Abstract
Free full text

Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches
Abstract
While haematopoietic stem cells (HSCs) are commonly assumed to reside within a specialized microenvironment, or niche1, most published experimental manipulations of the HSC niche have also impacted the function of diverse restricted progenitors. This raises the fundamental question of whether HSCs1 and restricted progenitors2,3 reside within distinct, specialized niches or whether they share a common niche. Here we assess the physiological sources of the chemokine, CXCL12, for HSC and restricted progenitor maintenance. Cxcl12DsRed knock-in mice showed that Cxcl12 was primarily expressed by perivascular stromal cells and at lower levels by endothelial cells, osteoblasts, and some haematopoietic cells. Conditional deletion of Cxcl12 from haematopoietic cells or Nestin-cre-expressing cells had little or no effect on HSCs or restricted progenitors. Deletion of Cxcl12 from endothelial cells depleted HSCs but not myeloerythroid or lymphoid progenitors. Deletion of Cxcl12 from perivascular stromal cells depleted HSCs and certain restricted progenitors and mobilized these cells into circulation. Deletion of Cxcl12 from osteoblasts depleted certain early lymphoid progenitors, but not HSCs or myeloerythroid progenitors and did not mobilize these cells into circulation. Different stem/progenitor cells thus occupy distinct cellular niches in bone marrow: HSCs in a perivascular niche and early lymphoid progenitors in an endosteal niche.
Using SLAM family markers that isolate quiescent HSCs4–8 we found that most HSCs localize adjacent to sinusoidal blood vessels in the bone marrow4,9,10. Using independent approaches others obtained similar results11–13. We therefore hypothesized that there is a perivascular niche for HSC maintenance4. Consistent with this, Stem Cell Factor (SCF) is primarily or exclusively expressed in the bone marrow by endothelial cells and perivascular stromal cells10. Conditional deletion of Scf from endothelial cells or Leptin receptor (Lepr)-expressing perivascular stromal cells depleted HSCs10. Combined deletion of Scf from both endothelial cells and perivascular stromal cells caused severe HSC depletion and anemia. In contrast, conditional deletion of Scf from osteoblasts or haematopoietic cells did not affect HSC frequency or function. This proves there is a perivascular niche for HSC maintenance and raises the question of whether other haematopoietic progenitors reside in distinct niches.
CXCL12 is a chemokine required for HSC maintenance and retention in the bone marrow11,14–17. Global conditional deletion of Cxcl12, or the gene that encodes its receptor, Cxcr4, depletes HSCs from adult bone marrow11,14. CXCL12 also promotes the proliferation and maintenance of B lineage progenitors15,18 and common lymphoid progenitors (CLPs)19. CXCL12 is expressed by perivascular stromal cells, endothelial cells and osteoblasts11,20,21. Some have proposed that the physiological source of CXCL12 for HSC maintenance is osteoblasts14 while others have proposed perivascular stromal cells11; however, Cxcl12 has not yet been conditionally deleted from any candidate niche cell. Thus, the cellular sources of CXCL12 for the maintenance of HSCs and lymphoid progenitors remain uncertain.
To systematically examine the Cxcl12 expression pattern we generated Cxcl12DsRed knock-in mice by recombining DsRed-Express2 (DsRed) into the endogenous Cxcl12 locus (Supplementary Fig. 1a–c). Cxcl12 was primarily expressed by cells surrounding sinusoids throughout the bone marrow, irrespective of proximity to the endosteum (Fig. 1a–c; Supplementary Fig. 1d). Cxcl12-DsRed expression overlapped with endothelial marker staining, suggesting that endothelial cells were one source of CXCL12 (Fig. 1a–c); however, perivascular stromal cells also appeared to produce CXCL12 (Fig. 1a–c).
