Abstract
Free full text

GATA-3 deficiency abrogates the development and maintenance of T helper type 2 cells
Abstract
T helper type 2 (Th2) cells secrete IL-4, IL-5, IL-10, and IL-13 and mediate allergic and asthmatic disease. GATA-3 is a Th2-specific transcription factor that appears in overexpression studies and transgenic systems to function as a Th2 lineage determinant. Because GATA-3 is also crucial for development of the T lineage and throughout thymic development, direct demonstration that GATA-3 is required for Th2 development by targeted deletion has been lacking. Using a conditional knockout approach, we found that GATA-3 is required for optimal Th2 cytokine production in vitro and in vivo. Our data also show that GATA-3 expression must be sustained to maintain the Th2 phenotype.
Naïve CD4 T cells in immunocompetent animals are poised to differentiate into T helper (Th) effector cell populations with distinct functions. These Th subtypes, termed Th type 1 (Th1) or Th type 2 (Th2) cells, have long been characterized by their differential expression of cytokines, high levels of IFN-γ, IL-2 and lymphotoxin production in the case of Th1 and IL-4, IL-5, IL-13, and IL-10 in the case of Th2 (1–3). Establishment of these two lineages of Th cells can be influenced by a variety of factors, including genetic background, type of antigen and route of administration, mode of presentation, degree of costimulation, and, most importantly, the cytokine milieu (reviewed in refs. 4–6).
In our current model of Th2 lineage commitment, pluripotent Th precursor cells elaborate both IL-4 and IFN-γ early in response to TCR signals in a signal transducer and activator of transcription (STAT)-independent fashion (7). Sustained signals by means of IL-4/IL-4 receptor/STAT6 activation then drive commitment to the Th2 lineage by up-regulating GATA-3, a Th2-specific transcription factor (8, 9) whose expression could potentially activate its own expression in a positive feedback loop (10). High levels of GATA-3 would then instruct the cell to produce Th2 cytokines by inducing characteristic DNase hypersensitivity sites in the IL-4 locus and the IL-4/IL-13 intergenic regions locus (refs. 10–16; for review, see ref. 38) and direct binding to critical elements in the IL-5 (8, 17) and IL-13 (18) promoters. Potentially, GATA-3 could be involved in a recently identified locus control region (19). Forced expression of GATA-3 in developing Th1 cells is sufficient to drive Th2 cytokine expression and induce changes in chromatin in the absence of the upstream activator STAT6 (10, 12), further supporting the notion that GATA-3 is a master regulator of Th2 cytokine production. Similarly, antisense experiments (9, 20) and analysis of transgenic and mutant GATA-3 mice (21, 22) also support this model. However, direct demonstration that endogenous GATA-3 is required for Th2 development in a whole animal has been hampered by the fact that deletion of GATA-3 in the immune system results in a profound early block in commitment to the T lineage (23). It is also unclear whether GATA-3 is required for the expression of other Th2 cytokines, e.g., IL-10, which is not located in the IL-4/IL-5/IL-13 cluster. In addition, most of the data elucidating the role of GATA-3 in the production of Th2 cytokines have been derived from CD4+ Th cells; it is not clear whether GATA-3 is also needed for the production of Th2 cytokines by CD8 cells.
Although expression of GATA-3 is sufficient to drive Th2 lineage commitment, the plasticity of this process has not been fully investigated. Th2 lineage commitment requires multiple cell divisions (24), and developing Th2 cells that have undergone fewer than three to four divisions can still be diverted to the Th1 lineage by reculture in IL-12 (7). After four divisions have passed and remodeling of cytokine loci has occurred, a pattern of silencing and activation is established that stabilizes the Th2 phenotype (24–26). Therefore, once the program of Th2 cytokine production has been set into motion, GATA-3 may no longer be required to maintain commitment to the Th2 lineage; however, this question has yet to be directly studied.
