Abstract
Free full text

miR-7 suppresses brain metastasis of breast cancer stem-like cells by modulating KLF4
Abstract
Despite significant improvement in survival rates of breast cancer patients, prognosis of metastatic disease is still dismal. Cancer stem-like cells (CSCs) are considered to play a role in metastatic progression of breast cancer; however, the exact pathological role of CSCs is yet to be elucidated. In this report, we found that CSCs (CD24−/CD44+/ESA+) isolated from metastatic breast cell lines are significantly more metastatic than non-CSC populations in an organ specific manner. The results of our microRNA profile analysis for these cells revealed that CSCs that are highly metastatic to bone and brain expressed significantly lower level of miR-7 and that this microRNA was capable of modulating one of the essential genes for induced pluripotent stem cell, KLF4. Interestingly, high expression of KLF4 was significantly and inversely correlated to brain- but not bone-metastasis free survival of breast cancer patients, and we indeed found that the expression of miR-7 significantly suppressed the ability of CSCs to metastasize to brain but not to bone in our animal model. We also examined the expression of miR-7 and KLF4 in brain-metastatic lesions and found that these genes were significantly down- or up-regulated, respectively, in the tumor cells in brain. Furthermore, the results of our in vitro experiments indicate that miR-7 attenuates the abilities of invasion and self-renewal of CSCs by modulating KLF4 expression. These results suggest that miR-7 and KLF4 may serve as biomarkers or therapeutic targets for brain metastasis of breast cancer.
Introduction
Breast cancer is becoming a curable disease when it is diagnosed at an early stage; however, the majority of cancer death is still attributed to metastatic disease. Despite this clinical importance, the exact pathological process of tumor metastasis is as yet poorly understood, and deciphering the exact molecular mechanism of this process is of paramount importance in order to identify specific therapeutic targets for this devastating disease. It is generally believed that a series of mutations in multiple genes are needed for cancer cells to become metastatic; however, recent cancer stem cell theory which still remains as a hypothesis predicts that metastatic tumor cells are by definition cancer stem cells and that these cells may exist in tumor mass even at an early stage. This hypothesis well explains the occurrence of metastatic disease which occasionally is observed in early stage patients. CSCs are a minor population of tumor mass and considered to have abilities of tumor –initiation, differentiation and chemo-resistance (1). In breast cancer, CSCs or tumor –initiating cells were first identified by using a combination of cell surface markers, CD24−/CD44+/ESA (EpCAM)+ (2). More recently, aldehyde dehydrogenase 1 (ALDH1) has been identified as a new CSC marker, and the cells isolated by this marker partially overlap with previous identified CSC population (3). CD44+/CD24− cells are readily detectable in metastatic pleural effusions, and signature generated from CD44+/CD24− cells suggests a metastatic ability of this population (4, 5). Interestingly, CD44+/CD24− population can be generated by induction of epithelial-mesenchymal transition (EMT) by treating breast epithelial cells with EMT-inducing agents such as transforming growth factor beta 1 (TGF-β1) (6). It should be noted that EMT is considered to be a crucial event in the metastatic process that involves the acquisition of a migratory mesenchymal phenotype (7).
Recent active research in microRNA identified a series of this type of molecules that are involved in tumor progression in various tumors as oncogenes and tumor suppressors (8, 9). In breast cancer, miR-21, 155, 27, 96, 182 and128 were identified as oncogenes, while miR-125, 205, 27, 17, 206 and 145 were found to be tumor suppressor genes, and their corresponding target genes were also defined (10–12). Therefore, microRNA network is considered to play critical roles in tumor initiation and progression. However, much less information is available for microRNA in cancer stem cells, particularly in relation to the metastatic ability of CSCs in breast cancer, and therefore, it is of significant interest to identify such microRNAs which may serve as novel therapeutic targets for metastatic breast cancer. Toward this goal, we isolated CSCs population (CD24−/CD44+/ESA+) from metastatic breast cells and performed microRNA profile analysis. We found that miR-7 was significantly down-regulated in metastatic CSCs and specifically blocked brain metastasis in our animal model by modulating the KLF4 expression.
Materials and Methods
Cells and cell culture
We purchased MDA-MB-231 (MB231) and MCF7 breast cancer cell lines from ATCC. The 231BoM-1833, 231BrM-2a, CN34, CN34-BoM2d, CN34-BrM2c and MCF7-BoM2d cells were a kind gift from Dr. Joan Massagué (Memorial Sloan-Kettering Cancer Center). Firefly luciferase-labeled cells were generated by lentivirus infection. 293TN cells were obtained from System Bioscience. MB231, MB231 variant cells, MCF7, MCF7-BoM2d and 293TN cells were cultured in DMEM medium supplemented with 10% FBS and antibiotics. CN34 and its variant cells were cultured in Medium199 supplemented with 2.5% FBS, 10 μg/ml insulin, 0.5 μg/ml hydrocortisone, 20 ng/ml EGF, 100 ng/ml cholera toxin and antibiotics. The immortalized mouse brain microvascular endothelial cells (mBrEC) were supplied by Dr. Isaiah J. Fidler (M.D. Anderson Cancer Center) (13). mBrEC was maintained at 8% CO2 at 33 °C in DMEM with 10% FBS, 2 mM of L-glutamine, 1 mM of sodium pyruvate, 1% of non-essential amino acids and 1% of vitamin mixture. Breast tumor cells that were directly transplanted to animal only one generation without in vitro culture were obtained from conversant biologics, Stem cell populations were isolated as described previously (14). Isolated CSCs were cultured in DMEM/F12 medium with 2%B27, 20ng/ml EGF, 4μg/ml insulin and 0.4% of BSA.
MicroRNA microarray profiling
Total RNAs were isolated from CSCs from MB231, 231BoM and 231BrM cells using miRNeasy RNA isolation kit (Qiagen). Micro RNA expression profiling was determined by miRNA microarray analysis by using the human miRNA chip (LC Science, miRBase version 14.0). Clustering and its visualization were performed using Cluster3.0 and TreeView softwares.
