mM Mg-ATP and were carried out at 37°C. Error bars are SDs (n=3). Insert: biotinylation of MacBwt and MacBD. Proteins 0.5 μg each were separated by 12% SDS-PAGE, electroblotted onto a PVDF membrane and probed with streptavidin conjugated alkaline phosphatase. Biotinylated MacBD was visualized by NBT/BCIP reagents.
B. Binding of 0.25 μM MacA to 794 RU (Dlow) and 1978 RU (Dhigh) densities of immobilized MacBD in MES (pH 6.0) running buffer. In all experiments, MacA was injected at the constant rate 50 μl/min.
C. Binding of 0.25 μM MacA to 998 RU MacBD surface in MES (pH 6.0) and HEPES (pH 7.0) running buffers.
D. Sensorgrams (red lines) of two-fold dilutions of 31.3-1000 nM MacAwt injected over MacBD (1377 RU) surface in MES (pH 6.0) buffer supplemented with 8 mM MgCl2. Sensorgrams are fit globally into TS model (black lines).
MacB variants were next treated with biotin-maleimide to determine whether the intrinsic and introduced cysteines are accessible to thiol-reactive reagents. As shown on Fig. 1A (insert), MacBD643C mutant but not MacBwt was labeled with biotin. Thus, the intrinsic C56 residue of MacBwt is not accessible to biotin-maleimide and will not be engaged during immobilization. Therefore, for binding experiments the fully functional MacBD643C (MacBD) was biotinylated and immobilized onto a streptavidin-coated SA chip. To establish the specificity of interactions, the protein was immobilized at two different densities ~1978 RU (Dhigh) and 794 RU (Dlow), which correspond to ~100 μM and ~40 μM concentrations of MacBD, respectively (Tikhonova et al., 2009). Purified MacA protein (MacAwt) was injected over MacBD surfaces in buffers of pH 7.0 and pH 6.0 containing 0.2% TX-100. Although both conditions supported MacA-MacB interactions, the specific binding response was more than twice higher at pH 6.0 than at pH 7.0 (Fig. 1C). MacAwt binding response was higher at the higher MacBD density indicating that MacAwt binding is specific to MacBD (Fig. 1B).
At pH 6.0, MacBD was functional and, similar to MacBwt, hydrolyzed about four molecules of ATP per minute (Fig. 2A, Table 1). To confirm that immobilized MacBD can bind nucleotides, doubling concentrations of ATP, ADP and AMP-PNP spanning a concentration range from 0.25 to 8 mM were injected over MacBD and protein-free surfaces (Fig. 2B). Binding of ATP generated the largest SPR signal, which decreased with decreasing densities of immobilized MacBD (data not shown). At all nucleotide concentrations, the response reached steady-state rapidly and nucleotides dissociated from MacBD completely within 5 s, indicating that the reaction is readily reversible (Fig. 2B). The shapes of binding curves and absolute responses were similar for ATP and ADP (Fig. 2C). These nucleotide-binding isotherms could not be fit into commonly used kinetic models but their shapes suggest the presence of at least two binding steps. The equilibrium of the high-affinity step is in the 0.5-2.0 mM range for both ATP and ADP, whereas the low affinity step is in a high millimolar range. The high-affinity step is comparable to 1.7-2.5 mM KM for ATP determined from ATPase activities of MacBD (Table 1). Therefore, at pH 6.0 immobilized MacBD is fully competent in binding of nucleotides and MacA.
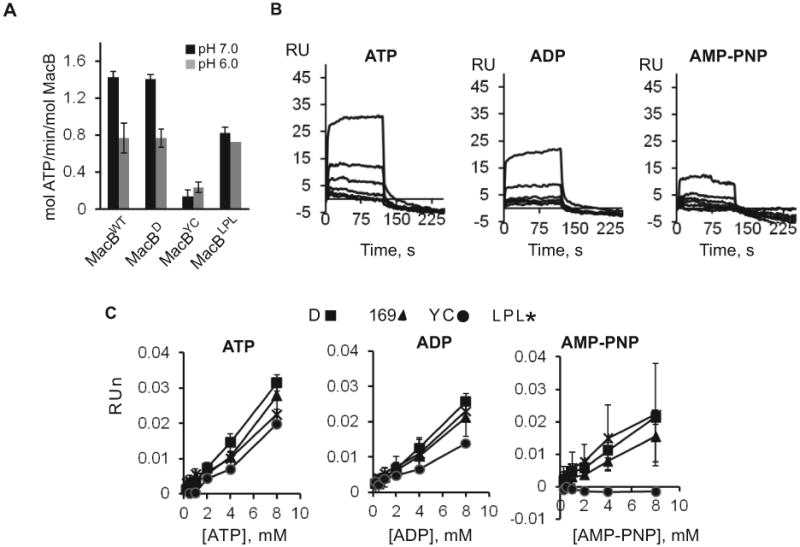
A. Specific ATPase activity of MacBwt and its derivatives in MES (pH 6.0) and HEPES (pH 7.0) running buffers measured at 20°C and 37°C. The composition of reactions was the same as in Fig. 1A.
B. Sensorgrams of doubling 0.25-8.00 mM concentrations of indicated nucleotides flowed over MacBD (1377 RU) surface in MES plus 8 mM MgCl2 running buffer (pH 6.0).
C. Equilibrium binding responses of nucleotides to MacBD (1377 RU) MacB169 (1462 RU), MacBYC (1340 RU) and MacBLPL (800 RU) plotted as functions of nucleotide concentrations. Binding responses were normalized to differences in surface densities and molecular masses of MacB variants.
