Abstract
Free full text

Dnmt3a-Dependent Nonpromoter DNA Methylation Facilitates Transcription of Neurogenic Genes
Abstract
DNA methylation at proximal promoters facilitates lineage restriction by silencing cell type–specific genes. However, euchromatic DNA methylation frequently occurs in regions outside promoters. The functions of such nonproximal promoter DNA methylation are unclear. Here we show that the de novo DNA methyltransferase Dnmt3a is expressed in postnatal neural stem cells (NSCs) and is required for neurogenesis. Genome-wide analysis of postnatal NSCs indicates that Dnmt3a occupies and methylates intergenic regions and gene bodies flanking proximal promoters of a large cohort of transcriptionally permissive genes, many of which encode regulators of neurogenesis. Surprisingly, Dnmt3a-dependent nonproximal promoter methylation promotes expression of these neurogenic genes by functionally antagonizing Polycomb repression. Thus, nonpromoter DNA methylation by Dnmt3a may be used for maintaining active chromatin states of genes critical for development.
De novo DNA methyltransferases Dnmt3a and Dnmt3b and maintenance methyltransferase Dnmt1 covalently modify mammalian genomes by cytosine methylation, an epigenetic mark that is essential for normal development and primarily occurs at CpG dinucleotides (1). Most CpG-rich regions (CpG islands) overlap with proximal promoters, where DNA methylation is linked to gene silencing (2). However, unbiased genome-wide analyses have shown that DNA methylation is widespread across the euchromatic portion of mammalian genomes and predominantly takes place in regions outside proximal promoters, including intergenic regions and gene bodies (2, 3), where tissue-specific DNA methylation is frequently localized (4). Deletion of Dnmt1 or treatment with Dnmt inhibitors in dividing somatic cells can lead to global DNA hypomethylation, chromosomal instability, and compromised cell-cycle progression, thereby hindering functional analysis of DNA methylation in self-renewing tissue-specific stem cells. Unlike Dnmt1 or Dnmt3b (the deletion of which, in mice, leads to early embryonic lethality), Dnmt3a is not required for maintaining methylation at heterochromatic repeat regions, and Dnmt3a-null mice appear to be grossly normal at birth (5, 6). However, mice lacking Dnmt3a do acquire developmental defects postnatally and die prematurely (5), suggesting a specific role for Dnmt3a in regulating euchromatic methylation and postnatal development.
The function of DNA methylation in the postnatal brain is of particular interest, because brain-specific deletion of DNA methylation–related machineries results in postnatal neurodevelopmental abnormalities and premature death in mice (7, 8). Multiple isoforms of Dnmt3a/3b are expressed in mouse embryonic stem (ES) cells, but only the Dnmt3a full-length variant is expressed in diverse regions of the postnatal brain (7, 9), including subependymal/subventricular zones (SEZ/ SVZ) of the forebrain and the hippocampal dentate gyrus (fig. S1), two neurogenic regions in which neural stem cells (NSCs) persist throughout life (10). In Dnmt3a-null mice, quantification of doublecortin (Dcx)–positive immature neurons demonstrated that postnatal neurogenesis at both neurogenic zones was impaired (Fig. 1, A and B, and fig. S2) (11). However, the decrease in the number of newborn neurons was unlikely to be caused by changes in proliferation and/or survival of postnatal NSCs (fig. S3, A and B). To further evaluate the function of Dnmt3a in NSCs, we derived monolayer NSC cultures from wild-type (WT) and Dnmt3a-null [knockout (KO)] mice. Though Dnmt3a was dispensable for maintaining NSCs in an undifferentiated and proliferating state (fig. S3, C to F), upon mitogen withdrawal, differentiating Dnmt3a-null cells produced more than 10-fold fewer Tuj1+/Map2+ neurons (Fig. 1, C and D) and more glial cells, as compared with WT cells (fig. S4). To exclude the possibility that impaired neurogenic potential of Dnmt3a-null NSCs resulted from secondary effects after long-term Dnmt3a-deletion, postnatal NSCs were isolated from mice homozygous for floxed alleles of Dnmt3a (Dnmt3aflox/flox) (12) and were infected with control or Cre-expressing lentiviruses (fig. S5, A and B). Differentiation analyses indicated that acute Dnmt3a-deletion (5-day) also impaired neuronal differentiation (fig. S5C). Moreover, the neurogenesis deficit in Dnmt3a-null cultures was rescued by re-expressing physiological levels of WT Dnmt3a, but not catalytically inactive mutant Dnmt3a (Fig. 1D and fig. S6, A and B) (13). Expression of other Dnmt proteins and global DNA methylation patterns remained largely unchanged in Dnmt3a-null and rescued NSCs (fig. S6, B to D). Thus, specific and cell-autonomous functions of Dnmt3a are required for neurogenesis from postnatal NSCs.