To address these questions, we used the Cre/lox system (27) to generate mice conditionally deficient in GATA-3 (28). CD4+ T cell development in GATA-3-deficient animals was abnormal with evidence of in vivo activation. GATA-3-deficient CD4+ and CD8+ T cells failed to produce optimal Th2 cytokines both in vitro and in response to antigen injection in vivo. These findings were confirmed when developing Th2 cells were rendered GATA-3-deficient by retroviral transduction. Finally, we found that persistent expression of GATA-3 was required for established Th2 cells to maintain their Th2 cytokine production.
Materials and Methods
Animals. The generation of homozygous flox (FF) and knockout (FF cre) animals and genotyping procedures have been described (28). Animals used in these experiments were backcrossed onto the BALB/c background at least four to six generations. Animals were housed in specific pathogen-free conditions at either the Dana-Farber Cancer Institute or the Harvard School of Public Health under protocols approved by their respective institutional review boards.
Cell Culture, Flow Cytometry, and Retroviral Transduction. CD4 and CD8 T cells were purified from lymph node and spleen by using magnetic-activated cell sorting to >95% purity (Miltenyi Biotec, Auburn, CA). For primary stimulation, purified T cell subsets were stimulated with 1 μg/ml plate-bound anti-CD3 (145-2C11) and 2 μg/ml soluble anti-CD28 (PV1). Cells were then cultured in 1 ng/ml murine IL-12 (PeproTech, Rocky Hill, NJ) and anti-murine IL-4 (11B11, National Cancer Institute Preclinical Repository, Bethesda) for Th1 or Tc1 conditions or in 10 ng/ml murine IL-4 (PeproTech) and anti-murine IFN-γ (R4-682) for Th2 or Tc2 conditions. Cells were expanded in recombinant human IL-2 (National Cancer Institute Preclinical Repository) and analyzed for cytokine production at days 6–8. All antibodies except anti-IL-4 were processed by Bioexpress (Kaysville, UT).
Surface staining was performed by using fluorescent-tagged antibodies to CD3 (17A2), B220 (RA3-6B2), CD4 (RM4-5), CD8α (53-6.7), and CD62L (MEL-14) (BD Biosciences) and analyzed on FACSCalibur or FACSSort machines (BD Biosciences). Cell sorting was performed on MoFlo machinery (Cytomation, Fort Collins, CO).
A bicistronic retroviral construct containing murine stem cell virus backbone, Cre recombinase, and enhanced GFP driven by a phosphoglycerate kinase promoter was the kind gift of Daniel Littman (Skirball Institute, New York University, New York) (29). Cre construct and control vector containing GFP alone were transfected into Phoenix helper-free packaging cell line to derive retrovirus-containing supernatants as described (gift of Garry P. Nolan, Stanford University, Palo Alto, CA, www.stanford.edu/group/nolan). CD4 cells were transduced with retroviral supernatants 30 h after CD3/CD28 stimulation in the presence of polybrene and centrifugation as described (30). Th2 effector cells were restimulated with 0.1 μg/ml platebound anti-CD3 antibody and soluble anti-CD28 antibody in Th2 conditions for 30 h before retroviral transduction.
Measurement of Cytokine Production. Cells were stimulated with 50 ng/ml phorbol 12-myristate 13-acetate and 1 μM ionomycin for 4 h (in the presence of 3 μM monensin for the last 2 h), fixed with 4% paraformaldehyde in PBS, and analyzed for intracellular cytokine production by fluorescence-activated cell sorter analysis as described (18). For measurement of secreted cytokines, cells were washed and then restimulated with 1 μg/ml plate-bound anti-CD3. Supernatants were harvested at 24 h, and sandwich ELISA was performed for IL-4, IL-5, IFN-γ, and IL-10 (BD Biosciences) and for IL-13 (R & D Systems).
PCR Analysis. The proportion of deleted and nondeleted GATA-3 alleles was detected as described (28). For retrovirally infected cells, primers 5, 6, and 5′-CCCTTCTGATCTCAATGCAT-3′ were used to detect floxed and deleted alleles in genomic DNA. After 35 cycles of amplification (94°C for 30 seconds, 58°C for 30 seconds, and 72°C for 60 seconds), PCR products were separated on a 2% agarose gel, and 856-bp floxed and 641-bp deleted bands were visualized by ethidium bromide staining.