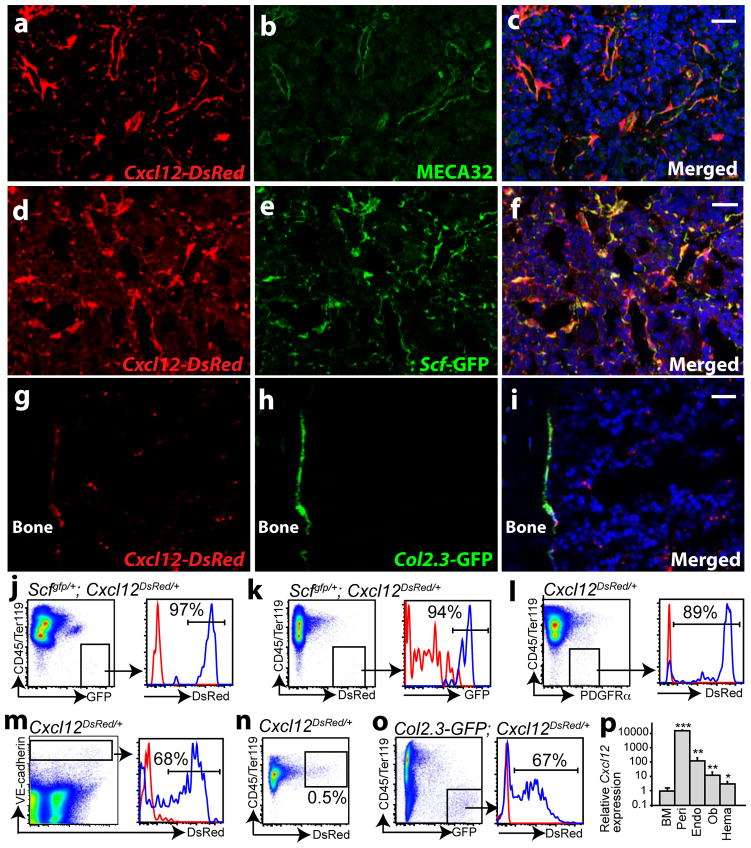
a–c, In Cxcl12DsRed/+ mice, DsRed was primarily expressed by perivascular cells throughout the bone marrow. d–f, Cxcl12-DsRed and Scf-GFP expression strongly overlapped around sinusoids throughout the bone marrow of Scfgfp/+; Cxcl12DsRed/+ mice. g–i, Col2.3-GFP+ bone-lining osteoblasts expressed Cxcl12-DsRed in the bone marrow of Col2.3-GFP+; Cxcl12DsRed/+ mice. Nuclei were stained with 4′, 6-diamidino-2-phenylindole (DAPI, blue) in c, f and i. j, CD45/Ter119−Scf-GFP+ perivascular stromal cells expressed Cxcl12-DsRed in Scfgfp/+; Cxcl12DsRed/+ (blue histogram) but not in Scfgfp/+ control marrow (red histogram). k, CD45/Ter119−Cxcl12-DsRedhigh cells expressed Scf-GFP in Scfgfp/+; Cxcl12DsRed/+ (blue) but not in Cxcl12DsRed/+ control marrow (red). l, CD45/Ter119−PDGFRα+ perivascular stromal cells expressed Cxcl12-DsRed in Cxcl12DsRed/+ (blue) but not control (+/+; red) marrow. m, VE-cadherin+ endothelial cells expressed Cxcl12-DsRed in Cxcl12DsRed/+ (blue) but not control (+/+; red) marrow. n, 0.5% of CD45/Ter119+ haematopoietic cells expressed Cxcl12-DsRed in Cxcl12DsRed/+ but not in control marrow (Supplementary Fig. 1g). o, CD45/Ter119−Col2.3-GFP+ osteoblasts from enzymatically dissociated bone expressed Cxcl12-DsRed in Col2.3-GFP; Cxcl12DsRed/+ (blue) but not Col2.3-GFP control (red) mice. p, Cxcl12 transcript levels by qRT-PCR in different subpopulations of bone marrow cells (n=3–6). BM, whole bone marrow cells. Peri, EYFP+ perivascular stromal cells from Lepr-cre; loxpEYFP mice. Endo, VE-cadherin+ bone marrow endothelial cells. Ob, Col2.3-GFP+ osteoblasts. Hema, CD45/Ter119+Cxcl12-DsRed+ haematopoietic cells. All data represent mean±s.d. from at least three independent experiments. Two-tailed student’s t-tests were always used to assess statistical significance (*P<0.05, **P<0.01, ***P<0.001). Scale bars are 20 um.