Real-time PCR and Western blotting
Total RNA was isolated from cells using TRIzol reagent (Invitrogen) and from paraffin-embedded human tissue samples using miRNeasy FFEP RNA purification kit (Qiagen), respectively. We used TaqMan MicroRNA Reverse Transcription kit and High Capacity RNA-to-cDNA kit for microRNA and mRNA reverse-transcription, respectively (Applied Biosystems). qPCR was performed by using TaqMan Universal Master Mix II and TaqMan microRNA assays or TaqMan gene expression assays (Applied Biosystems). A human RNU48 and actin TaqMan probes were used as endogenous controls for micro RNA and regular gene expressions, respectively. Western blotting was carried out using a general method with KLF4 antibody (Santacruz). qRT-PCR was performed using the SYBR Green qPCR Kit (Fermentas) by using following primers: Actin F: TGAGACCTTCAACACCCCAGCCATG, Actin R: CGTAGATGGGCACAGTGTGGGTG, KLF4 F: GTCTTGAGGAAGTGCTGAGC, KLF4 R: ATCGTCTTCCCCTCTTTGGC.
Plasmids
The plasmids expressing hsa-mir-7-1 and hsa-mir-7-2 precursors in lentiviral pCDH-CMV-MCS-EF1-copGFP vector (System Bioscience) were supplied by Yin-Yuan Mo. pSIN-KLF4-puro lentiviral plasmid was constructed by subcloning from pLM-mCherry-2A-KLF4 purchased from Addgene. KLF4 reporter plasmid, pMir-Report-KLF4-3′-UTR was a kind gift from Dr. Kenneth S. Kosik (University of California) (15). Psin-miR7-2 plasmid was constructed by subcloning PCR product from pCDH-CMV-MCS-EF1-copGFP-miR7-2. Deletion of miR-7 binding core sequence on 3′-UTR region of KLF4 gene was achieved using overlap extension PCR methods (16) by using following primers: pMIR-REPORT-F: GTCCAAATTGCTCGA GTGAT, pMIR-REPORT-R: AGGCGATTAAGTTGGGTA, KLF4 mut1 F: ACTTTTCACAC TCGATGAGGGAAG, KLF4 mut1 R: CTTCCCTCATCGAGTGTGAAAAGT, KLF4 mut2 F: AAATCTATATTTGATCAACATTTA, KLF4 mut2 R: TAAATGTTGATCAAATAT AGATTT,
3′UTR Reporter assay
10 ng of reporter plasmid and 200ng of miR7 expression plasmid were co-transfected with 1 ng of phRG-TK Renilla luciferase internal control plasmid (Promega) into 293TN cells using Lipofectamine 2000 reagent (Invitrogen). After 24 hours, luciferase activities were measured by using dual-luciferase reporter assay system (Promega).
Transfection
For reporter assay, cells were transfected with Lipofectamine 2000. For knock down of miR7, cells were transfected with Locked nucleic acid (LNA™) targeting miR-7 (Exiqon) using RNAifectin reagent (Applied Biological Materials).
Matrigel invasion and transmigration assays
For Matrigel-invasion assay, CSCs were labeled with Cell tracker green (Invitrogen) and fifty-thousand cells were seeded into Matrigel-coated trans-well insert (Corning) supplemented with DMEM with 10% serum. The bottom side of trans-wells was filled with DMEM with 20% serum. For transmigration assay, one-hundred-thousand of mBrEC were seeded into trans-well insert (Corning, pore size 3 μm) and allowed to grow to confluence for 1 day. CSCs were labeled with Cell tracker green and fifty-thousand of cells were seeded into trans-well inserts supplemented with DMEM with 10% serum. The bottom side of trans-wells was filled with DMEM with 20% serum. After 24 hours, labeled cells were counted under the fluorescent microscope.
Animal experiments
For experimental metastasis assay, nude mice (7–8 weeks) were injected with fifty-thousand luciferase-labeled CSCs in PBS into left cardiac ventricle in a total volume of 100 μl. To confirm a successful injection, the photon flux from whole body of the mice was immediately measured using IVIS Xenogen bioimager (Caliper). The brain metastasis progression was monitored and the luminescence was quantified. At the endpoint of this study, whole brain was removed, incubated in RPMI-1640 medium with 0.6 mg/ml luciferin for 15 min and photon flux was monitored.
Sphere formation assay
Metastatic variant of MCF7, MCF7-BoM2d cells, were suspended in DMEM-F12 medium supplemented with 2% of B27 supplement, 0.4% bovine serum albumin (BSA), 4 μg/ml insulin, 20 ng/ml basic fibroblast growth factor (bFGF), and 20 ng/ml epidermal growth factor (EGF) (Invitrogen). Cells were then seeded in 96-well Ultra-low attachment plates (Corning) as a density of 500 per well. 8 days later, mammospheres in the plate were counted under the microscope. For passage culture, MCF7-BoM2d cells were seeded in low-attachment 10cm dish. After 8 days, mammospheres were collected by using 40μm mesh cell strainer, trypsinized and seeded in another 10cm dish. This passage culture was repeated 4 times.
MTS assay
Two-thousand CSCs were seeded in a 96-well plate with DMEM medium with 1% FBS for 72 hours. After the incubation, cell proliferation was measured by the MTS dye method (Promega).
Statistical analysis
For in vitro experiments, T-test or one-way ANOVA was used to calculate the p-values. Wilcoxon rank sum test was used to calculate the p-value for ex vivo bioluminescence from brain and expression level of miR-7 and KLF4 in human specimens. The Kaplan-Meier method was used to calculate the survival rates and was evaluated by the log-rank test.
Results
miR-7 is down-regulated in metastatic CSCs
To identify microRNAs which are specific to metastatic CSCs, we first isolated CSCs population using well established markers, CD24−, CD44+ and ESA+, from human breast cancer cell line MDA-MB231, and also from its variants, 231BoM and 231BrM. The latter two cell lines were established by Massague’s group as highly metastatic variant to bone and brain, respectively. These cells were examined for their tumor initiating ability by injecting them into mammary fat pad of nude mice. The results of our limiting dilution analysis confirmed that the isolated CSCs (CD24−/CD44+/ESA+) population has significantly stronger ability of generating tumors compared to non-stem cell population (Table 1). We then examined the metastatic ability of these cells by implanting various doses of CSCs into mice via intracardiac injection. As shown in Table 2, we found that CSCs from 231BoM and 231BrM were more metastatic to bone and brain, respectively, compared to non-stem cells. These results strongly support our notion that metastatic cells derive from CSCs and they metastasize in an organ specific manner.