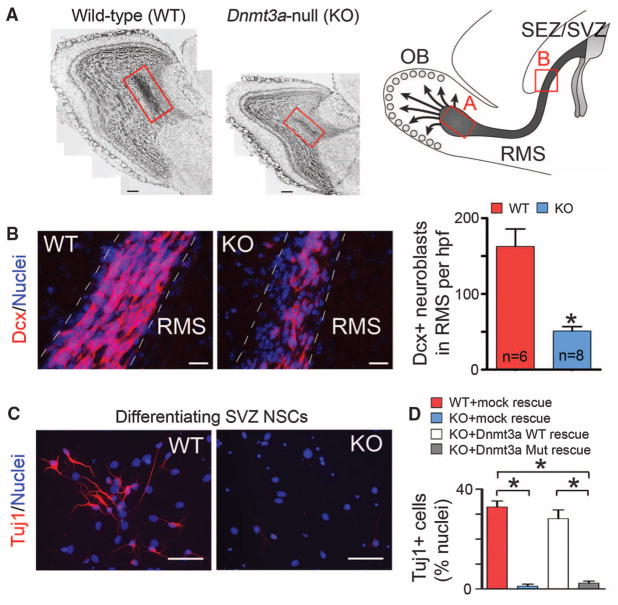
Essential roles of Dnmt3a in postnatal neurogenesis. (A) Cell nuclei staining of olfactory bulb (OB) in WT and KO mice at postnatal day (P) 24. The schematic on the right outlines the postnatal neurogenesis from SEZ/SVZ NSCs, and the red boxes denote the portion of the rostral migratory stream (RMS) with newborn neurons entering the OB. Note that both the OB size and the number of newborn neurons in KO mice (red box) are reduced. Arrows indicate radially migrating immature neurons in the postnatal OB. Scale bars, 200 μm. (B) Immunohistochemistry and quantification of Dcx+ neuroblasts in WT and KO RMS (P21 to P24). Hpf, high-power field; n, number of fields from three pairs of littermate mice. Error bars indicate SEM (*P < 0.01). Scale bars, 50 μm. (C) Immunocytochemistry of the immature neuronal marker Tuj1 in differentiating WT and KO SEZ/SVZ cultures. Scale bars, 50 μm. (D) Quantification of Tuj1+ neurons in differentiating WT, KO, and rescued NSC cultures. Error bars indicate SEM (*P < 0.01, n = 10 to 15).