Table 1
| Protein | KM (mM) | Vmax (μM/min) | kcat (s−1) | kcat/KM (M−1s−1) |
|---|---|---|---|---|
|
| ||||
| MacBWT | 1.74±0.1 | 1.82±0.05 | 0.072±0.001 | 41.5±0.57 |
| MacABWT | 1.84±0.16 | 2.29±0.08 | 0.091±0.002 | 49.4±1.08 |
| MacBD | 2.41±0.15 | 1.88±0.05 | 0.074±0.002 | 30.9±1.24 |
| MacABD | 2.11±0.14 | 2.47±0.06 | 0.098±0.001 | 46.5±0.47 |
| MacBYC | 2.5±0.32 | 0.25±0.014 | 0.01±0.0003 | 3.97±0.12 |
| MacABYC | 4.7±0.43 | 0.52±0.028 | 0.021±0.0007 | 4.39±0.15 |
To characterize kinetics of MacA-MacB association, increasing concentrations of MacAwt were injected over MacBD surface (Fig. 1D). Quantitative analysis of the curves showed that the association and dissociation phases of MacAwt could be well approximated using double-exponential rate equations, indicating multiple reaction events. The best fit of MacAwt-MacBD binding curves was obtained for the model that assumes a possible conformational change upon association. The initial association event was characterized by a moderate on-rate ~104 M−1s−1 (Table 2). The stability of the complex with the k−1 value ~10−2 s−1 exceeds by about ten folds the turnover number of MacABD ATPase (Table 1), suggesting that MacAwt-MacBD interactions remain stable during ATP hydrolysis. Derivation of the equilibrium dissociation constant given in Table 2 yields a high ~10−8 M affinity for MacAwt-MacBD complex. Thus, MacA binds MacB directly and with high affinity.
Table 2
| Surface | k1(M−1·s−1) | k−1 (s−1) | k2 (s−1) | k−2 (s−1) | KD (M) |
|---|---|---|---|---|---|
| MacBD | (2.97±0.08)×104 | (2.95±2.40) ×10−2 | (2.06±0.13)×10−2 | (10.7±1.86)×10−4 | 5.18×10−8 |
| MacBD+ATP | (4.17±0.12)×104 | (3.56±0.26)×10−2 | (1.89±0.11)×10−2 | (3.55±3.55)×10−4 | 1.61×10−8 |
| MacBD+ADP | (2.79±0.11)×104 | (2.79±0.37)×10−2 | (2.28±0.24)×10−2 | (9.29±2.93)×10−4 | 4.07×10−8 |
| MacBD+AMPPNP | (2.8±0.11)×104 | (2.64±0.37)×10−2 | (2.18±0.23)×10−2 | (8.54±2.84)×10−4 | 3.6×10−8 |
| MacBYC | (2.69±0.01)×104 | (2.59±0.15)×10−2 | (1.9±0.10)×10−2 | (8.21±1.85)×10−4 | 4.17×10−8 |
| MacBYC+ATP | (3.64±0.09)×104 | (2.92±0.20)×10−2 | (1.85±0.10)×10−2 | (4.34±1.6)×10−4 | 1.88×10−8 |
| MacBYC+ADP | (2.32±0.08)×104 | (1.91±0.27)×10−2 | (2.2±0.25)×10−2 | (11.9±2.56)×10−4 | 4.46×10−8 |
| MacBYC+AMPPNP | (2.54±0.10)×104 | (2.43±0.37)×10−2 | (2.32±0.27)×10−2 | (9.36±3.08)×10−4 | 3.86×10−8 |
| MacBD169N | (2.5±0.06)×104 | (2.48±0.24)×10−2 | (1.86±0.14)×10−2 | (7.4±2.35)×10−4 | 3.94×10−8 |
| MacBD169N+ATP | (3.56±0.10)×104 | (2.84±0.24)×10−2 | (1.9±0.13)×10−2 | (4.25±2.07)×10−4 | 1.78×10−8 |
| MacBD169N+ADP | (2.33±0.10)×104 | (2.15±0.37)×10−2 | (2.24±0.31)×10−2 | (9.48±3.68)×10−4 | 3.89×10−8 |
| MacBD169N+AMPPNP | (2.99±0.06)×104 | (3.68±0.15)×10−2 | (2.61±0.08)×10−2 | (9.29±1.02)×10−4 | 4.37×10−8 |
| MacBLPL | (3.96±0.05)×104 | (2.27±0.12)×10−2 | (1.82±0.17)×10−2 | (15.3±3.93)×10−4 | 4.81×10−8 |
| MacBLPL+ATP | (8.86±0.12)×104 | (4.64±0.16)×10−2 | (1.97±0.09)×10−2 | (9.2±1.67)×10−4 | 2.44×10−8 |
| MacBLPL+ADP | (5.17±0.01)×104 | (2.90±0.004)×10−2 | (3.31±0.15)×10−2 | (26.3±2.50)×10−4 | 4.41×10−8 |
| MacBLPL+AMPPNP | (5.07±0.09)×104 | (4.79±0.06)×10−2 | (2.27±0.05)×10−2 | (11.5±0.21)×10−4 | 4.80×10−8 |
Data were fit globally using the TS model. k−1, k−2, k1 and k2 are microscopic rate constants. Equilibrium dissociation constants (KD) were calculated from the ratio of the dissociation and association rate constants.
Construction and characterization of MacB mutants
Previous studies suggested that interactions between MacA and MacB on both sides of the inner membrane are important for stimulation of ATP hydrolysis (Tikhonova et al., 2007). To control the activity of MacB ATPase, we constructed MacB variants with alterations in both the periplasmic and cytoplasmic domains of the protein disrupting different stages of ATP hydrolysis.
A highly conserved D169 is located in the Walker B region of NBD and is proposed to play a central role in the ATP hydrolysis reaction by acting as a general base (Geourjon et al., 2001). We previously showed that D169N substitution in MacB inactivates macrolide efflux in vivo and the ATPase activity in vitro (Tikhonova et al., 2007). In agreement, MacB169 (D643C/D169N) failed to complement macrolide susceptibility of E. coli W4680AD (ΔacrAB ΔacrD) cells (Table 3) and lacked a basal ATPase activity in vitro (data not shown). In the SPR assays however, this mutant bound nucleotides with affinities comparable to those of MacBD (Fig. 2C). Thus, inactivation of the catalytic activity in MacB169 did not affect its affinity toward nucleotides.