Antigen-Specific Immune Response Assay. FF and FF cre mice were injected with 100 μg of 2,4,6,-trinitrophenyl–keyhole limpet hemocyanin (TNP-KLH; Bioresearch Technologies, Novato, CA) in alum at the base of the tail on day 0. On day 7, inguinal lymph nodes (draining) and axillary lymph nodes (nondraining) were harvested, crushed whole, and cultured in T cell medium containing 250 μg/ml TNP-KLH. Supernatant from cultures was harvested at 60 h, and cytokine production was measured by sandwich ELISA.
Results
GATA-3-Deficient Mice Have a Deficiency of Naïve CD4+ T Cells in the Periphery. The generation of mice conditionally deficient in GATA-3 using the Cre/lox system has been described (28). Animals in which the exons encoding the zinc finger regions of GATA-3 were flanked by loxP sites (FF animals) were generated and were tested to ensure that full-length functional GATA-3 protein was expressed (28). The size of the lymphoid organs, T and B cell percentage, and CD4/CD8 percentages in FF animals were comparable to those of animals carrying two wild-type alleles (data not shown). After several rounds of backcrossing, CD4+ T cells from wild-type (WW) and FF littermate controls also made the same amount of Th1 and Th2 cytokines by ELISA (Fig. 1A); thus, we considered WW and FF animals to be equivalent.
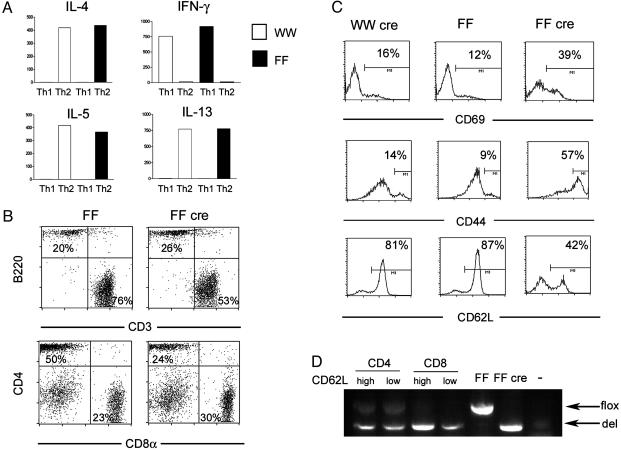
Characterization of FF and FF cre animals. (A) Production of cytokines (in ng/ml) by in vitro differentiated Th1 and Th2 cells from wild-type animals (WW, open bars) and animals bearing two floxed GATA-3 alleles with no Cre transgene (FF, filled bars). (B) Fluorescence-activated cell sorter (FACS) analysis of lymph node cells from FF and knockout (FF cre) animals. (C) FACS analysis of expression of activation markers in CD4+ lymph node cells from wild-type mice expressing Cre (WW cre), FF, and FF cre animals. (D) PCR analysis showing the relative proportions of floxed (flox) and deleted (del) alleles in sorted cell populations from FF cre mice. Lanes marked FF and FF cre are thymocyte DNA from FF and FF cre mice, respectively. The lane marked with a dash is a no template control.
Subsequently, mice bearing two floxed GATA-3 alleles (FF mice) were crossed with mice carrying a transgene encoding lck-driven Cre recombinase, generating FF cre knockout animals. The proximal lck-cre transgene is active in double negative stage 3 thymocytes (31, 32), but deletion of the GATA-3 gene in FF cre mice is highly variable in extent (28). In severely affected mice, in which efficient deletion of the GATA-3 gene in the thymus was documented by PCR analysis of genomic DNA, the CD4/CD8 ratio was reversed because of deficiency of single-positive CD4 thymocytes; the severity of CD4 deficiency in the thymus correlated with the lack of GATA-3 protein (28). All GATA-3-deficient mice analyzed in the data presented here were therefore screened for deletional efficiency by analysis of the DNA by PCR, CD4/CD8 ratio, or both. Using this method, about half of the FF cre mice have clear reversal of the CD4/CD8 ratio in the periphery (data not shown). Even in severely affected animals, undeleted flox alleles were always detected in the CD4 single-positive thymocytes (28) and in CD4+ T cells of FF cre animals (Fig. 2A), thus demonstrating that a varying mixture of wild-type, heterozygous deficient, and homozygous deficient cells is likely present in most animals.