To investigate the mechanism underlying Dnmt3a-dependent neurogenesis, we mapped genome-wide Dnmt3a occupancy in postnatal NSCs by chromatin immunoprecipitation (ChIP) (N′ Ab in fig. S1B) combined with whole-genome tiling microarrays (ChIP-chip). We identified 30,417 Dnmt3a binding-sites with high-confidence (false discovery rate < 1%). Quantitative polymerase chain reaction (qPCR) analysis confirmed that all 20 randomly selected loci showed significant enrichment for Dnmt3a (fig. S7A). Control ChIP-chip experiments in Dnmt3a-null NSCs did not yield specific enrichment (fig. S8A), confirming the specificity of Dnmt3a ChIP results. Most Dnmt3a binding sites were located in gene-rich euchromatic regions, as 80.7% of all binding sites were within gene bodies or 50-kb intergenic regions up- and downstream of annotated genes (fig. S9A). Mapping Dnmt3a binding sites to regions flanking transcription start sites (TSSs) revealed two categories of Dnmt3a targets (Fig. 2A). The first group (n = 2449 targets) was characterized by the presence of CpG islands, which generally harbor high levels of trimethylation of lysine 4 on histone H3 (H3K4me3), a histone mark indicative of transcriptionally permissive chromatin states, within their proximal promoter regions (hereafter referred to as CpG-rich/H3K4me3-high targets) (Fig. 2A and fig. S8, A to C). On these targets, Dnmt3a was generally excluded from H3K4me3-high, CpG-rich proximal promoters, but was enriched in inter-and intragenic regions flanking CpG islands (black in Fig. 2B), possibly because of the inhibitory effect of H3K4 methylation on Dnmt3a binding to chromatin (14). The other group of Dnmt3a targets (n = 1096) was associated with CpG-poor promoters and low levels of H3K4me3 (Fig. 2A and fig. S8, A to C). Dnmt3a binding sites on these H3K4me3-low targets frequently overlapped with proximal promoters (orange in Fig. 2B). Subsequent genome-wide methylated DNA immunoprecipitation assays (MeDIP-chip) revealed that Dnmt3a-dependent DNA methylation was restricted to Dnmt3a binding sites (Fig. 2C and fig. S9B). Locus-specific methylation assays (MeDIP-qPCR, bisulphite sequencing, or mass spectrometry) further demonstrated that, on H3K4me3-high targets (for instance, Dlx2, Gbx2), Dnmt3a-dependent DNA methylation was present in regions encompassing proximal promoters, but not within core promoter regions (Fig. 2, C and D, and figs. S7; S10, A and B; and S11, B and C). In contrast, Dnmt3a-mediated methylation was enriched at proximal promoters of H3K4me3-low targets (e.g., Gfap) (Fig. 2, C and D, and fig. S7C). Although DNA methylation changes have been reported after long-term cell culture (15), short-term in vitro culturing in our experiments did not significantly alter promoter DNA methylation levels, either globally or on Dnmt3a targets (fig. S10, B and C). Collectively, these data indicate that Dnmt3a may occupy and methylate defined genomic regions associated with both transcriptionally active and inactive genes in postnatal NSCs.
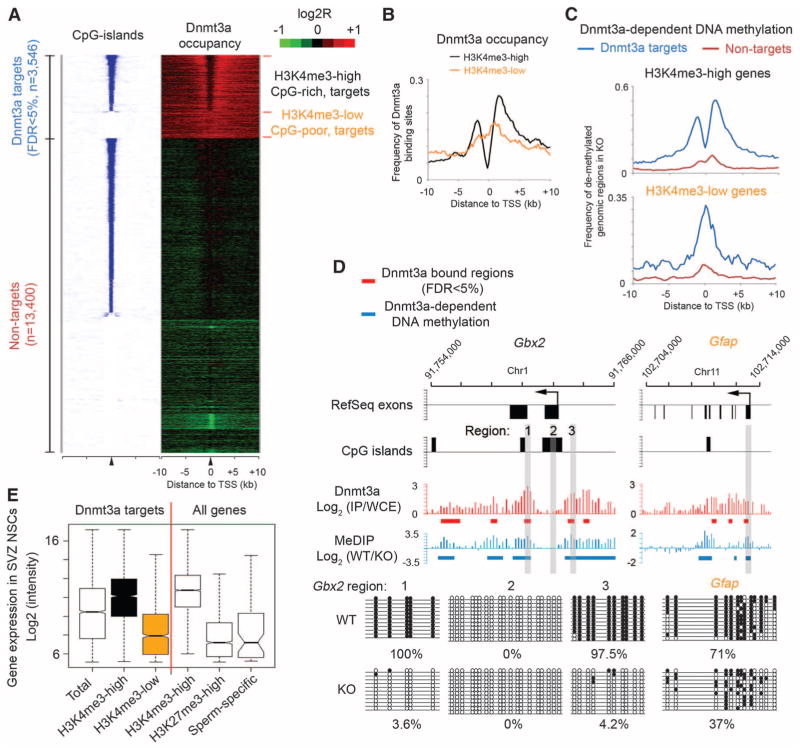
Dnmt3a occupies and methylates defined regions at both transcriptionally active and inactive genes. (A) Heat map representations of CpG islands (blue/white) and Dnmt3a (red/green) are shown for both Dnmt3a targets and nontargets. Red/blue represents enrichment, whereas green/white represents no enrichment. Genes were rank-ordered by CpG-island length within 4-kb regions flanking TSSs. FDR, false discovery rate; n, number of genes. (B) Distribution of Dnmt3a binding-sites within regions flanking TSSs of H3K4me3-high/CpG-rich and H3K4me3-low/CpG-poor targets. (C) Distribution of Dnmt3a-dependent DNA methylation within regions flanking TSSs of CpG-rich and CpG-poor genes. (D) Dnmt3a occupancy and Dnmt3a-dependent DNA methylation within representative CpG-rich/H3K4me3-high (Gbx2) and CpG-poor/ H3K4me3-low (Gfap) targets. Regions validated by bisulphite sequencing are shown in gray. IP, immunoprecipitation; WCE, whole-cell extract. (E) Box plots of expression levels in WT postnatal NSCs are shown for genes associated with Dnmt3a, H3K4me3, H3K27me3, and proximal promoter methylation (sperm-specific genes). Middle bars, medians; notches, standard errors; boxes, interquartile ranges; and whiskers, 10th and 90th percentiles.