Table 1
Limiting dilution analysis for tumor incidence and metastasis of CSCs in nude mice
| Incidence of tumors/Number of injections Cells per injection | ||||||
|---|---|---|---|---|---|---|
| Strain | Population | 104 | 103 | 102 | 10 | CSC frequency (95% confidence interval) |
| MB231 | Stem cells | 6/6 | 5/6 | 2/6 | (1/183–1/1,097) *** | |
| Non-stem cells | 1/2 | 0/2 | 0/2 | (1/2,356–1/118,284) | ||
|
| ||||||
| 231BrM | Stem cells | 4/4 | 5/6 | 2/6 | (1/19–1/110)*** | |
| Non-stem cells | 5/6 | 1/6 | 0/6 | (1/236–1/1,374) | ||
|
| ||||||
| 231BoM | Stem cells | 7/7 | 7/7 | 0/1 | (1/7–1/89) *** | |
| Non-stem cells | 2/4 | 0/4 | 0/4 | (1/419–1/6,668) | ||
CSCs isolated from MB231, 231BoM and 231BrM were injected subcutaneously into the mammary fat pad of nude mice, and the growth of tumors was monitored by BLI.
Table 2
| Incidence of brain metastasis/Number of injections Cells per injection | frequency of brain metastatic cell (95% confidence interval) | |||
|---|---|---|---|---|
| 231BrM | 105 | 104 | 103 | |
| Stem Cells | 4/4 | 6/6 | 3/6 | 1/1425 (1/4,282–1/474)*** |
| Non-Stem Cells | 3/4 | 3/6 | 0/5 | 1/40,529 (1/112,528–1/14,598) |
| Incidence of bone metastasis/Number of injections Cells per injection | frequency of bone metastatic cell (95% confidence interval) | |||
|---|---|---|---|---|
| 231BoM | 105 | 104 | 103 | |
| Stem Cells | 6/6 | 4/5 | 1/5 | 1/5,802(1/15,427–1/2,182)*** |
| Non-Stem Cells | 2/6 | 0/5 | 0/5 | 1/274,471 (1/1,094,044–1/68,859) |
CSCs isolated from 231BoM and 231BrM were injected intracardially into the nude mice, and the growth of brain and bone metastasis were monitored by BLI.
Next, we performed microRNA profile analysis of CSCs prepared from MDA231, 231BoM and 231BrM by extracting RNAs from these cells followed by hybridizing them to a microRNA chip (LC Science) which contains 894 human microRNAs. The results of our array analysis revealed that there are eight microRNAs whose expressions were significantly altered among these CSCs, and that two microRNAs, miR-7 and miR-21, were significantly reduced in both 231BoM and 231BrM compared to MDA-MB231 (Figure 1A). We then confirmed the expression of miR-7 and miR-21 in CSCs of these cells by Taqman qPCR and found that only miR-7 was significantly down-regulated in CSCs in both 231BoM and 231BrM, suggesting that miR-7 may contribute to the metastatic ability of CSCs (Figure 1B). To further clarify this notion, we examined the specificity of miR-7 expression in CSCs and non-stem cell population from MDA-MB231, 231BoM and 231BrM. As shown in Fig. 1C, the expression of miR-7 was significantly reduced in CSCs compared to non-stem cells in both 231BoM and 231BrM, but not in CSCs of MDA-MB231, strongly suggesting that miR-7 is specific to metastatic CSCs. The generality of this observation was further confirmed by examining the expression of miR-7 in two different cell lines, CN34 and MCF7 and their metastatic variants, CN34BoM, CN34BrM and MCF7BoM. Again, we found that the expression of miR-7 was significantly suppressed in the bone and brain metastatic variants of these cells in a CSC-specific manner (Figure 1C). We also found that the expression of miR-7 was significantly lower in mammospheres of MCF7BoM compared to the parental cells, suggesting that miR-7 plays a role of self-renewal of CSCs (Figure 1D). Indeed, we found that ectopic expression of miR-7 significantly suppressed the ability of sphere formation of MCF7BoM cells, while miR7-specific LNA enhanced the sphere forming ability (Figure 1E).
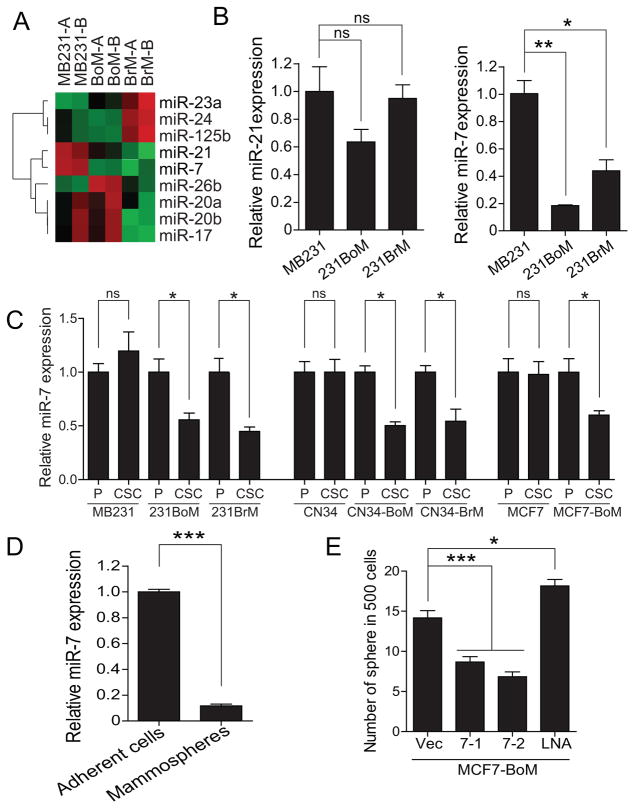
(A) CSCs from MB231, 231BoM and 231BrM were isolated, and their RNAs were subjected to microRNA array analysis. A heatmap was generated for the MicroRNAs that were significantly up- or down-regulated in CSCs among 231BrM and 231BoM compared to MB231. (B) The expression of miR-21 and miR-7 were examined by TaqMan PCR in CSCs prepared from MB231, 231BoM and 231BrM. (C) miR-7 expressions in both parental cells and CSCs from different cell lines were measured by TaqMan PCR. P; Unsorted parental cells, CSCs; cancer stem cell. (D) The expression of miR-7 was examined in adherent cells and mammospheres derived from MCF7-BoM cells by TaqMan PCR. (E) Sphere forming ability was measured in MCF7BoM cells that are ectopically expressing miR-7 or miR7-LNA. Data are represented as mean±SEM (n=3). *** indicates p<0.0001, ** indicates p<0.001, * indicates p<0.05, ns indicates not significant.