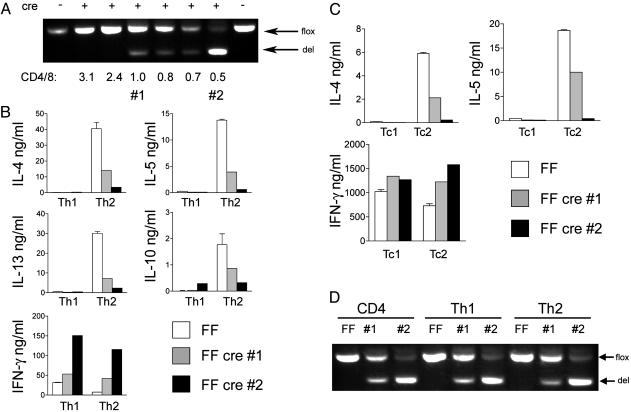
Cytokine production from differentiated CD4 and CD8 T cells. (A) PCR analysis showing the relative proportions of floxed (flox) and deleted (del) alleles in a panel of Th2 cells from FF animals positive (+) or negative (–) for the Cre transgene. The CD4/CD8 ratio in each animal is indicated. Lanes 1 and 2 represent samples from cells used for ELISA shown in B and C. (B) Cytokine production from CD4 T cells of FF (white bar), FF cre animal 1 (gray bar), and FF cre animal 2 (black bar) differentiated in vitro under Th1 and Th2 conditions. (C) Cytokine production as in B using CD8 T cells differentiated in vitro under Tc1 and Tc2 conditions. (D) PCR analysis as in A using genomic DNA purified from freshly isolated CD4 cells, differentiated Th1 and Th2 cells from a FF control animal (FF), FF cre animal 1 (#1), and FF cre animal 2 (#2).
Spleen and lymph nodes in FF cre mice analyzed at 4–8 weeks of age were grossly normal in appearance and in total cell numbers (data not shown). In contrast, FF cre animals had a mild reduction in the percentage of CD3+ cells in lymph node (Fig. 1B). The expression of CD3 as well as of the α/β TCR was slightly but consistently lower in FF cre animals (Fig. 1B and data not shown) although the TCR repertoire was normal in diversity (data not shown). FF cre mice were also deficient in CD4+ T cells (Fig. 1B). On average, the yield of CD4+ T cells from GATA-3-deficient mice was 30–50% of that of FF mice. In addition, the percentage of CD4 T cells positive for CD44 and CD69 was markedly increased whereas the percentage of naïve CD62Lhigh cells in lymph nodes was severely reduced in FF cre mice (Fig. 1C). All of the above findings were also seen in splenocytes from FF cre mice (data not shown). Because of the variegation in lck-cre observed in CD4+ thymocytes (28), we examined the degree of deletion of the GATA-3 locus in both naïve (CD62Lhigh) and activated (CD62Llow) cells. As seen in Fig. 1D, a PCR assay that simultaneously detects floxed and deleted alleles showed that the relative proportions of these alleles, and therefore the extent of deletion, was identical in these two populations. We also found that the degree of deletion in CD8 cells was in general more complete that in CD4 cells (Fig. 1D). Thus, GATA-3-deficient mice were deficient in CD4+ T cells, and the cells present manifested a preactivated phenotype.