To gain insights into the biological function of Dnmt3a targets, we performed gene ontology analysis with Dnmt3a targets. Genes functionally related to nervous system development (P = 1.5 × 10−30, hypergeometric test) and neurogenesis (P = 6.5 × 10−19) were significantly enriched within H3K4me3-high Dnmt3a targets, whereas genes involved in development of non-neuronal lineage were enriched in H3K4me3-low Dnmt3a targets (fig. S8D). H3K4me3-high targets were transcribed at medium-to-high levels in postnatal NSCs, significantly higher than expression levels of H3K4me3-low targets, or genes marked by an inactive histone modification H3K27me3 or a group of sperm-specific genes with proximal promoter DNA methylation (Fig. 2E).
To establish a direct link between Dnmt3a binding and transcriptional regulation, we performed gene-expression profiling and identified a total of 2275 genes (1253 up-regulated and 1022 down-regulated in Dnmt3a-null mice) that are differentially expressed between undifferentiated WT and KO NSCs. Dnmt3a binding sites were detected within 10-kb up- and downstream of the TSSs of 942 differentially expressed transcripts (41.4% of 2275 genes, P = 1.36 × 10−122, Fisher’s exact test) (Fig. 3A), indicating that a significant fraction of Dnmt3a-dependent expression changes represent a direct effect of Dnmt3a binding. Although DNA methylation has been primarily associated with repression, Dnmt3a targets were significantly enriched in both up- and down-regulated genes in KO NSCs (536 up-regulated, P = 3.3 × 10−71; 406 down-regulated, P = 1.9 × 10−43). Notably, genes with known roles in postnatal neurogenesis (for example, Dlx2, Sp8, and Neurog2) were among the down-regulated Dnmt3a targets in KO NSCs (fig. S11A) (16, 17). In contrast, several Dnmt3a targets involved in astroglial and oligodendroglial differentiation (e.g., Sparcl1, Nkx2-2) were up-regulated in KO cultures. Gene-expression analyses with differentiating NSCs also supported the conclusion that Dnmt3a promotes transcription of neurogenic targets while repressing glial differentiation genes (table S1 and supporting online material text). Further in vivo analysis revealed fewer Dlx2+ and Sp8+ progenitors/neuroblasts and their subtype-specific inter-neuron progenies in KO mice (fig. S12, A to E), and re-expression of Dlx2 in KO cells substantially rescued the neurogenesis defect (fig. S12F). Taken together, these results indicate that Dnmt3a is not only involved in transcriptional repression but also required by postnatal NSCs to promote expression of many direct targets, including key neurogenic factors, thereby contributing to postnatal neurogenesis.