miR-7 suppresses the expression of KLF4 gene
To understand the functional role of miR-7 in CSCs, we first searched its potential target genes that may be involved in stem cell physiology using multiple database including TargetScan, PicTar, miRanda and SLOAN-Kettering. We found that KLF4 which is one of the IPS genes is among the highest scored genes on the target list and that there are two potential miR7 target sites in 3′ UTR region of this gene. The target site 1 is conserved in mammals while the target site 2 is well conserved among vertebrates (Figure 2A). To examine the effect of miR-7 on the expression of KLF4, we ectopically expressed miR-7-1 or miR-7-2 in 231BoM and 231BrM cells by infecting corresponding lentivirus expression vector followed by qRT-PCR analysis. As shown in Figure 2B, we found that both miR-7-1 and miR-7-2 significantly suppressed the expression of KLF4 in these cells, and the similar suppressive effect of miR-7 was observed when we used CN34BoM, CN34BrM and MCF7BoM cells. These results were also confirmed by Western blot analysis (Figure 2C). To further validate our results, we co-transfected lentiviruses carrying miR-7 expression vector and the luciferase reporter plasmid with 3′-UTR of KLF4 into 293TN cells. As shown in Figure 2D, the reporter activity of 3′UTR of KLF4 was significantly suppressed by miR-7, while co-transfection of LNA targeting miR-7 stimulated the expression of this gene. We then introduced mutations in the two target sequences of 3′UTR, either alone or in combination, and examined their abilities to respond to miR-7. We found that the deletion of target site 1(MT1) partially inhibited the suppressive effect of miR-7, while the deletion of the target site 2 (MT2) or a combination of both sites almost completely attenuated the effect of miR-7 (Figure 2E). These results strongly suggest that miR-7 is capable of suppressing the expression of KLF4 by binding to these target sequences of 3′-UTR.
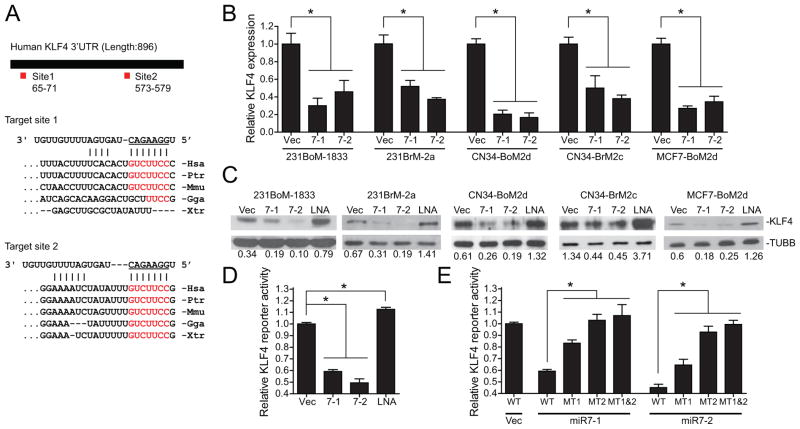
(A) Schematic representation of the KLF4 3′-UTR with two miR-7 binding sites and sequence alignment of predicted miR-7 binding sites on KLF4 3′-UTR. Complementary sequences of miR7 to mammalian KLF4 3′-UTR are shaded red. Hsa, human; Ptr, chimp; Mmu, mouse; Gga, chicken; Xtr, frog. (B) 231BoM-1833, 231BrM-2a, CN34-BoM2d, CN34-BrM2c and MCF7-BoM2d were infected with lentivirus expressing pri-miR-7-1, pri-miR-7-2 or vector only and the KLF4 mRNA levels were measured by RT-PCR. (C) Western blotting analysis of KLF4 in the same set of CSCs as described in (B). (D) KLF4 3′-UTR luciferase reporter plasmid was transfected in 293TN cells with indicated microRNAs or LNA. The luciferase activities were then measured after 24hrs of transfection. (E) Constructs carrying KLF4 3′ UTR luciferase reporter or deletion mutant of miR7 binding sites (site 1; MT1, site 2; MT2) were transfected in 293TN cells with indicated miRNAs. Cells were harvested and luciferase activities were measured after 24hrs transfection. * indicates p<0.05
MiR-7 suppresses brain metastasis of CSCs in vivo
The results of our in vitro experiments strongly suggest that miR-7 plays a role in metastatic CSCs by modulating the expression of KLF4. Interestingly, when we analyzed the relationship between KLF4 and metastatic status of breast cancer using the existing GEO data base for combined 710 patients(GSE12276, GSE2034, GSE2603, GSE5327, and GSE14020), we found that the expression of KLF4 was significantly and inversely correlated with brain but not bone metastasis-free survival (Figure 3A). To validate this observation, we next examined the effect of miR-7 in metastatic ability of CSCs in vivo. CSCs were prepared from 231BrM and CN34BrM cells that were infected with lentivirus carrying with or without miR-7-2, and they were transplanted into nude mice through intracardiac injection followed by monitoring metastatic tumor growth in the brain. As shown in Figure 3B and C, bioluminescent image right after the tumor injection showed distribution of tumor cells in the whole body indicating a successful intracardiac injection. After 5 weeks, metastatic tumor growth in the brain was prominent in all mice that received CSCs carrying vector only. On the other hand, animals that received CSCs carrying miR-7-2 showed significantly less signals in the brain, suggesting that miR-7-2 was indeed capable of suppressing brain metastasis of CSCs (Figure 3B, C). We then performed a similar experiment using the CSCs prepared from the bone metastatic cell line, 231BoM and MCF7BoM which carried lentivirus with or without miR-7-2. Interestingly, however, we found that miR-7-2 did not affect the incidence or growth of bone metastasis (Figure 3D and Supplementary Figure 1 A). We also found that overexpression of miR7 in 231BrM cells decreased the tumor size by around 30%, and this effect was blocked by ectopic expression of KLF4, indicating that miR7 does affect primary tumor but not to the level of metastasis (Supplementary Figure 1B). Ectopic expression of KLF4 alone in 231BrM did not affect the primary tumor growth in vivo and this is due to the fact that the endogenous level of KLF4 in 231BrM cells is already quite high. These observations are consistent with the results of clinical cohort analysis in Fig. 3A and strongly support the notion that miR-7-2 specifically suppresses brain metastasis by down-regulating KLF4.