The perivascular Cxcl12 expression pattern was very similar to the Scf expression pattern10. In Scfgfp/+; Cxcl12DsRed/+ mice we found a strong overlap in Cxcl12-DsRed and Scf-GFP expression by perivascular cells throughout the bone marrow (Fig. 1d–f). By flow cytometry, virtually all Scf-GFP+ stromal cells were positive for Cxcl12-DsRed (Fig. 1j) and virtually all Cxcl12-DsRed+ stromal cells were positive for Scf-GFP (Fig. 1k). Perivascular HSC niche cells therefore produce both SCF and CXCL12.
To quantitate Cxcl12 expression in perivascular stromal cells we sorted CD45/Ter119−PDGFRα+ mesenchymal stem/stromal cells from enzymatically dissociated bone marrow. The Lepr-Cre-expressing perivascular stromal cells that contribute to the HSC niche by secreting SCF are uniformly positive for PDGFRα10 and virtually all CD45/Ter119−PDGFRα+ bone marrow cells express Scf-GFP (Supplementary Fig. 1e, f). Approximately 90% of CD45/Ter119−PDGFRα+ perivascular stromal cells expressed Cxcl12-DsRed (Fig. 1l). By flow cytometry, approximately 70% of VE-cadherin+ endothelial cells and 0.5% of CD45/Ter119+ haematopoietic cells in the bone marrow also expressed Cxcl12-DsRed (Fig. 1m, n and Supplementary Fig. 1g).
In contrast to Scf, which is not detectably expressed by osteoblasts10, we did observe very low levels of Cxcl12-DsRed expression by bone-lining cells (Fig. 1g–i). Almost 70% of Col2.3-GFP+ osteoblasts isolated from enzymatically digested bone from Col2.3-GFP; Cxcl12DsRed/+ mice expressed low levels of Cxcl12-DsRed (Fig. 1o).
By quantitative reverse transcription polymerase chain reaction (qRT-PCR), EYFP+ perivascular stromal cells from Lepr-cre; loxpEYFP mice expressed Cxcl12 at ~15,000-fold the level observed in unfractionated bone marrow (Fig. 1p). VE-cadherin+ endothelial cells, Col2.3-GFP+ osteoblasts, and Cxcl12-DsRed+ haematopoietic cells expressed significantly lower levels of Cxcl12 at ~120 fold, ~13 fold, and ~3 fold the levels observed in bone marrow cells (Fig. 1p).
We generated a floxed allele of Cxcl12 (Cxcl12fl) that led to a frameshift upon recombination (Supplementary Fig. 2a–c). The floxed allele itself did not appear to have any phenotype as unrecombined Cxcl12fl/fl mice were born and matured into adulthood in normal numbers with normal HSC frequency and haematopoiesis (Supplementary Fig. 2d–f). We recombined Cxcl12fl in the germline with CMV-cre mice to generate a Cxcl12− predicted null allele. Cxcl12+/− mice were born in expected numbers (Supplementary Fig. 2g) with normal cellularity, B cell frequency, and HSC frequency in the bone marrow and spleen (Supplementary Fig. 2h–j). In contrast, Cxcl12−/− progeny were not born alive (Supplementary Fig. 2g) consistent with the known perinatal lethal phenotype of Cxcl12 deficient mice15.
Global deletion of Cxcl12 by administering tamoxifen to 8-week old adult Ubc-creER; Cxcl12fl/fl mice significantly reduced white blood cell counts (Supplementary Fig. 4a), lymphocyte frequencies (Supplementary Fig. 4b), bone marrow cellularity (Supplementary Fig. 4c) and CD150+CD48−Lineage−Sca1+cKit+ HSC4 frequency (Supplementary Fig. 4d). Bone marrow cells from Ubc-creER; Cxcl12fl/fl mice also gave significantly lower levels of donor cell reconstitution in all major haematopoietic lineages upon transplantation into irradiated mice (Supplementary Fig. 4e). Consistent with an independently targeted Cxcl12fl allele14, these results demonstrate CXCL12 promotes adult HSC maintenance and lymphopoiesis.