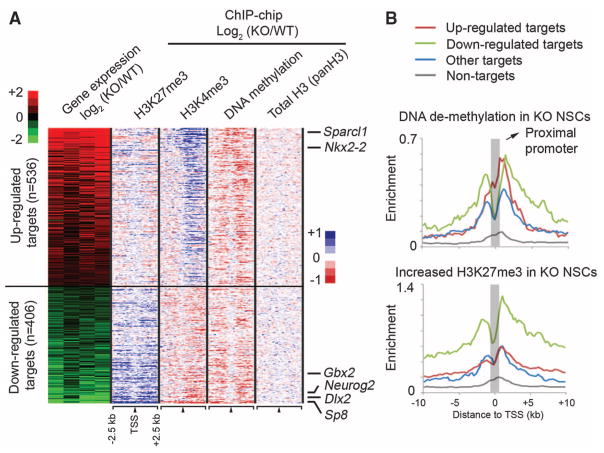
Dnmt3a promotes transcription through nonpromoter methylation. (A) Heat map representations of relative changes in expression (from four experiments), DNA methylation, H3K4me3, H3K27me3, and total histone H3 levels (panH3) between KO and WT NSCs are shown for differentially expressed Dnmt3a targets, which are ranked by changes in expression between KO and WT NSCs. n, number of targets. (B) Distributions of DNA demethylation and H3K27me3 increase in KO NSCs are shown for up-regulated targets (red), down-regulated targets (green), targets without expression changes (blue), and nontargets (gray).
The percentage of H3K4me3-high promoters in down-regulated targets (65.4% of 406 genes) was significantly higher than that expected by chance (P = 7.7 × 10−8), and DNA demethylation was predominantly detected outside proximal promoters of these down-regulated targets in KO NSCs (Fig. 3A and fig. S11, B and C). In contrast, DNA demethylation on up-regulated targets in KO NSCs (red in Fig. 3B) was more enriched at proximal promoters compared with down-regulated targets (green compared with red in Fig. 3B; P = 0.0018) or targets without expression changes (blue in Fig. 3B; P = 6.8 × 10−9). Although proximal promoter DNA methylation may repress transcription by antagonizing active histone marks (Fig. 3A) (15, 18, 19), growing evidence suggests that non-promoter DNA methylation positively correlates with transcription of tissue-specific genes or genes on the active X chromosome (2, 20). DNA methylation also inversely correlates with the presence of H3K27me3 in human cancer cell lines or on the transcriptional regulatory region for the imprinting locus Rasgrf1 (21, 22). Therefore, we hypothesized that Dnmt3a-dependent nonpromoter DNA methylation may facilitate transcription by antagonizing H3K27me3. To test this idea, we examined genome-wide H3K27me3 occupancy in WT and KO NSCs. Our analysis revealed that Dnmt3a-deficiency resulted in an increase in H3K27me3 levels across large genomic regions associated with many down-regulated targets (green in Fig. 3B and fig. S11, B and C), whereas the increase in H3K27me3 levels in KO NSCs was observed less often on up-regulated targets (red in Fig. 3B; P = 1.8 × 10−20) or targets without changes in expression (blue in Fig. 3B; P = 1.6 × 10−28). Acute ablation of Dnmt3a also led to a significant increase in H3K27me3 levels on down-regulated targets, suggesting that the antagonism between Dnmt3a and H3K27me3 is unlikely due to nonspecific compensation after long-term Dnmt3a deletion (fig. S5D). Thus, Dnmt3a-dependent non-promoter DNA methylation may facilitate the expression of many H3K4me3-high targets by reducing H3K27me3 levels.
Because Dnmt3a deficiency did not change the overall levels of H3K27me3 or Polycomb repression complex 2 (PRC2) that catalyzes trimethylation of H3K27 (fig. S13, A and B), we considered a model in which Dnmt3a functioned to antagonize PRC2 binding to chromatin. ChIP-chip analysis of Suz12 and Ezh2, two core subunits of PRC2, revealed that Dnmt3a deficiency led to a significant increase in Suz12 occupancy on Dnmt3a targets with a marked increase in H3K27me3 in KO NSCs (P < 2.2 × 10−16); similar results were obtained for Ezh2 (Fig. 4A and fig. S11, D and E). The increase of PRC2/H3K27me3 levels on these Dnmt3a targets was specific, as other PRC2 targets that were not targeted by Dnmt3a (e.g., Hoxb9 in Fig. 4A) were associated with similar levels of PRC2/H3K27me3 in WT and KO NSCs. To evaluate whether Dnmt3a enzymatic activity is required for opposing PRC2 binding, we analyzed Dnmt3a-null NSCs rescued with physiological levels of WT or catalytically inactive mutant Dnmt3a. In rescued KO NSCs, both WT and mutant Dnmt3a re-occupied Dnmt3a targets (fig. S14A), but only WT Dnmt3a restored DNA methylation levels (fig. S14, B and C). WT, but not mutant Dnmt3a, reversed aberrantly elevated levels of PRC2/H3K27me3 on down-regulated Dnmt3a targets in KO NSCs (Fig. 4B), indicating that Dnmt3a-dependent DNA methylation, but not Dnmt3a binding, is incompatible with PRC2 occupancy and may directly antagonize the PRC2 binding. Therefore, we tested recruitment of PRC2 complexes to in vitro DNA-methylated chromatin arrays consisting of recombinant Xenopus octamers (23, 24), which are free of any other epigenetic marks existing on endogenous chromatin that may also regulate PRC2 recruitment (fig. S15). This analysis indicated that DNA methylation alone partially inhibited PRC2 binding to chromatin arrays (Fig. 4C). Furthermore, PRC2 deficiency in Dnmt3a-null NSCs could substantially restore the expression of neurogenic targets and rescue neuronal differentiation defects (Fig. 4D and fig. S13, C and D). Dnmt3a-dependent DNA methylation may, therefore, down-regulate H3K27me3 levels on transcriptionally active targets by antagonizing binding of PRC2 to chromatin.