Table 3
| Plasmid | Erythromycin (μg/ml) | Oleandomycin (μg/ml) |
|---|---|---|
| MacABWT | 32 | 78 |
| pUC18 | 1 | 2.44-4.88 |
| MacABYC | 4 | 9.75-19.5 |
| MacABLPL | 1 | 4.88 |
| MacAB169 | 1 | 2.44-4.88 |
The large periplasmic loop (LPL) between TM1 and TM2 of MacB is proposed to be the periplasmic site of interactions with MacA and possibly TolC. MacB mutant lacking this loop, MacBLPL (D643C/Δ302-512), failed to complement the macrolide hypersusceptibility phenotype of W4680AD (ΔacrAB ΔacrD) cells (Table 3). Purified MacBLPL however retained its ATPase activity in vitro (Fig. 2A). The specific activity of MacBLPL was similar to that of MacBD at pH 6.0, and was only ~40% lower than that of MacBD at pH 7.0 (Fig. 2A). In agreement, immobilized MacBLPL bound nucleotides with efficiency comparable to that of MacBD (Fig. 2C).
Finally, to disrupt binding of nucleotides, we mutated a highly conserved Y14 residue in the A-loop of MacB. In well-characterized ABC transporters, this tyrosine residue stacks against the adenine ring of ATP and provides essential contribution to the nucleotide-binding affinity of NBDs (Carrier et al., 2007, Smith et al., 2002, Zaitseva et al., 2005, Geourjon et al., 2001). W4680AD cells producing MacBYC (Y14C/C56A) mutant were partially resistant to macrolide antibiotics, suggesting that the protein retained some of its activity (Table 3). Indeed, the purified MacBYC hydrolyzed ATP, albeit about six times slower than MacBD (Fig. 2A). In SPR assays, this mutant was notably deficient in nucleotide binding. Approximately two times less ATP and ADP were bound to MacBYC than to MacBD. Furthermore, we found no AMP-PNP binding to the MacBYC surface (Fig. 2C).
We previously found that in the presence of nucleotides MacB is protected from trypsin digest, presumably due to the closure of NBDs upon nucleotide binding (Modali & Zgurskaya, 2011). We next compared proteolytic profiles of the constructed MacB variants. As shown on Fig. 3, nucleotide binding notably protected MacBD, MacB169 and MacBLPL from trypsin. At the concentration 1 μg/ml of trypsin, significant amounts of whole length proteins could be seen in the presence but not in the absence of ATP (Fig. 3A, 3B and 3D). Addition of either ADP or AMP-PNP led to similar protection of these MacB variants from trypsin, suggesting that binding of nucleotides produces the same changes in proteins (data not shown). The proteolytic pattern of MacBYC was different. This protein was more resistant than other MacB variants to trypsin digest even without ATP, as seen from the presence of the whole length MacBYC and its 60 and 50 kD fragments at 1 μg/ml trypsin (Fig. 3C). Interestingly, the proteolytic profile of MacBYC in the absence of nucleotides was similar to that of MacBD and MacB169 in the nucleotide bound states. This result suggested that NBDs of MacBYC are in a semi-open conformation, which has a reduced affinity toward nucleotides.

Trypsin (T) digestion patterns of MacBD (A), MacB169 (B), MacBYC (C) and MacBLPL (D). Purified MacB mutants, 0.42 μg, were mixed with increasing concentrations of trypsin (0.01, 0.1, 1.0, 10 μg/ml) in MES plus 8 mM MgCl2 running buffer (pH 6.0), supplemented with 4 mM Mg-ATP when indicated. Reactions were incubated 15 min at 37°C and stopped by addition of SDS-sample buffer and boiling for 5 min. MacB fragments were resolved on 12% SDS-PAGE followed by anti-MacB immunoblotting analysis. The numbers on the right are MWs of digestion products in kD calculated based on the mobility of fragments as compared to the standard protein marker (indicated on the left).
Taken together, the above results showed that MacBLPL, MacB169 and MacBYC represent the three different states of MacB transporter. The ATPase activity and nucleotide binding of MacBLPL is comparable to that of MacBD but are uncoupled from macrolide efflux. MacB169 can bind but does not hydrolyze ATP, whereas MacBYC is defective in nucleotide binding and adopts a different conformation.
Periplasmic loop of MacB is essential for MacA-dependent stimulation of MacB ATPase
We next investigated how interactions and assembly of MacAB complex are affected by the functional state of MacB ATPase. To analyze the assembly of complexes in vivo, the 6His-tagged MacB mutants were co-expressed with MacA and purified using a metal affinity chromatography. After purification, MacB fractions were probed by immunoblotting with anti-MacA antibodies to determine the presence of MacA. As shown on Fig. 4A, similar amounts of MacA are co-purified with MacBWT, MacB169, MacBYC and MacBLPL from E. coli membranes, suggesting that none of the mutations disrupted MacA-MacB interactions. In agreement, the immobilized fully functional MacBD and its compromised variants bound MacAwt with similar affinities and kinetics (Fig. 4B). Normalized binding responses of MacAwt varied depending on MacB surfaces. However, the kinetics of MacAwt –MacB interactions was very similar on all four surfaces with the variability in the rate constants and derived equilibrium dissociation constants within a ~30% range (Table 2). Thus, MacAwt binding to MacB require neither the LPL nor the catalytic activity of MacB. This result further suggests that the affinity of MacA to MacB is largely defined by interactions between TM domains of the two proteins.

A. Co-purification analysis. MacA and 6His-tagged MacB variants were co-expressed from plasmids in W4680AD (ΔacrAB ΔacrD) cells. After purification on metal affinity columns, MacB samples were separated by 12% SDS-PAGE and analyzed by immunoblotting with anti-MacB (top), anti-MacA (middle) and anti-TolC antibodies (bottom panel). MacABwt and MacBLPL dimers are indicated.
B. Sensorgrams of MacAwt (0.5 μM) injected over MacBD (1377 RU), MacB169 (1462 RU), MacBYC (1340 RU) and MacBLPL (800 RU) surfaces in MES plus 8 mM MgCl2 running buffer (pH 6.0). Sensorgrams were normalized to the differences in surface densities and molecular masses of MacB variants.
C. Folds of increase in the specific activity of MacB in the presence of the three fold molar excess of MacAwt. All reactions contained 0.7 μM MacB and 1 mM Mg-ATP and were carried out at 20°C in MES running buffer (pH 6.0). Error bars are SDs (n = 3).