CD4 and CD8 T Cells from GATA-3-Deficient Animals Have Defective Th2 Cytokine Production in Vitro. We next examined the production of Th1 and Th2 cytokines from CD4 and CD8 T cells differentiated in vitro. First, CD4+ T cells from GATA-3-deficient animals were stimulated with anti-CD3 and anti-CD28 in Th1 or Th2 conditions. After culture in IL-2 for 6–7 days, cells were washed and restimulated with anti-CD3, and supernatants were collected for analysis of cytokines by ELISA. Because of the variability of lck-cre-driven deletion of the GATA-3 gene, the same cells were used to purify DNA and the degree of deletion measured by PCR analysis. In Fig. 2 A, the proportion of floxed and deleted alleles in a panel of differentiated Th2 cells from knockout animals is shown, with the CD4/CD8 ratio denoted beneath. As we found in the thymus, the degree of CD4 deficiency and reduction in CD4/CD8 ratio correlated with the efficiency of deletion (data not shown and Fig. 2 A).
In Fig. 2B the production of IL-4, IL-5, IFN-γ, IL-13, and IL-10 from Th1 and Th2 cells is shown from two different FF cre animals with different degrees of deletion (see Fig. 2 A) and their FF littermate control. Th2 cells from both knockout animals showed reduction in IL-4, IL-5, IL-13, and, interestingly, IL-10 production; conversely, IFN-γ production was increased even under Th2 conditions. In the experiment shown in Fig. 2B, FF cre CD4 cells grown under Th1 conditions made more IFN-γ than FF cells did, but this was not a consistent finding; in other assays FF cre Th1 cells made similar amounts of IFN-γ compared with FF Th1 cells (data not shown). FF cre 2, which had a lower CD4/CD8 ratio and more efficient deletion (Fig. 2 A), had a more severe defect in Th2 cytokine production, confirming the dose dependence of Th2 cytokine production on GATA-3.
We conducted a similar analysis with CD8 T cells, which express moderate levels of GATA-3 when grown under Tc2 conditions (33). Tc1 and Tc2 differentiated cells were obtained by growing under Th1 and Th2 conditions. FF cre Tc2 cells, similar to Th2 cells, failed to make IL-4 and IL-5 (Fig. 2C). In addition, FF cre cells failed to exhibit even the modest degree of suppression of IFN-γ seen in Tc2 cells compared with Tc1 cells. Thus, the optimal production of Tc2 cytokines and the suppression of Tc1 cytokines also depended on GATA-3.
In some experiments, PCR analysis of CD4+ T cells from FF cre animals that initially showed good deletion showed a loss of deleted alleles and a high proportion of flox alleles by the end of Th2 differentiation, likely because of overgrowth of normal cells; in these cases, Th2 cytokine production was not surprisingly normal (data not shown). That GATA-3-deficient CD4 cells sometimes fail to expand relative to normal cells is not surprising given that complete deletion of GATA-3 driven by CD4-cre resulted in a profound loss of CD4 cells (28). However, in other experiments the proportions of floxed and deleted alleles did not change after culturing (Fig. 2D), and in these same animals Th2 cytokine production was clearly defective (Fig. 2B). Thus, we cannot ascribe the lack of IL-4 production in such cases to a gross defect in proliferation. In summary, the production of all Th2 cytokines in both CD4 and CD8 required GATA-3 in a dose-dependent fashion.
GATA-3 Is Required for both Th2 Cell Development and Maintenance. CD4 and CD8 T cells that had matured in GATA-3-deficient animals were deficient in Th2 cytokine production in vitro. The fact that these cells, known to be more activated in vivo (Fig. 1C), produced more Th1 cytokines even under Th2 culture conditions could imply that perhaps some Th cells had already fully differentiated into Th1 cells in vivo. To verify that GATA-3 is required for development of Th2 cells in a system not subject to these limitations, we transduced FF CD4+ T cells with retrovirus encoding Cre recombinase 30 h after activation under Th2 conditions, expanded the cells with IL-2, and measured cytokine production by intracellular staining on day 6. Analysis of sorted GFP+ cells from control (GFP-RV) and Cre-infected (GFP-Cre) cultures showed that the majority of Cre-infected cells had indeed deleted the GATA-3 gene (Fig. 3A). After restimulation with phorbol 12-myristate 13-acetate and ionomycin, GFP+ cells in GFP-RV-infected controls made IL-4 and IL-5 but very little IFN-γ, as expected. In contrast, GFP+ cells in GFP-Cre-infected FF cells had on average a 30% reduction in IL-4 production and a 50% reduction in IL-5 production, whereas the percentage of IFN-γ producing cells, although small, increased 2-fold (Fig. 3B). A difference in expansion between the GFP+ and GFP– populations could explain a defect in cytokine production. However, the percentage of GFP-positive cells detected at the time of restimulation was identical to that found shortly after infection, implying that the growth of the GFP-positive and GFP-negative populations was similar (data not shown). Thus, developing Th2 cells required GATA-3 for optimal Th2 differentiation.