HSCs do not express Cxcl12 by flow cytometry (Supplementary Fig. 4f). However, since some other haematopoietic cells expressed Cxcl12-DsRed (Fig. 1n, p), we conditionally deleted Cxcl12 from all haematopoietic cells in Vav1-cre; Cxcl12fl/fl mice. Recombination was highly efficient in Vav1-cre; Cxcl12fl/fl HSCs (Supplementary Fig. 5a). Adult Vav1-cre; Cxcl12fl/fl mice had normal cellularity, HSC frequency (Supplementary Fig. 4g, h), lineage composition in the bone marrow and spleen (Supplementary Fig. 5b), and reconstituting potential in irradiated mice (except for a modest decline in T cell reconstitution; Supplementary Fig. 4i). Cells from the bone marrow, spleen and blood of Vav1-cre; Cxcl12fl/fl mice formed normal numbers of haematopoietic colonies in culture (Supplementary Fig. 5c). We were unable to detect HSCs in the blood of Vav1-cre; Cxcl12fl/fl mice when we competitively transplanted 600,000 mononucleated blood cells into irradiated mice (data not shown).
Adult Vav1-cre; Cxcl12fl/fl mice also had normal frequencies of CD150−CD48−LSK multipotent progenitors (MPPs; see Supplementary Figure 3 for flow cytometry gates and references for each cell population), Flt3+LSK lymphoid-primed MPPs (LMPPs), CD34+FcγR−Lineage−Sca1−cKit+ common myeloid progenitors (CMPs), Lineage−Sca1lowcKitlowFlt3+IL7Rα+ common lymphoid progenitors (CLPs), CD34−FcγR−Lineage−Sca1−cKit+ megakaryocytic/erythroid progenitors (MEPs), and CD34+FcγR+Lineage−Sca1−cKit+ granulocyte/macrophage progenitors (GMPs) (Supplementary Fig. 4j, k). Committed B lineage progenitors including B220+sIgM−CD43+CD24− pre-pro B cells, B220+sIgM−CD43+CD24+ pro B cells, B220+sIgM−CD43− pre B cells, and B220+sIgM− B cells were also similar to controls (Supplementary Fig. 4l). Cxcl12 expression by haematopoietic cells is therefore not required for the maintenance or retention of HSCs or restricted progenitors in adult bone marrow.
Nestin-cre; Cxcl12fl/fl mice also had normal bone marrow and spleen cellularity, HSC frequency, and haematopoietic lineage composition (Supplementary Fig 6a, b). Bone marrow cells from Nestin-cre; Cxcl12fl/fl mice were indistinguishable from control cells in their capacity to give long-term multilineage reconstitution of irradiated mice (Supplementary Fig. 6c). The bone marrow of Nestin-cre; Cxcl12fl/fl mice had normal frequencies of MPPs, LMPPs, CMPs, CLPs, MEPs, GMPs, committed B lineage progenitors (Supplementary Fig. 6d–f). Nestin-cre; Cxcl12fl/fl mice also had normal frequencies of colony-forming progenitors in the bone marrow, spleen, and blood (Supplementary Fig. 6g–i). We were unable to detect HSCs in the blood of these mice in competitive reconstitution assays (data not shown). Nestin-cre expressing cells are therefore not a physiologically important source of CXCL12 for the maintenance of HSCs or restricted progenitors in the bone marrow. While HSC niche cells do not express endogenous Nestin, Nestin-cre, or Nestin-creER, they do appear to express the Nestin-GFP transgene10, consistent with previously published results12.
Given that Cxcl12 from haematopoietic cells was not required by HSCs or restricted progenitors (Supplementary Fig. 4g–l), Tie2-cre; Cxcl12fl/fl mice allowed us to test whether Cxcl12 from endothelial cells is physiologically important. Adult Tie2-cre; Cxcl12fl/fl mice had normal bone marrow and spleen cellularity (Fig. 2a), blood cell counts, and lineage composition in the bone marrow and spleen (Supplementary Fig. 7a, b). However, the frequency of CD150+CD48−Lineage−Sca1+cKit+ HSCs in the bone marrow of Tie2-cre; Cxcl12fl/fl mice was significantly lower than in littermate controls (Fig. 2b). Bone marrow cells from Tie2-cre; Cxcl12fl/fl mice also gave significantly lower levels of donor cell reconstitution in all major hematopoietic lineages upon transplantation into irradiated mice (Fig. 2c). Endothelial cells are therefore a physiologically important source of CXCL12 for HSC maintenance. This likely reflects a requirement for CXCL12 produced by postnatal bone marrow endothelial cells because Cxcl12 deficient mice do not exhibit HSC depletion during fetal development, only in postnatal bone marrow17,22. HSCs from Tie2-cre; Cxcl12fl/fl mice had normal frequencies of BrdU+ and Annexin V+ cells, suggesting that HSCs may be depleted in these mice through premature differentiation (Supplementary Fig. 7c, d).