D. Dependence of MacB ATPase activity on MacA. All reactions (10 μL total volume) contained 0.7 μM MacB and 1 mM Mg-ATP and were carried out at 20°C in MES running buffer (pH 6.0). Error bars are SDs (n = 3).
The mutant MacB ATPases however differed dramatically in their response to the stimulation by MacA (Fig. 4C-D). In agreement with previous data, co-reconstitution of MacBWT and MacA into proteoliposomes resulted in ~25 fold increase in the specific ATPase activity (Fig. 4C). MacBYC activity was also stimulated by MacA, both in detergent solution and in proteoliposomes (Fig. 4C-D). In contrast, less than two fold stimulation of ATP hydrolysis by MacA was detected in MacBLPL proteoliposomes (Fig. 4C). Since MacBLPL interacts with MacAwt both in vitro and in vivo, this result suggests that the LPL of MacB coordinates the MacA-dependent stimulation of ATP hydrolysis and the macrolide efflux activity of the transporter.
ATP binding increases MacA-MacB affinity
To investigate how binding and hydrolysis of ATP affect kinetics of MacA-MacB interactions, 0.5 μM MacAwt were pre-mixed with increasing concentrations of ATP, AMP-PNP and ADP and injected over MacBD, MacBLPL, MacB169 and MacBYC surfaces. Binding curves were normalized by subtracting SPR responses generated by nucleotides alone and then fitted locally to obtain a kobs parameter, which describes both the on- and off-rates (see Experimental Procedures). AMP-PNP had no effect on MacA-MacB interactions (data not shown). The presence of ADP decreased the absolute binding response of MacAwt by ~20% (Fig. 5A-B). In addition, on all MacB surfaces, the kobs of MacAwt decreased by ~15% at 1 mM concentration of ADP and then remained the same at up to 8.0 mM of ADP. In contrast, kobs values increased when MacAwt was injected along with ATP (Fig. 5A). At 0.25-1.0 mM ATP concentrations, kobs increased monotonously by ~30% and then slightly decreased at higher concentrations of ATP. The same changes in kobs values were observed on the ATPase proficient MacBD and MacBLPL as well as ATPase deficient MacB169 and MacBYC surfaces suggesting that the increase in rate constants is due to ATP binding not hydrolysis.
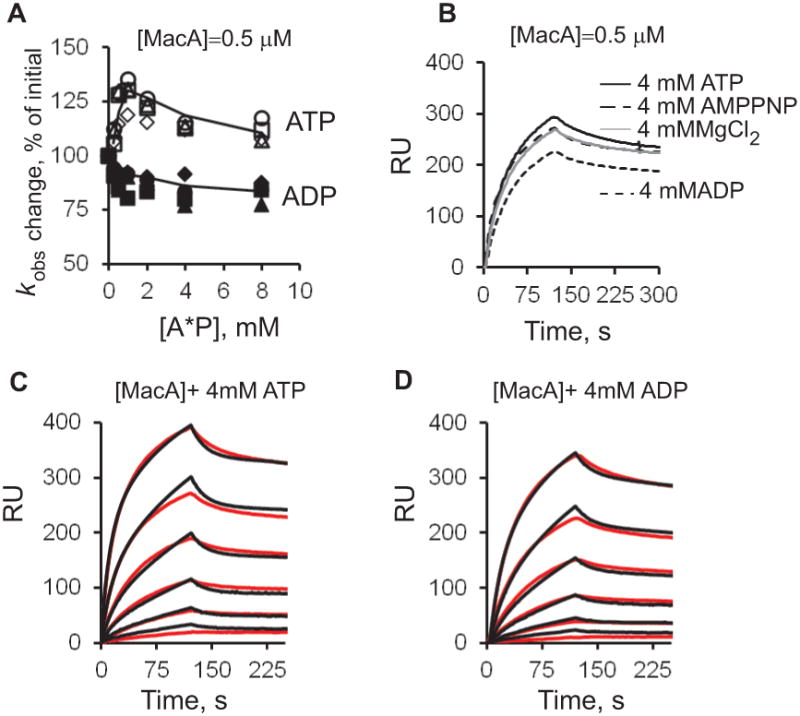
A. MacAwt (0.5 μM) was pre-mixed with two-fold dilutions 0.25-8.0 mM of ATP (open symbols) or ADP (closed symbols) and injected over MacBD (1377 RU, squares), MacB169 (1462 RU, triangles), MacBYC (1340 RU, circles) and MacBLPL (800 RU, diamonds) surfaces in MES plus 8 mM MgCl2 running buffer (pH 6.0). Sensorgrams were fit locally using the BIAevaluation software, and the kobs values expressed as percents of kobs of MacAwt without a nucleotide, are plotted against the concentration of nucleotides.
B. Sensorgrams of MacAwt pre-mixed with 4.0 mM of indicated nucleotides and injected over MacBD (1377 RU) surface.
C. Sensorgrams (red lines) of two-fold dilutions 0.031-1.00 μM of MacAwt pre-mixed with 4 mM of ATP and injected over MacBD (1377 RU). Sensorgrams are fit globally into TS model (black lines).
D. The same as D but MacAwt was pre-mixed with 4 mM of ADP.
To characterize kinetics of MacAB interactions in the presence of nucleotides, binding experiments were carried out at the constant 4 mM concentrations of ATP, ADP or AMP-PNP and increasing concentrations of MacAwt (Fig. 5B). Quantitative analysis of binding curves showed complex reactions that could be fit by double-exponential rate equations. As in the absence of nucleotides (Fig. 1D), the best fit of MacAwt-MacB binding curves was obtained for the TS model that assumes a possible conformational change upon association. However, some deviations from this model could be seen in curve alignments (Fig. 5C) indicating that the reaction could be more complex. Derivation of the equilibrium dissociation constants given in Table 2 yields the same high ~10−8 M affinity for MacAwt complex with all MacB variants in the presence of nucleotides. However, a significant up to three fold increase in affinity of the MacA-MacB interaction was found in the presence of ATP, but not other nucleotides. As with variable concentrations of nucleotides, the effect of ATP can be seen on all four MacB surfaces. Thus, ATP binding not hydrolysis increases the affinity of MacA-MacB complex. This result further suggests that MacA has a higher affinity to MacB in its closed conformation.