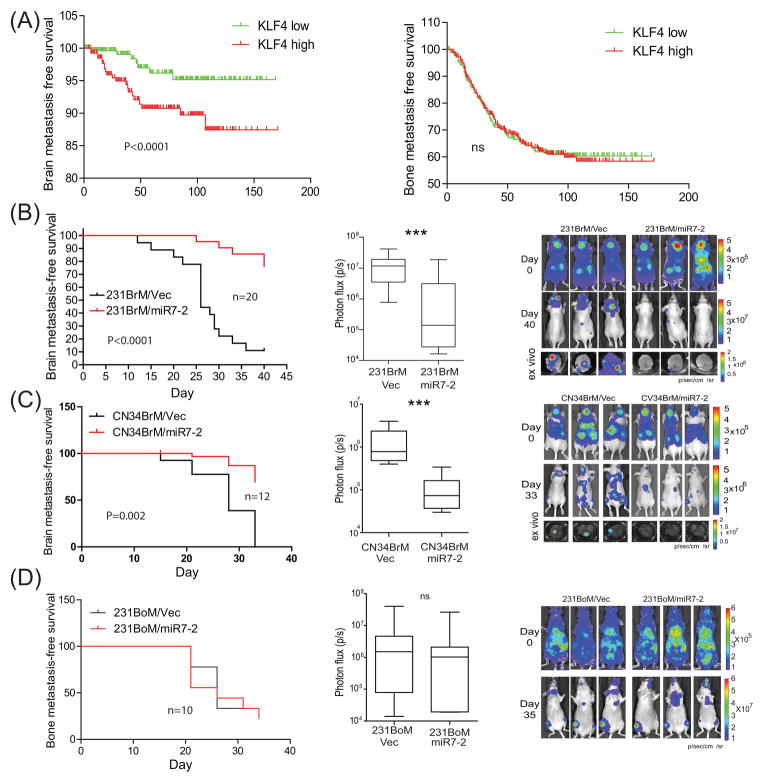
(A) Kaplan-Meier analysis for brain and bone metastasis-free survival of 710 breast cancer patients in GEO data bases (GSE12276, GSE2034, GSE2603, GSE5327, and GSE14020). Patients were divided into two groups based on the expression status of KLF4 in their primary tumors. (B) Left panel; Kaplan-Meier analysis for brain metastasis-free survival of mice inoculated with CSCs that were isolated from 231BrM/Vec or 231BrM/miR7-2. Middle panel; total photon flux of brain metastatic lesions was measured by BLI at the end point. Right panel; BLI images of brain metastatic lesions of three representative mice from each experimental group. (C) Left panel; Kaplan-Meier analysis for brain metastasis-free survival of mice inoculated with CSCs that were isolated from CN34BrM/Vec or CN34BrM/miR7-2. Middle panel; total photon flux of brain metastatic lesions was measured by BLI at the end point. Right panel; BLI images of brain metastatic lesions of three representative mice from each experimental group.(D) Left panel; Kaplan-Meier analysis for brain metastasis-free survival of mice inoculated with CSCs that were isolated from 231BoM/Vec or 231BoM/miR7-2. Middle panel; total photon flux of bone metastatic lesions was measured by BLI at the end point. Right panel; BLI images of bone metastatic lesions of three representative mice from each experimental group. *** indicates p<0.0001
miR-7 blocks invasion and proliferation of CSCs
To further understand the effect of miR-7 and KLF4 on tumor metastasis, we examined the transmigrating ability of CSCs of 231BrM through brain endothelial cells that were originated from mouse brain blood vessel. As shown in Figure 4A, we found that miR-7 significantly suppressed transmigration of CSCs of 231BrM, while LNA that was targeted to miR-7 significantly stimulated the transmigrating ability of CSCs from MB231. We also examined the invasive ability of these cells using Matrigel invasion chamber and found that miR-7 was also able to block the invasion of CSCs through Matrigel (Figure 4A right panel). We then tested the effect of KLF4 on transmigrating and invasive ability of CSCs using the same assay systems. As shown in Figure 4B, suppressive ability of miR-7 on both transmigration through endothelial cells and Matrigel was significantly attenuated by ectopic expression of KLF4, suggesting that miR-7 suppresses invasive ability of CSCs by blocking KLF4 expression. Next, because KLF4 is involved in self-renewal of stem cell, we also examined the effect of both miR-7 and KLF4 on proliferation of CSCs by MTS assay and found that miR-7 was capable of blocking the growth of CSCs, while ectopic expression of KLF4 significantly mitigated this effect of miR-7 (Figure 4C).