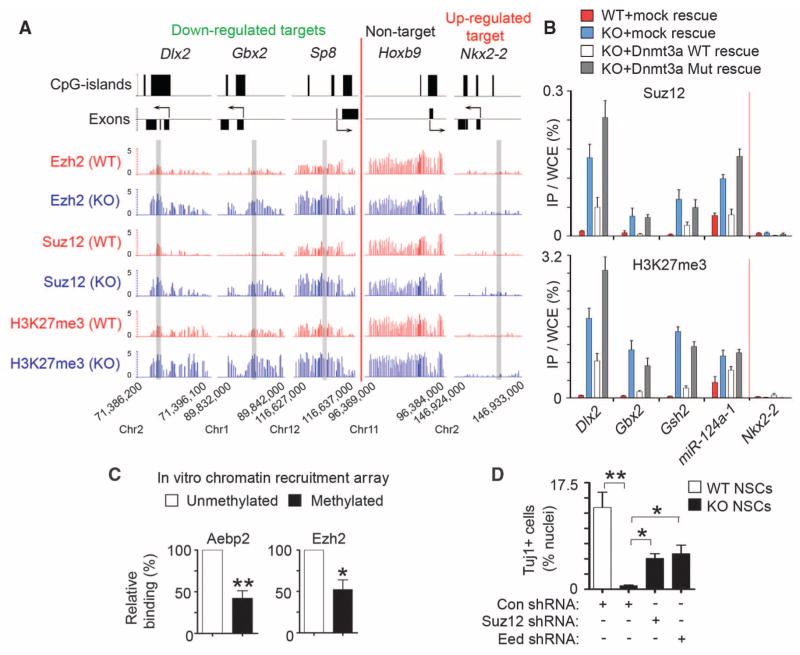
Dnmt3a promotes transcription by antagonizing Polycomb repression. (A) Ezh2, Suz12, and H3K27me3 occupancy at representative genes in WT and KO NSCs. Regions analyzed by ChIP-qPCR (in fig. S11E) are shaded in gray. (B) Quantitative PCR analysis of Suz12 and H3K27me3 at representative targets in WT, KO, and rescued NSCs. (C) Relative binding of PRC2 subunits on methylated or unmethylated chromatin arrays was quantified by densitometry (*P = 0.027, n = 4 experiments; **P = 0.0013, n = 6). (D) Quantification of Tuj1+ neurons in control and PRC2-deficient differentiating NSCs. Error bars indicate SEM of randomly selected fields from two independent cultures (*P < 0.05; **P < 0.01).