MacB contributes to recruiting of TolC
During ATP hydrolysis, conformational changes in MacAB are believed to be coupled to transport of substrates across the outer membrane through TolC channel. MacA is thought to play the major role in this process. We next used the affinity co-purification and SPR approaches to determine how different functional states of MacB transporter affect TolC recruitment by MacA. We previously found that chemical cross-linking is not required to detect MacAB-TolC complexes in vivo (Tikhonova et al., 2007). Therefore, the purified MacAwt expressed alone or in combination with MacB variants was probed by immunoblotting with anti-TolC antibodies. In agreement with previous results, TolC was clearly detected in MacAwt and MacA-MacBWT samples, supporting the notion that interactions with MacA bring TolC into the MacAB-TolC complex (Fig. 4A). The amounts of the co-purified TolC remained the same in MacA-MacB169 and MacA-MacBYC samples. Surprisingly, no TolC was co-purified with MacA-MacBLPL (Fig. 4A). Since MacAwt alone, but not in the complex with MacBLPL, pulls down TolC, this result suggests that the LPL is required not only for the MacA-dependent stimulation of MacB ATPase but also for the proper assembly of MacAB-TolC complex. In contrast, the inactivation of MacB ATPase does not significantly affect MacAB-TolC assembly.
To determine whether TolC can bind MacB directly, TolC mutant containing a single A269C substitution in its extracellular loop was immobilized onto a biochip surface using a direct thiol-coupling protocol (Tikhonova et al., 2009). As shown on Fig. 6A, no binding response was detected when increasing concentrations of MacBwt or MacBLPL were injected over this surface. In contrast, MacAwt bound immobilized TolC with a nanomolar affinity (Fig. 6B). Similarly, in the reverse assay, in which MacB was immobilized onto a surface and TolC was injected as an analyte, no specific interaction between MacB and TolC was detected (data not shown). Consistent with in vivo co-purification studies (Tikhonova et al., 2007), this result demonstrate that MacB and TolC are unlikely to directly interact with each other.

A. Sensorgrams of doubling concentrations of MacBwt (125-1000 nM) injected over TolC (790 RU) surface in MES plus 8 mM MgCl2 (pH 6.0) running buffer.
B. The same as in A, but doubling concentrations of MacAwt (125-2000 nM) were injected over TolC surface.
C. MacAwt (250 nM) was pre-incubated with the equimolar concentrations of either MacBwt or MacBLPL in MES plus 8 mM MgCl2 (pH 6.0) running buffer and injected over TolC surface.
D. A proposed mechanism of MacAB-TolC. Similar to other ABC transporters, MacB ATPase cycles at least through four steps: binding of ATP and substrate (1), transport of substrate (2), hydrolysis of ATP (3) and release of ADP and Pi (4). MacA has a higher affinity to MacB in the ATP-bound state and promotes MacB transition into a closed conformation for hydrolysis of ATP. The MacA-dependent stimulation of MacB and recruitment of TolC by MacA are linked together because both require interactions between MacA and the large periplasmic loop of MacB. Amino acid residues of MacB targeted in this study are indicated.
To mimic the in vivo conditions, MacA-MacBwt and MacA-MacBLPL complexes pre-assembled in various stoichiometries were injected over immobilized TolC. Since MacB does not bind TolC (Fig. 6A) an increase in MacA binding response in the presence of MacB would indicate that MacB is retained by MacA on the TolC surface. In contrast, a decrease in the binding signal would indicate that MacB interferes with MacA-TolC interactions. We found that kobs values and binding signals increased when MacA-MacBwt complexes were injected over immobilized TolC (Fig. 6C). In contrast, MacA binding to TolC was inhibited by increasing concentrations of MacBLPL.
Taken together, these results show that the large periplasmic loop of MacB has a dual function in the assembly of the functional MacAB-TolC complex. This loop is required for the MacA-dependent stimulation of MacB ATPase and is needed to assemble the tri-partite MacAB-TolC complex. This finding further suggests that the MacA-dependent stimulation of MacB ATPase is coupled to recruitment of TolC channel.
Discussion
MFP-dependent transporters are broadly represented in both gram-positive and gram-negative bacteria (Zgurskaya et al., 2009). Furthermore, in addition to ABC transporters such as MacAB, the MFP dependency is also found among members of RND (Resistance-Nodulation and cell Division) and MF (Major Facilitator) superfamilies of proteins. All three families contain transporters that do not require accessory proteins for their transport activities. Why some of the transporters became dependent on the MFP function and the molecular mechanism of this dependency remain unclear.
The main features of the catalytic cycles of ABC transporters are believed to be highly conserved (Davidson et al., 2008). These transporters cycle through at least four conformational states: (i) binding of substrate and ATP, (ii) closing of NBD dimers and transport of substrate, (iii) hydrolysis of two ATP molecules, (iv) release of ADP and Pi and transition into the resting state (Higgins & Linton, 2004) (Fig. 6D). Numerous studies of ABC importers and exporters have merged into a view that ABC transporters adopt an outward-facing conformation in the ATP-hydrolysis transition state and an inward-facing conformation in the resting state (the “alternate access model”) (Oldham et al., 2008, Dawson et al., 2007). We previously reported that the ATPase activity of MacB is stimulated by MacA and that this stimulation is linked to the ability of MacA to promote MacB transition into a closed conformation needed for hydrolysis of ATP (Modali & Zgurskaya, 2011, Tikhonova et al., 2007). The results of this study show that during the entire ATP hydrolysis cycle MacA likely remains bound to MacB. Under all tested conditions, the life-time of MacA-MacB complex exceeded the turnover number of MacB ATPase by at least ten fold (Fig. 1). The affinity remained the same for complexes with MacB mutants that are catalytically inactive or defective in binding of nucleotides, suggesting that neither ATP hydrolysis nor conformational changes of MacB during reaction affect interactions between MacA and MacB. However, ATP binding seems to increase the affinity of MacA-MacB complex by about threefold. In addition, ADP had a negative effect on kinetics of MacAB interaction, albeit only modest (Fig. 5 and Table 2). Taken together with previous studies, this result suggests that binding of nucleotides brings MacB into a conformation with a higher affinity toward MacA, whereas interactions with MacA bring transporter into a closed conformation for hydrolysis of ATP. The modest effects of nucleotides on the equilibrium and rate constants are in line with the 30-50% stimulation of MacB ATPase by MacA in detergent (Table 1 and ref. (Lin et al., 2009, Tikhonova et al., 2007)). However, in proteoliposomes, the presence of MacA increases the ATPase activity of MacB by more than 30 fold (Fig. 2A and (Tikhonova et al., 2007)). It is possible that the effects of nucleotides on kinetics of MacA-MacB interactions could be significantly amplified in the lipid environment of membranes.