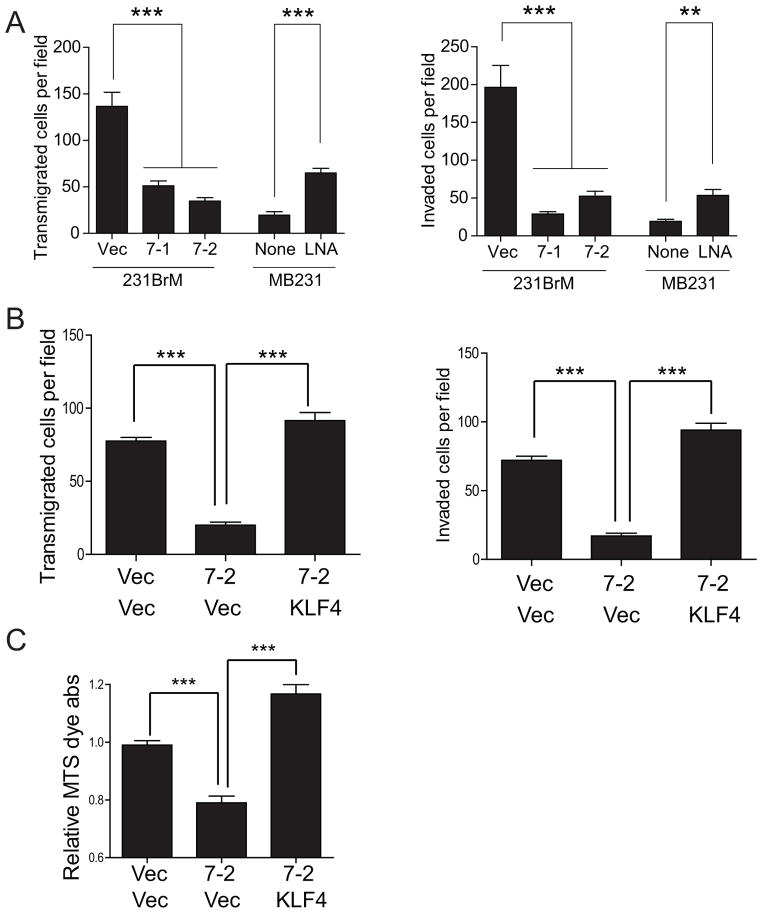
(A) CSCs isolated from 231BrM and MB231 with the ectopic expression of miR7 and miR7-LNA were seeded on top of mouse brain endothelial cells or matrigel. The number of invaded cells was counted after 24hrs. (B) CSCs isolated from 231BrM infected with lentivirus of miR7 or KLF4 expression plasmids were seeded on top of mouse brain endothelial cells or matrigel. The number of invaded cells was counted after 24hrs. (C) Cell growth was measured by MTS assay in the CSCs isolated from 231BrM infected with lenti virus of miR7 or KLF4 expression plasmids. *** indicates p<0.0001, ** indicates p<0.001
KLF4 promotes self-renewal of CSCs
One of the most important features of CSCs is their ability to maintain undifferentiated status and self-renewal. To evaluate whether KLF4 regulates the self-renewal of CSCs, we performed FACS analysis of 231BrM cells with or without ectopic expression of KLF4 followed by sorting the cells using the CSCs markers (CD24, CD44 and ESA). As shown in Figure 5A, ectopic expression of KLF4 indeed significantly increased CSCs population in 231BrM cells. We also found that KLF4 significantly enhanced the mammosphere forming ability of 231BrM cells, which is another hallmark of CSCs (Figure 5B). To further validate our result in clinical samples, we obtained primary tumor from advanced breast cancer patients and the tissue was passaged once in NOD/SCID mouse without in vitro culture. The tumor cells were dissociated and the cells were infected with KLF4 lentivirus and cultured in low-attachment plates. We then measured CSCs population by FACS after 72hrs as well as their mammosphere forming ability by counting the number of spheres after 10 days. As shown in Figure 5 C and D, we again found that KLF4 significantly enriched the CSCs populations and mammosphere forming ability. On the other hand, over expression of miR7 significantly decreased the mammosphere forming ability of primary breast tumor cells (Figure 5D).
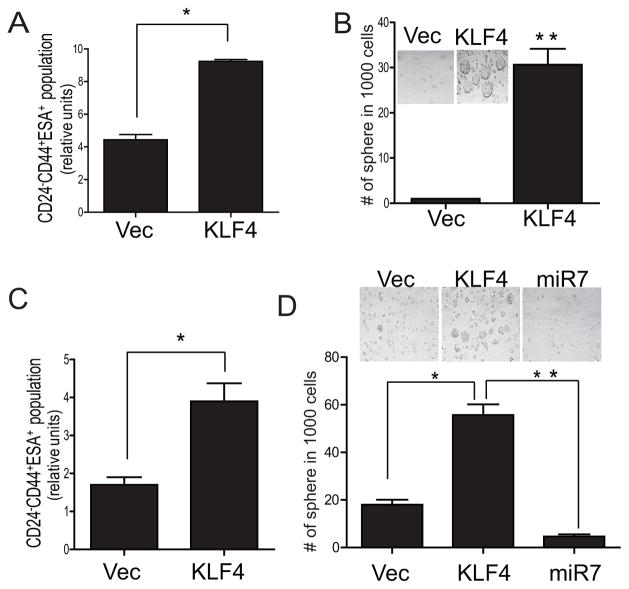
(A) 231BrM was infected with KLF4 or control lenti virus and the CSCs population (CD24− CD44+ ESA+) were measured by FACS after 72hrs. (B) CSCs were isolated from 231BrM infected with KLF4 or control lenti virus and cultured in low-attachment plates. The number of mammospheres was counted after 10 days. (C) Primary breast cancer cells isolated from advanced cancer patients were directly infected with KLF4 or control lenti virus, and the CSCs population (CD24− CD44+ ESA+) were measured by FACS after culturing the cells for 72 hrs in low-attachment plates. (D) The same primary breast cancer cells infected with KLF4 or control lenti virus were cultured in low attachment plates and the number of mammosphere was counted after 10 days. * indicates p<0.05, ** indicates p<0.001
The expressions of miR-7 and KLF4 are inversely correlated in brain metastatic lesions of breast cancer patients
To further validate the clinical significance of miR-7 and KLF4 in brain metastasis of breast cancer, we microdissected tumor tissues from both primary and brain metastatic lesions followed by performing TaqMan qRT-PCR. We found that the expression of miR-7 was significantly lower in brain metastatic tumors than that in primary tumor tissues (Figure 6A left panel), while the expression of KLF4 was significantly higher in brain metastatic tissue compared to primary tumors (Figure 6A right panel). The results of co-relation analysis between miR-7 and KLF4 indicate that there was a significant inverse correlation between these two genes in brain metastatic tissues (Figure 6B). Furthermore, we examined the expression of KLF4 in brain metastatic tissues by immunohistochemistry and found that KLF4 was expressed more in the nucleus and significantly less in cytosol in brain metastatic tissues compared to primary tumors (Figure 6C,D). These clinical data are consistent with our notion that miR-7 suppresses brain metastasis through inhibition of KLF4 expression.