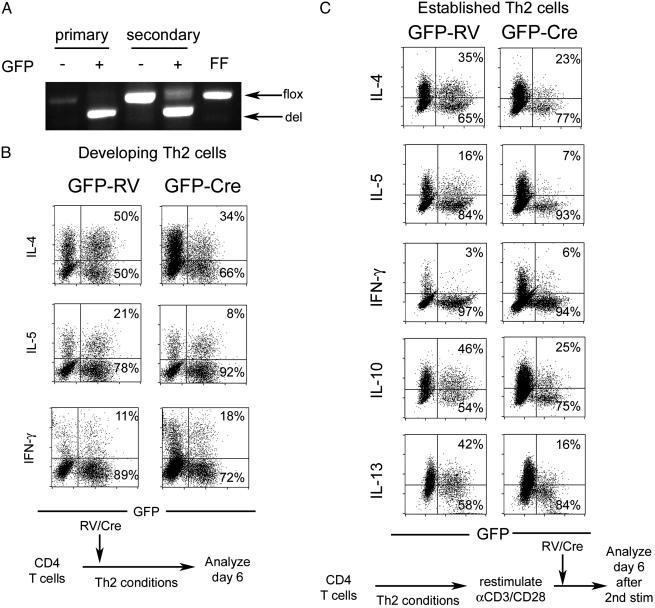
Retroviral transduction of developing and established Th2 cells. (A) PCR analysis showing the proportion of floxed (flox) and deleted (del) alleles in sorted cell populations after infection of developing (primary) or established (secondary) Th2 cells. FF denotes control thymocyte DNA from a FF animal. (B) Intracellular cytokine staining of cells from FF animals infected with Cre (GFP-Cre) or control (GFP-RV) virus during Th2 differentiation according to the schema depicted beneath. Percentages of infected GFP+ cells that express or do not express the indicated cytokines are indicated in top right and bottom right corners, respectively. (C) Analysis as in B of FF cells infected with Cre virus or control after 1 week of Th2 differentiation, as depicted in the schema beneath.
Whether GATA-3 plays a critical role in maintenance of the Th2 phenotype after full differentiation has occurred is unknown. To address this question, FF CD4+ T cells were differentiated under Th2 culture conditions for 1 week. At the end of polarization, the cells appropriately produced Th2 cytokines and had an effector phenotype, as manifested by high expression of CD44 (data not shown). These established Th2 cells were then restimulated and infected with GFP-Cre or control virus, and cytokine production was measured 6–8 days after infection. Th2 cytokine production, including IL-10 and IL-13, clearly required the continual presence of GATA-3, and, similar to infection of developing Th2 cells, the production of IFN-γ was slightly, but consistently, increased (Fig. 3C). Interestingly, the reduction in Th2 cytokine production was similar to the decrement seen in developing Th2 cells; IL-4 and IL-10 were decreased ≈30%, IL-5 and IL-13 were decreased ≈50%, and IFN-γ was increased ≈2- to 3-fold (Fig. 3C). Analysis of sorted cells revealed a detectable amount of nondeleted alleles remaining in the population infected after full Th2 differentiation (Fig. 3A).