The MacA-dependent stimulation of MacB ATPase appears to be directly linked to recruitment of TolC channel. MacBLPL lacking the large periplasmic loop bound MacA with the same high affinity as MacBwt (Table 2) but its ATPase activity was no longer stimulated by MacA (Fig. 4D). Furthermore, MacA bound to MacBLPL failed to recruit TolC in vivo (Fig. 4A) and less efficiently bound immobilized TolC in vitro (Fig. 6C). This result is particularly intriguing because MacA binds TolC when produced alone in the absence of MacB (Fig. 4A). This finding implies that binding to MacB changes the conformation of MacA, so that productive recruitment of TolC is possible only under certain conditions, such as MacA interaction with the LPL of MacB. In turn, binding to TolC changes conformation of the membrane proximal domain of MacA (Modali & Zgurskaya, 2011). This domain of MacA contacts the LPL of MacB and stimulates MacB ATPase (Fig. 6D).
At present the only other MFP-dependent transporter characterized in a biochemical detail is the multidrug efflux pump AcrAB-TolC (Nikaido, 2011). In this complex, AcrB belongs to the RND superfamily of transporters and functions as a trimer driven by the proton-motive force, whereas AcrA is a structural and functional homolog of MacA. The two AcrAB and MacAB efflux complexes share the same outer membrane channel TolC to export their substrates across the outer membrane. Interestingly, the equilibrium nanomolar affinities of MacA toward MacB are very similar to those of AcrAB complex (Tikhonova et al., 2011). Thus, in cells both MacB and AcrB transporters exist in complexes with their cognate MFPs. In both complexes, MFPs stimulate activities of their respective transporters (Tikhonova et al., 2007, Zgurskaya & Nikaido, 1999). Although the two complexes differ in their interactions with TolC, some mechanistic parallels could also be seen. AcrB transporter binds TolC directly and possibly participates in opening of TolC channel (Tikhonova et al., 2011 Tikhonova et al., 2000, Weeks et al., 2010). MacB does not directly interact with TolC (Fig. 6) but its LPL controls MacA-TolC interactions and links conformational changes in the transporter to the transport of substrate through TolC channel (Figs 4 and and66).
Experimental procedures
ATP hydrolysis assay
The rate of ATP hydrolysis of MacBwt and MacB mutants in detergent were measured using 0.3-0.7 μM (0.2-0.5 mg) MacB. Reactions were performed either under conditions previously used in analyses of MacB ATPase (pH 7.7 and 37°C) (Tikhonova et al., 2007) or under conditions used in the SPR experiments described below (pH 6.0 and 20°C). In the latter, the reaction buffer contained 20 mM MES-KOH (pH 6.0), 150 mM NaCl, 0.2% Triton X-100 (TX-100) and 2 mM or 8 mM MgCl2. The 32P-γ-phosphate labeled ATP was pre-mixed with unlabelled ATP-MgCl2 and then added to the reaction to 1 mM final concentration. At time points of 0, 4, 8, 12, 16 min, 1 μl aliquots were removed from the reaction and mixed with 10 μl of the stop buffer containing 50 mM Tris-HCl (pH 8.0), 20 mM EDTA (pH 8.0), 0.5% SDS, 200 mM NaCl, 0.5 mg/mL proteinase K. The samples were incubated for at least 20 min at 55°C to stop the reaction and analyzed by thin-layer chromatography as described before (Tikhonova et al., 2007). The amounts of the free Pi were quantified using Storm PhosphoImager and ImageQuant Software (Molecular Dynamics). MacAWT was pre-mixed with MacB variants and incubated for 15 min before adding into the reaction mixture.
Mutagenesis
Point mutations were introduced into MacB and TolC using a QuikChange XL site-directed mutagenesis kit (Stratagene). The pBBhis plasmid was used as a template for PCR. Substitutions were confirmed by DNA sequencing (Oklahoma Medical Research Foundation Sequencing Facility). To construct MacBLPL mutant lacking the large periplasmic loop, DNA sequences encoding the N-terminal 1-301 amino acid residues and the C-terminal 518-648 amino acid residues of MacB were amplified by PCR using pBBHis as a template. The two fragments were ligated by PCR and the DNA product was cloned into NcoI and PstI restriction sites of pBAD/Myc-HisC.
Protein purification and analyses
All MacB mutants and MacAwt containing the C-terminal six histidine (6His) affinity tags were expressed and purified from E. coli ET103 strains carrying pBBhis and pBAhis respectively as described before (Tikhonova et al., 2007). Purified proteins were separated by electrophoresis on SDS-polyacrylamide gels and visualized by Coomassie Brilliant Blue (CBB), silver nitrate staining or immunoblotting with polyclonal anti-MacB antibody (Tikhonova et al., 2007). The protein concentration was determined using a Protein DC Assay (Bio-Rad) and Bradford protein assay (Bio-Rad). Reconstitution of proteoliposomes was carried out as described before (Tikhonova et al., 2007). Previous structural studies suggested that MacA exists as an oligomer (Yum et al., 2009). In agreement, a gel filtration analysis showed that purified MacAwt is eluted from a Sephacryl S-300 column as a single symmetric peak with the apparent molecular weight exceeding that of 440 kD ferritin (data not shown). Thus, purified MacAwt used in the above experiments is a homogeneous, stable oligomer. Similarly, the gel filtration analysis showed that the purified MacBD protein is a homogeneous dimer (data not shown). MgCl2 in concentration 8 mM did not affect oligomerization of MacA and MacB.