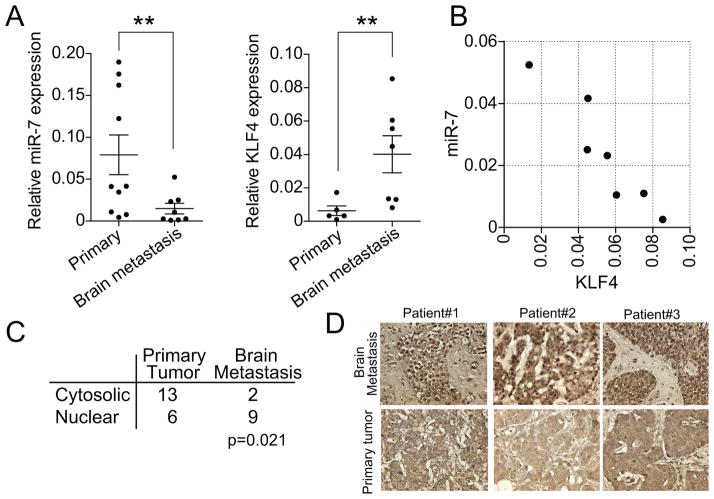
(A) Expression of miR-7 and KLF4 was measured by TaqMan PCR in primary and brain metastasis samples after micro-dissection. The relative expression level of miR-7 to RNU48 (n=8–10 per group) and the expression level of KLF4 to ACTB were plotted (n=7–10 per group). (B) Correlation of the miR-7 and KLF4 expression in each sample. (C) Immunohistochemical analysis for KLF4 expression was performed for primary breast tumors and brain metastatic tumors, and the numbers of nuclear or cytosolic staining were assessed under microscope. (D) Representative photos of KLF4 staining in primary tumor and brain metastasis samples. ** indicates p<0.001
Discussion
According to the recent cancer stem cell theory, metastatic cells are considered to have the typical characteristics of CSCs including self-renewal capability in addition to their invasiveness, and they also need to adapt themselves to the distant organs that have totally different microenvironment from the primary tumors (17, 18). Therefore, it is plausible that there are distinct populations of metastatic CSCs that can proliferate in the niche of specific distant organs. In this report, we indeed demonstrated that CSCs isolated from both 231BoM and 231BrM have strong tumor-initiating abilities; however, they preferentially metastasize to bone and brain, respectively, despite the fact that these cells were isolated using the same markers (CD24−/CD44+/ESA+). In addition, these metastatic CSCs showed significantly lower level of miR-7 and higher expression of KLF4 compared to CSCs from less metastatic parental cells. These results suggest an existence of wide range of heterogeneous population of CSCs although they still share the self-renewal ability and same surface markers. This is also exemplified by the fact that CSCs from different organs have been isolated by using different markers (1, 19). While these observations add further complexity to cancer stem cell biology, it opens a window of opportunity to identify specific targets for metastatic and recurrent diseases.
Mature miR-7 is individually transcribed and processed from three different gene locus in human genome, and it is highly expressed in parts of the brain, eye and pancreas, suggesting its role in the development of these organs (20– 22). On the other hand, aberrant expression of miR-7 was observed in glioblastomas, and it has been characterized as a putative tumor suppressor by targeting EGFR and AKT(23, 24). In breast cancer, miR-7 was shown to block invasiveness and tumorigenic potential by targeting PAK, which is consistent with the idea of tumor suppressor function of miR-7 (25). Interestingly, Bos et al. recently reported that EGFR and its ligand were highly expressed in brain metastatic cells and knock down of EGFR ligands significantly attenuated cell motility and invasion in vitro and in vivo. Notably, inhibition of EGFR with cetuximab also inhibited brain metastasis in mice (35). However, another report found that miR-7 expression correlated with poorer prognosis of breast cancer, and these apparently conflicting results suggest that miR-7 may be differentially expressed in a specific cell population of breast cancer (26). We found that miR-7 is indeed down-regulated specifically in highly metastatic CSCs to brain and bone but not in CSCs isolated from the parental cell (MDA-MB231), suggesting that miR-7 has a specific function in metastatic CSCs in addition to its tumor suppressor activity. In fact, we found that miR-7 was down-regulated in mammosphere, a hallmark of self renewal of CSCs, and that ectopic expression of miR-7 significantly reduced the ability in mammosphere formation. How miR-7 attenuates CSCs is a clinically relevant question. A range of potential targets of miR-7 were reported including EGFR, IRS-1, PAK-1, RAF-1 and SATB1 that are involved in key signaling of tumor progression (23, 25). In this report, we identified KLF4 as a critical downstream target of miR-7 in CSCs. KLF4 is one of IPS genes that are required for maintenance of stemness of progenitor cells (27, 28). Interestingly, Yu et al. recently demonstrated that knockdown of KLF4 decreased the proportion of cancer stem cells, while overexpression of this gene led to an increase in the cancer stem cell population, suggesting that KLF4 plays a strong oncogenic role in mammary tumorigenesis likely by maintaining stem cell-like futures of breast cancer cells (29). In support of this notion, KLF4 has been found to be frequently overexpressed in 70% of breast cancers, and its localization in the nucleus of breast cancer cells has been identified as a marker of aggressive phenotype in early-stage infiltrating ductal carcinoma (30). On the contrary, the expression level of KLF4 was down regulated in the gastrointestinal cancer due to the promoter methylation, and overexpression of this gene was shown to attenuate the tumorigenicity of colonic and gastric cancer cells in vivo (31, 32). Therefore, the clinical picture of KLF4 is complex and context dependent. It should be noted that KLF4 is a transcription factor that can both activate and suppress genes that are related to the proliferation and differentiation. In fact, KLF4 blocks senescence and apoptosis by repressing transcription of P53, while it can activate P21-dependent cell cycle arrest, and therefore, KLF4 can function both as a tumor suppressor and an oncogene(33, 34).