GATA-3 Is Required for Th2 Cytokine Production in Vivo. To address whether the defect in Th2 cytokine production would occur in response to specific antigens in the intact animal, GATA-3-deficient animals and FF controls were injected s.c. with TNP-KLH antigen suspended in alum to elicit a Th2-biased response. Draining and nondraining lymph nodes were harvested 7 days later and restimulated with or without TNP-KLH in vitro, and cytokine production was measured by ELISA. As expected, draining lymph node cells from FF animals restimulated with antigen in vitro made IL-4 but very little IFN-γ in response to antigen (Fig. 4). In contrast, lymph node cells from knockout animals made 3- to 8-fold more IFN-γ in comparison with controls and made nearly nondetectable amounts of IL-4 (Fig. 4). Draining lymph nodes cultured in the absence of antigen and nondraining lymph nodes cultured in the presence of antigen did not produce detectable amounts of either IL-4 or IFN-γ (Fig. 4), demonstrating that lymphocytes from FF cre animals, while preactivated, did not show aberrant, antigen-independent cytokine production. Thus, GATA-3 was clearly required for induction and maintenance of Th2 responses in isolated cells in culture and in whole animals.
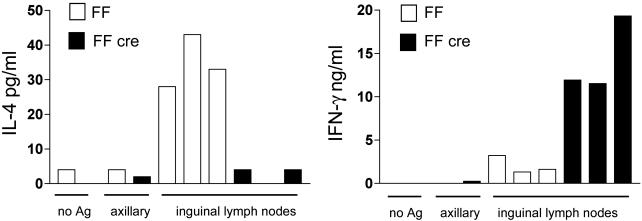
Antigen-specific recall responses in FF and FF cre animals. The amount of IL-4 (Left) and IFN-γ (Right) produced by cells from draining (inguinal) lymph nodes of FF and FF cre animals immunized with TNP-KLH is shown. As controls, cytokine production from cells grown without antigen (no Ag) and cells from nondraining (axillary) lymph node is also shown.
Discussion
While overexpression, antisense, and transgenic models have indicated that GATA-3 can induce Th2 differentiation, we demonstrate definitively here that GATA-3 is indeed required both in vitro and in vivo for optimal Th2 differentiation. CD4 T cells from GATA-3-deficient animals were highly deficient in Th2 cytokine production when differentiated in vitro (Fig. 2B). The variability of Cre-mediated excision we observed fortuitously brought out the exquisite dose dependence of the Th2 phenotype on the extent of GATA-3 deletion. Therefore, it is not surprising to us that a substantial proportion of developing Th2 cells rendered GATA-3-deficient in vitro by retroviral transduction of Cre continued to express Th2 cytokines (Fig. 3B). This likely reflects residual GATA-3 protein persisting after gene deletion, both because cells were not transduced until 30 h after stimulation (during which time GATA-3 would have already induced heritable epigenetic changes to cytokine loci) and also because the fall in the level of GATA-3 message and protein may lag behind excision of the gene. In a system with timely and complete loss of GATA-3 protein, perhaps all Th2 cytokine production would be abolished.
Alternatively, the residual Th2 cells we observed may reflect Th2 differentiation that is truly GATA-3-independent. Recently, it has been demonstrated that overexpression of signal transducer and activator of transcription 5a in developing Th1 cells induces the production of IL-4 and potentially other Th2 cytokines in the absence of GATA-3 activation (34); because IL-2 was present during differentiation in the data presented here, this pathway would be expected to be active in our system. How GATA-3-independent pathways of Th2 cell differentiation might function remains to be investigated.
The IL-4/IL-5/IL-13 locus is subject to chromatin remodeling in Th2 differentiated cells and also in the setting of GATA-3 overexpression (10–12, 14–16, 25, 35). Whether GATA-3 is also required for the remodeling of the IL-4/IL-5/IL-13 locus is now under active investigation. IL-10, in contrast, is located outside of this locus, yet it is coordinately produced with the other Th2 cytokines. While cell-type nonspecific factors such as Sp1, Sp3 (36), cAMP response element binding protein/activating transcription factor, and CCAAT/enhancer binding protein (37) have been shown to be important for IL-10 promoter activity, T cell-specific factors or factors controlling the accessibility of this locus to the transcriptional machinery in Th2 cells have not been identified. Interestingly, production of IL-10 was also reduced in GATA-3-deficient Th2 cells (Fig. 3C), demonstrating that GATA-3 is a major orchestrator of coordinated Th2 responses. The mechanism by which GATA-3 controls IL-10 production is entirely unknown and could involve direct binding to the promoter and/or remodeling of IL-10 locus at the chromatin level.