Surface Plasmon Resonance
BIAcore 3000 biosensor and SensiQ Pioneer systems were used to characterize binding interactions. MacB mutants were biotinylated with maleimide-polyethylene glycol (PEG2)-biotin following the published procedures (Tikhonova et al., 2011). Labeled proteins were separated from the unreacted reagent by gel filtration on NAP-5 column (GE HealthCare, Sephadex G-25) followed by extensive dialysis in buffer containing 20 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.2 % TX-100 buffer. Biotinylation of MacB mutants was further confirmed by western blotting with streptavidin-alkaline phosphatase conjugate (Sigma) and nitro-blue tetrazolium chloride (NBT) and 5-bromo-4-chloro-3′-indolyphosphate p-toluidine (BCIP) salt reagents (Bio-Rad). Biotinylated proteins were immobilized onto SA biosensor chip (Biacore) in Hepes buffer (pH 7.4) as reported before (Tikhonova et al., 2011). The untreated SA surface was used as a control surface. TolCA269C was immobilized onto a COOH2 chip using a previously described thiol-coupling protocol (Tikhonova et al., 2009).
All experiments were performed in MES buffer (pH 6.0) containing 20 mM MES-KOH (pH 6.0), 150 mM NaCl, 0.2% TX-100 or Hepes buffer (pH 7.0) of the same composition except MES was substituted with 20 mM HEPES-KOH (pH 7.0). When indicated running buffers were supplemented with 8 mM MgCl2. Serial dilutions of analytes (proteins, nucleotides and MgCl2) were prepared in an indicated running buffer. The flow rate of injections was 50 μl/min. The surface was regenerated by a 2 min injection of 500 mM MgCl2 prepared in 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 0.2% TX-100 followed by a 1 min injection of the running buffer. All sensorgrams were normalized by subtracting responses from the control surface and responses of buffer injections.
The data analysis was carried out using the BIAevaluation (GE Healthsciences), Origin (OriginLab) or Excel (Microsoft) software. The best global fit for MacA-MacB interactions was obtained using the Two-State reaction (TS) model. Schematic representation and equations relevant to this model are given below:
The equilibrium dissociation constant for this model is:
where [A] is the concentration of analyte, [B] is the ligand binding capacity, [A·B] and [A·B*] are intermediate and final complexes, respectively, k−1, k−2, k1 and k2 are microscopic rate constants, and Rt is the total SPR response, which is directly proportional to [A·B] + [A·B*].
The observed rate constant kobs = ka·[A]+kd, where ka is the rate constant for association, kd is the rate constant for dissociation and [A] is the analyte concentration was determined by fitting sensorgrams locally into a simple bimolecular interaction model using BIAevaluation software.
Acknowledgments
This work was supported by National Institutes of Health Grant R21 AI092486 to HIZ. Authors declare no conflict of interest. We thank Abigail Ntreh for construction and immobilization of TolCA269C mutant.
References
- Drug resistance threatens to reverse medical progress. World Health Organizatiton; Geneva, Switzerland: 2000. [Google Scholar]
- Carrier I, Urbatsch IL, Senior AE, Gros P. Mutational analysis of conserved aromatic residues in the A-loop of the ABC transporter ABCB1A (mouse Mdr3) FEBS Lett. 2007;581:301–308. [Abstract] [Google Scholar]
- Davidson AL, Dassa E, Orelle C, Chen J. Structure, Function, and Evolution of Bacterial ATP-Binding Cassette Systems. Microbiol Mol Biol Rev. 2008;72:317–364. [Europe PMC free article] [Abstract] [Google Scholar]
- Dawson RJP, Hollenstein K, Locher KP. Uptake or extrusion: crystal structures of full ABC transporters suggest a common mechanism. Molecular Microbiology. 2007;65:250–257. [Abstract] [Google Scholar]
- Dubern JF, Coppoolse ER, Stiekema WJ, Bloemberg GV. Genetic and functional characterization of the gene cluster directing the biosynthesis of putisolvin I and II in Pseudomonas putida strain PCL1445. Microbiology. 2008;154:2070–2083. [Abstract] [Google Scholar]
- Geourjon C, Orelle C, Steinfels E, Blanchet C, Deleage G, Di Pietro A, Jault JM. A common mechanism for ATP hydrolysis in ABC transporter and helicase superfamilies. Trends Biochem Sci. 2001;26:539–544. [Abstract] [Google Scholar]
- Higgins CF, Linton KJ. The ATP switch model for ABC transporters. Nat Struct Mol Biol. 2004;11:918–926. [Abstract] [Google Scholar]
- Kobayashi N, Nishino K, Hirata T, Yamaguchi A. Membrane topology of ABC-type macrolide antibiotic exporter MacB in Escherichia coli. FEBS Lett. 2003;546:241–246. [Abstract] [Google Scholar]
- Kobayashi N, Nishino K, Yamaguchi A. Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J Bacteriol. 2001;183:5639–5644. [Europe PMC free article] [Abstract] [Google Scholar]
- Lin HT, Bavro VN, Barrera NP, Frankish HM, Velamakanni S, van Veen HW, Robinson CV, Borges-Walmsley MI, Walmsley AR. MacB ABC transporter is a dimer whose ATPase activity and macrolide-binding capacity are regulated by the membrane fusion protein MacA. J Biol Chem. 2009;284:1145–1154. [Europe PMC free article] [Abstract] [Google Scholar]
- Modali SD, Zgurskaya HI. The periplasmic membrane proximal domain of MacA acts as a switch in stimulation of ATP hydrolysis by MacB transporter. Mol Microbiol. 2011;81:937–951. [Europe PMC free article] [Abstract] [Google Scholar]