How the miR7-7/KLF4 pathway specifically links to brain metastasis is an intriguing question. The result of our cohort data analysis including 710 breast cancer patients clearly indicates significant correlation of KLF4 expression with brain- but not bone-metastasis free survival. Our TaqMan PCR analysis also revealed that the expression of miR-7 and KLF4 are significantly down- and up-regulated, respectively, in brain metastatic lesions of breast cancer patients and that their expressions are inversely correlated. Importantly, ectopic expression of miR-7 significantly blocked the brain metastasis but not bone metastasis in our animal model. The down-regulation of MiR-7 expression and hence activation of KLF4 gene is specific to metastatic CSCs but not to organ tropism of metastasis. Therefore, the brain specificity of miR-7/KLF4 is likely due to the downstream targets of this pathway or related to microenvironmental factors of brain. In this context, it should be noted that Bos et al. recently identified a brain-metastatic signature which includes 17 specific genes based on comprehensive expression array analysis of breast cancer patients (35). Interestingly, the results of our promoter analysis for all these genes revealed that 9 out of the 17 genes (COL13A1, CSF3, FSCN1, HG18, LTBP1, PELI1, PLOD2, SCNN1A, and TNFSF10) have perfectly matched consensus sequences of KLF4 binding site on the promoter regions, suggesting a potential possibility of controlling these genes by KLF4 although it is not clear whether these genes are cause or consequence of brain metastasis at this point. Several other targets of KLF4 have been identified including TGF-β and Notch (36, 37) and these genes are known to be involved in stem cell self-renewal and tumor progression. Our results also indicate that miR-7 significantly inhibits mammosphere formation while KLF4 promotes self-renewal of CSCs, suggesting that miR-7/KLF4 pathway is critical in CSC physiology in conjunction with TGF-β and Notch. Furthermore, it is becoming clear that CSCs require niche for their growth and they need to either adapt themselves to the existing niche or to generate their own niche in the distant organs that have totally different microenvironment from the primary tumor (38). The brain consists of highly specialized cells such as neuron, glia and astrocytes. Therefore, miR-7/KLF4 pathway may facilitate a reciprocal interaction of CSCs and these brain cells that generates a suitable niche for CSCs, although this hypothesis needs to be experimentally tested. Nevertheless, our results indicate that miR-7/KLF4 axis may serve as a potential therapeutic target for brain metastasis of breast cancer, which is almost always incurable at the present time.
Acknowledgments
Grant support
This work was supported by the National Institutes of Health (grants R01CA124650, R01CA129000 and R01CA124650-04S1 to K. Watabe), the US Department of Defense (BC096982 to K. Watabe), and McElroy Foundation (to K. Watabe).
References
Full text links
Read article at publisher's site: https://doi.org/10.1158/0008-5472.can-12-2037
Read article for free, from open access legal sources, via Unpaywall:
https://cancerres.aacrjournals.org/content/canres/73/4/1434.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Competing endogenous RNAs regulatory crosstalk networks: The messages from the RNA world to signaling pathways directing cancer stem cell development.
Heliyon, 10(15):e35208, 26 Jul 2024
Cited by: 0 articles | PMID: 39170516 | PMCID: PMC11337742
Review Free full text in Europe PMC
Crosstalk between breast cancer-derived microRNAs and brain microenvironmental cells in breast cancer brain metastasis.
Front Oncol, 14:1436942, 08 Aug 2024
Cited by: 0 articles | PMID: 39175471 | PMCID: PMC11338853
Review Free full text in Europe PMC
Comprehensive analysis of microRNAs modulated by histone deacetylase inhibitors identifies microRNA-7-5p with anti-myeloma effect.
Int J Hematol, 120(3):325-336, 02 Jul 2024
Cited by: 0 articles | PMID: 38954186
The occurrence and development of induced pluripotent stem cells.
Front Genet, 15:1389558, 18 Apr 2024
Cited by: 0 articles | PMID: 38699229 | PMCID: PMC11063328
Review Free full text in Europe PMC
Use of microRNAs as Diagnostic, Prognostic, and Therapeutic Tools for Glioblastoma.
Int J Mol Sci, 25(5):2464, 20 Feb 2024
Cited by: 2 articles | PMID: 38473710 | PMCID: PMC10931459
Review Free full text in Europe PMC
Go to all (186) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus (5)
- (2 citations) GEO - GSE2603
- (2 citations) GEO - GSE14020
- (2 citations) GEO - GSE12276
- (2 citations) GEO - GSE2034
- (2 citations) GEO - GSE5327
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
miR-509 suppresses brain metastasis of breast cancer cells by modulating RhoC and TNF-α.
Oncogene, 34(37):4890-4900, 09 Feb 2015
Cited by: 64 articles | PMID: 25659578 | PMCID: PMC4530094
Krüppel-like factor 4 acts as an oncogene in colon cancer stem cell-enriched spheroid cells.
PLoS One, 8(2):e56082, 13 Feb 2013
Cited by: 83 articles | PMID: 23418515 | PMCID: PMC3572033
miR-29a-KLF4 signaling inhibits breast tumor initiation by regulating cancer stem cells.
Int Immunopharmacol, 130:111797, 04 Mar 2024
Cited by: 0 articles | PMID: 38442582
MicroRNAs in cancer stem cells: current status and future directions.
Tumour Biol, 35(9):8395-8405, 26 Jun 2014
Cited by: 31 articles | PMID: 24964962
Review
Funding
Funders who supported this work.
NCI NIH HHS (7)
Grant ID: R01 CA154989
Grant ID: R01 CA151851
Grant ID: R01CA124650
Grant ID: R01 CA124650
Grant ID: R01CA124650-04S1
Grant ID: R01 CA129000
Grant ID: R01CA129000