Together, these results suggest that the DNA methyltransferase Dnmt3a not only mediates repression in self-renewing postnatal NSCs by methylating proximal promoters, but also promotes transcription of targets, including neurogenic genes, by antagonizing Polycomb repression through nonproximal promoter methylation (fig. S16). The positive correlation between nonpromoter DNA methylation and transcription on Dnmt3a targets is also observed during mouse ES cell differentiation into neural progenitors, as well as in vivo analysis of multiple adult tissues, suggesting that this finding may be generally applicable to other developmental processes (figs. S17 and S18). Embryonic neurogenesis is largely intact in Dnmt3a-null mice (10), perhaps because embryonic neuroblasts are primarily generated from neuroepithelial progenitors where Dnmt3b is still highly expressed (13). Furthermore, Polycomb repression can also be inhibited by H3K27me3 demethylase (e.g., Jmjd3), which may be recruited by H3K4 methyltransferases Mll to promoters of neurogenic factors during embryonic and/or postnatal neurogenesis (16, 25). Non-proximal promoter DNA methylation established by Dnmt3a may, therefore, represent an additional layer of epigenetic regulation of Polycomb repression in somatic cells. Although PRC2 may directly interact with Dnmt proteins (26, 27), it seems that DNA methyltransferases (for instance, catalytically inactive Dnmt3a) may coexist with high levels of PRC2 occupancy at the same genomic loci (27), only in the absence of de novo DNA methylation. Thus, Polycomb proteins may contribute to the initial recruitment of Dnmt, the accumulation of which leads to local de novo DNA methylation, thereby dampening Polycomb repression. Given that both DNA methylation and Polycomb pathways are indispensable for normal development and are implicated in diseases, including neurological disorders and cancer (21, 28), it will be of interest to fully elucidate mechanisms by which these two epigenetic machineries are targeted to specific genomic loci and are cross-regulated.
Acknowledgments
We thank M. Carey (UCLA) for technical assistance on in vitro chromatin recruitment assays and J. Rubenstein (University of California San Francisco) for providing the pCAG-Dlx2 expression plasmid. This work was supported by NIH RO1 grants to Y.E.S. All microarray data are deposited in Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/) under accession number GSE22476.
Footnotes
References and Notes
Full text links
Read article at publisher's site: https://doi.org/10.1126/science.1190485
Read article for free, from open access legal sources, via Unpaywall:
https://cdr.lib.unc.edu/downloads/4j03d7472
Subscription required at www.sciencemag.org
http://www.sciencemag.org/cgi/reprint/329/5990/444.pdf
Subscription required at www.sciencemag.org
http://www.sciencemag.org/cgi/content/full/329/5990/444
Free to read at www.sciencemag.org
http://www.sciencemag.org/cgi/content/abstract/329/5990/444
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/101919853
Article citations
The N-terminal region of DNMT3A engages the nucleosome surface to aid chromatin recruitment.
EMBO Rep, 11 Nov 2024
Cited by: 0 articles | PMID: 39528729
Injured inflammatory environment overrides the TET2 shaped epigenetic landscape of pluripotent stem cell derived human neural stem cells.
Sci Rep, 14(1):25186, 24 Oct 2024
Cited by: 0 articles | PMID: 39448736 | PMCID: PMC11502794
Genetic-epigenetic interplay in the determination of plant 3D genome organization.
Nucleic Acids Res, 52(17):10220-10234, 01 Sep 2024
Cited by: 1 article | PMID: 39149894 | PMCID: PMC11417408
Significance of targeting DNMT3A mutations in AML.
Ann Hematol, 30 Jul 2024
Cited by: 0 articles | PMID: 39078434
Review
Structural basis for the H2AK119ub1-specific DNMT3A-nucleosome interaction.
Nat Commun, 15(1):6217, 23 Jul 2024
Cited by: 0 articles | PMID: 39043678 | PMCID: PMC11266573
Go to all (402) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (1 citation) GEO - GSE22476
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Targeting of EZH2 to a defined genomic site is sufficient for recruitment of Dnmt3a but not de novo DNA methylation.
Epigenetics, 4(6):404-414, 29 Aug 2009
Cited by: 56 articles | PMID: 19717977
Dnmt3L antagonizes DNA methylation at bivalent promoters and favors DNA methylation at gene bodies in ESCs.
Cell, 155(1):121-134, 01 Sep 2013
Cited by: 121 articles | PMID: 24074865
Control of epigenetic states by WT1 via regulation of de novo DNA methyltransferase 3A.
Hum Mol Genet, 22(1):74-83, 05 Oct 2012
Cited by: 25 articles | PMID: 23042785 | PMCID: PMC6296327
The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape.
Nature, 573(7773):281-286, 04 Sep 2019
Cited by: 256 articles | PMID: 31485078 | PMCID: PMC6742567
Funding
Funders who supported this work.