Established Th2 cells rendered deficient in GATA-3 by retroviral transduction had a reduction in Th2 cytokine production (Fig. 3C). The sensitivity of established Th2 cells to a fall in GATA-3 levels is further underscored by the fact that the degree of deletion in cells infected with Cre virus during secondary stimulation was not always complete (Fig. 3A), yet the reduction in Th2 cytokine production was similar to that seen in developing Th2 cells transduced with Cre. Our findings suggest that high levels of GATA-3 protein must be continuously sustained to support Th2 cytokine production and that establishment of the Th2 lineage is a dynamic process that must be actively maintained. The mutability of the Th2 phenotype introduces an additional level of regulation that should allow for a more flexible immune response. These results also have profound implications for the development of drugs targeting GATA-3 or other Th1/Th2-specific transcription factors for the treatment of asthmatic, allergic, and autoimmune diseases even after the diseases have been well established.
Acknowledgments
We thank Drs. Garry P. Nolan and Daniel Littman for providing retroviral constructs and protocols, members of the laboratories of Dr. Laurie Glimcher and Dr. Michael Grusby for valuable discussions, and Dr. Michael B. Yaffe for critical review of the manuscript. This work was supported by K08 Award AI50601 (to S.-Y.P.) and R01 Award AI29673 from the National Institute of Allergy and Infectious Diseases and a Junior Award from the Sandler Program for Asthma Research (to I.-C.H.).
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.0308697100
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc357040?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Mef2d potentiates type-2 immune responses and allergic lung inflammation.
Science, 384(6703):eadl0370, 28 Jun 2024
Cited by: 1 article | PMID: 38935708 | PMCID: PMC7616247
Plasticity and lineage commitment of individual TH1 cells are determined by stable T-bet expression quantities.
Sci Adv, 10(23):eadk2693, 05 Jun 2024
Cited by: 1 article | PMID: 38838155
Deciphering the developmental trajectory of tissue-resident Foxp3+ regulatory T cells.
Front Immunol, 15:1331846, 28 Mar 2024
Cited by: 0 articles | PMID: 38605970 | PMCID: PMC11007185
Review Free full text in Europe PMC
MicroRNA-194 regulates parasitic load and IL-1β-dependent nitric oxide production in the peripheral blood mononuclear cells of dogs with leishmaniasis.
PLoS Negl Trop Dis, 18(1):e0011789, 19 Jan 2024
Cited by: 0 articles | PMID: 38241360 | PMCID: PMC10798644
Transcriptional regulation on effector T cells in the pathogenesis of psoriasis.
Eur J Med Res, 28(1):182, 03 Jun 2023
Cited by: 8 articles | PMID: 37270497 | PMCID: PMC10239166
Review Free full text in Europe PMC
Go to all (237) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Enforced expression of GATA-3 in transgenic mice inhibits Th1 differentiation and induces the formation of a T1/ST2-expressing Th2-committed T cell compartment in vivo.
J Immunol, 167(2):724-732, 01 Jul 2001
Cited by: 59 articles | PMID: 11441076
Inhibition of Th2 differentiation and GATA-3 expression by BCL-6.
J Immunol, 170(5):2435-2441, 01 Mar 2003
Cited by: 109 articles | PMID: 12594267
Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses.
Nat Immunol, 5(11):1157-1165, 10 Oct 2004
Cited by: 430 articles | PMID: 15475959
The function role of GATA-3 in Th1 and Th2 differentiation.
Immunol Res, 28(1):25-37, 01 Jan 2003
Cited by: 97 articles | PMID: 12947222
Review
Funding
Funders who supported this work.
NIAID NIH HHS (5)
Grant ID: K08 AI050601
Grant ID: R37 AI029673
Grant ID: R01 AI29673
Grant ID: AI50601
Grant ID: R01 AI029673