- Nikaido H. Structure and mechanism of RND-type multidrug efflux pumps. Adv Enzymol Relat Areas Mol Biol. 2011;77:1–60. [Europe PMC free article] [Abstract] [Google Scholar]
- Oldham ML, Davidson AL, Chen J. Structural insights into ABC transporter mechanism. Curr Opin Struct Biol. 2008;18:726–733. [Europe PMC free article] [Abstract] [Google Scholar]
- Smith PC, Karpowich N, Millen L, Moody JE, Rosen J, Thomas PJ, Hunt JF. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol Cell. 2002;10:139–149. [Europe PMC free article] [Abstract] [Google Scholar]
- Tikhonova EB, Dastidar V, Rybenkov VV, Zgurskaya HI. Kinetic control of TolC recruitment by multidrug efflux complexes. Proc Natl Acad Sci U S A. 2009;106:16416–16421. [Europe PMC free article] [Abstract] [Google Scholar]
- Tikhonova EB, Devroy VK, Lau SY, Zgurskaya HI. Reconstitution of the Escherichia coli macrolide transporter: the periplasmic membrane fusion protein MacA stimulates the ATPase activity of MacB. Mol Microbiol. 2007;63:895–910. [Abstract] [Google Scholar]
- Tikhonova EB, Yamada Y, Zgurskaya HI. Sequential Mechanism of Assembly of Multidrug Efflux Pump AcrAB-TolC. Chem Biol. 2011;18:454–463. [Europe PMC free article] [Abstract] [Google Scholar]
- Weeks JW, Celaya-Kolb T, Pecora S, Misra R. AcrA suppressor alterations reverse the drug hypersensitivity phenotype of a TolC mutant by inducing TolC aperture opening. Mol Microbiol. 2010;75:1468–1483. [Europe PMC free article] [Abstract] [Google Scholar]
- Xu Y, Song S, Moeller A, Kim N, Piao S, Sim SH, Kang M, Yu W, Cho HS, Chang I, Lee K, Ha NC. Functional implications of an intermeshing cogwheel-like interaction between TolC and MacA in the action of macrolide-specific efflux pump MacAB-TolC. J Biol Chem 2011 [Europe PMC free article] [Abstract] [Google Scholar]
- Yamanaka H, Kobayashi H, Takahashi E, Okamoto K. MacAB is involved in the secretion of Escherichia coli heat-stable enterotoxin II. J Bacteriol. 2008;190:7693–7698. [Europe PMC free article] [Abstract] [Google Scholar]
- Yum S, Xu Y, Piao S, Sim SH, Kim HM, Jo WS, Kim KJ, Kweon HS, Jeong MH, Jeon H, Lee K, Ha NC. Crystal structure of the periplasmic component of a tripartite macrolide-specific efflux pump. J Mol Biol. 2009;387:1286–1297. [Abstract] [Google Scholar]
- Zaitseva J, Jenewein S, Jumpertz T, Holland IB, Schmitt L. H662 is the linchpin of ATP hydrolysis in the nucleotide-binding domain of the ABC transporter HlyB. EMBO J. 2005;24:1901–1910. [Europe PMC free article] [Abstract] [Google Scholar]
- Zgurskaya HI, Nikaido H. Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc Natl Acad Sci USA. 1999;96:7190–7195. [Europe PMC free article] [Abstract] [Google Scholar]
- Zgurskaya HI, Yamada Y, Tikhonova EB, Ge Q, Krishnamoorthy G. Structural and functional diversity of bacterial membrane fusion proteins. Biochim Biophys Acta. 2009;1794:794–807. [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1111/mmi.12046
Read article for free, from open access legal sources, via Unpaywall:
https://onlinelibrary.wiley.com/doi/pdfdirect/10.1111/mmi.12046
Citations & impact
Impact metrics
Citations of article over time
Article citations
ABC Transporters in Bacterial Nanomachineries.
Int J Mol Sci, 24(7):6227, 25 Mar 2023
Cited by: 6 articles | PMID: 37047196 | PMCID: PMC10094684
Review Free full text in Europe PMC
Functional Annotation of Hypothetical Proteins From the Enterobacter cloacae B13 Strain and Its Association With Pathogenicity.
Bioinform Biol Insights, 16:11779322221115535, 06 Aug 2022
Cited by: 2 articles | PMID: 35958299 | PMCID: PMC9358594
From the soil to the clinic: the impact of microbial secondary metabolites on antibiotic tolerance and resistance.
Nat Rev Microbiol, 20(3):129-142, 16 Sep 2021
Cited by: 25 articles | PMID: 34531577 | PMCID: PMC8857043
Review Free full text in Europe PMC
Quantitative real-time analysis of the efflux by the MacAB-TolC tripartite efflux pump clarifies the role of ATP hydrolysis within mechanotransmission mechanism.
Commun Biol, 4(1):493, 22 Apr 2021
Cited by: 4 articles | PMID: 33888866 | PMCID: PMC8062640
Structure, Assembly, and Function of Tripartite Efflux and Type 1 Secretion Systems in Gram-Negative Bacteria.
Chem Rev, 121(9):5479-5596, 28 Apr 2021
Cited by: 85 articles | PMID: 33909410 | PMCID: PMC8277102
Review Free full text in Europe PMC
Go to all (20) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The periplasmic membrane proximal domain of MacA acts as a switch in stimulation of ATP hydrolysis by MacB transporter.
Mol Microbiol, 81(4):937-951, 04 Jul 2011
Cited by: 32 articles | PMID: 21696464 | PMCID: PMC3177148
MacB ABC transporter is a dimer whose ATPase activity and macrolide-binding capacity are regulated by the membrane fusion protein MacA.
J Biol Chem, 284(2):1145-1154, 27 Oct 2008
Cited by: 56 articles | PMID: 18955484 | PMCID: PMC2613632
Reconstitution of the Escherichia coli macrolide transporter: the periplasmic membrane fusion protein MacA stimulates the ATPase activity of MacB.
Mol Microbiol, 63(3):895-910, 04 Jan 2007
Cited by: 67 articles | PMID: 17214741
Antibiotic Resistance Mediated by the MacB ABC Transporter Family: A Structural and Functional Perspective.
Front Microbiol, 9:950, 28 May 2018
Cited by: 85 articles | PMID: 29892271 | PMCID: PMC5985334
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NIAID NIH HHS (2)
Grant ID: R01 AI052293
Grant ID: R21 AI092486