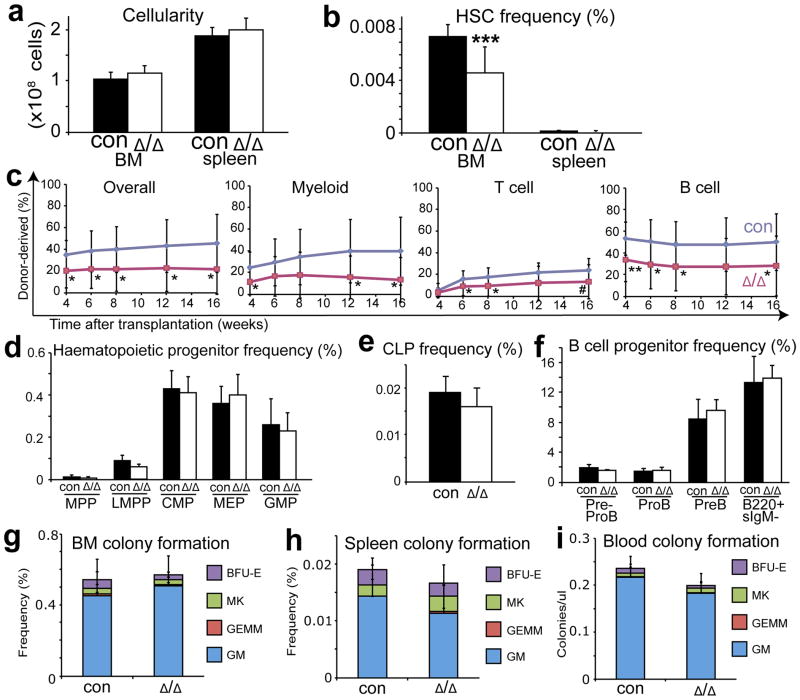
a, Bone marrow and spleen cellularity (n=8–9) and (b) HSC frequency in Tie2-cre; Cxcl12fl/fl mice versus littermate controls (n=10–11). c, 3×105 donor bone marrow cells from Tie2-cre; Cxcl12fl/fl mice gave significantly lower levels of donor myeloid, B, and T cell reconstitution in irradiated mice (three experiments with a total of 12–14 recipients per genotype). d–f, Tie2-cre; Cxcl12fl/fl mice had normal frequencies of MPPs, LMPPs, CMPs, MEPs, GMPs (d), CLPs (e), and committed B lineage progenitors in bone marrow (f) (n=3–4). g–i, Tie2-cre; Cxcl12fl/fl mice had normal frequencies of myeloerythroid colony-forming progenitors in bone marrow (g), spleen (h), and blood (i) (n=3–6). Δ, recombined Cxcl12fl allele; con, negative control mice with +/+ or fl/+ or fl/fl Cxcl12 genotypes (without cre). Data are mean±s.d. (*P<0.05, **P<0.01, ***P<0.001, #=0.057).
Tie2-cre; Cxcl12fl/fl mice had normal frequencies of MPPs, LMPPs, CMPs, CLPs, MEPs, GMPs, and B lineage progenitors in the bone marrow (Fig. 2d–f) and colony-forming progenitors in the bone marrow, spleen, and blood (Fig. 2g–i). We were unable to detect HSCs in the blood of these mice in competitive reconstitution assays (data not shown). CXCL12 produced by endothelial cells is therefore not required for the retention of HSCs or most restricted progenitors in bone marrow.
We deleted Cxcl12 from perivascular stromal cells in the bone marrow using Lepr-cre10. Lepr-cre; Cxcl12fl/− mice had normal bone marrow and spleen cellularity (Fig. 3a), normal blood cell counts, and normal lineage composition in the bone marrow and spleen (Supplementary Fig. 7e, f). CD150+CD48−Lineage−Sca1+cKit+ HSC frequency was normal in the bone marrow of Lepr-cre; Cxcl12fl/− mice (Fig. 3b) and the reconstituting capacity of bone marrow cells in irradiated mice was normal (Fig. 3c). The bone marrow of Lepr-cre; Cxcl12fl/− mice also had normal frequencies of MPPs, LMPPs, CMPs, CLPs, MEPs, GMPs, and B lineage progenitors (Fig 3d–f). However, the frequencies of HSCs and colony-forming progenitors were significantly elevated in the blood and spleen of Lepr-cre; Cxcl12fl/− mice (Fig. 3b, h–j). This demonstrates that CXCL12 expression by Lepr+ perivascular cells is required to retain HSCs and colony-forming progenitors in the bone marrow.
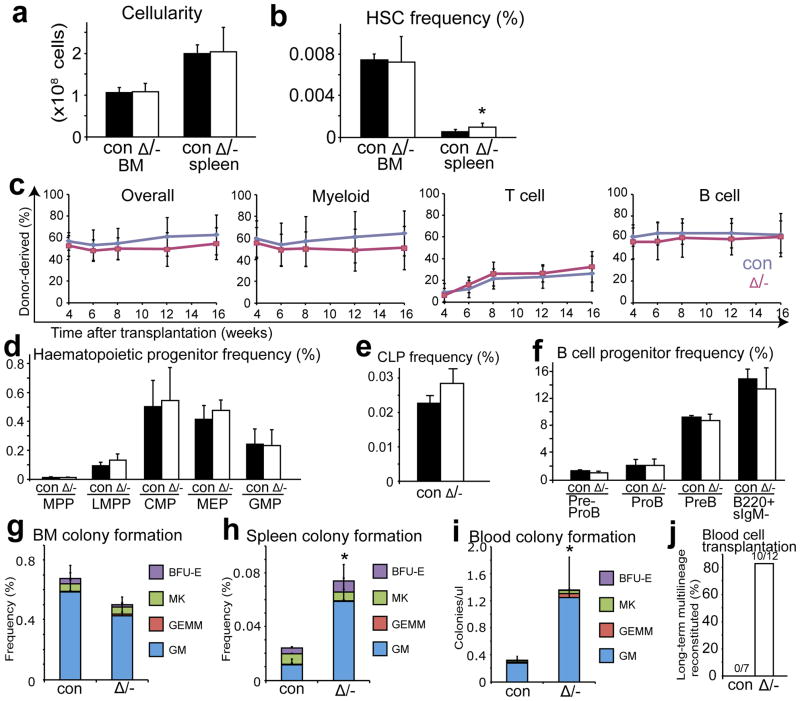
a, b, Cellularity (a, n=6) and HSC frequency (b, n=6–7) in the bone marrow and spleen of Lepr-cre; Cxcl12fl/− mice and littermate controls. c, 3×105 bone marrow cells from Lepr-cre; Cxcl12fl/− mice gave normal levels of donor myeloid, B, and T cell reconstitution in irradiated mice (three experiments with a total of 15 recipient mice per genotype). d–f, Lepr-cre; Cxcl12fl/− mice had normal frequencies of MPPs, LMPPs, CMPs, MEPs, GMPs (d), CLPs (e), and committed B lineage progenitors in bone marrow (f) (n=3). g–i, Lepr-cre; Cxcl12fl/− mice had normal frequencies of myeloerythroid colony-forming progenitors in bone marrow (g) but significantly increased frequencies in spleen (h), and blood (i) (n=3–5). j, 6×105 mononucleated blood cells from Lepr-cre; Cxcl12fl/− mice gave long-term multilineage reconstitution in irradiated mice, while blood cells from littermate controls did not. Δ, recombined Cxcl12fl allele; -, germline deleted Cxcl12 allele or Cxcl12DsRed allele; con, control mice. Data represent mean±s.d. (*P<0.05, **P<0.01, ***P<0.001).
Col2.3-Cre recombines genes in fetal and adult osteoblasts10,23. Col2.3-cre; Cxcl12fl/fl mice had normal bone marrow and spleen cellularity, normal CD150+CD48−Lineage−Sca1+cKit+ HSC frequency in the bone marrow and spleen (Fig. 4a, b), normal blood cell counts, normal lineage composition in the bone marrow and spleen (Supplementary Fig. 8a, b). Col2.3-cre; Cxcl12fl/fl bone marrow cells gave normal levels of donor myeloid reconstitution, but significantly lower levels of T and B cell reconstitution in irradiated mice relative to control cells (Fig. 4c). In contrast, purified HSCs from Col2.3-cre; Cxcl12fl/fl mice gave similar levels of reconstitution in all lineages as control HSCs (Fig. 4d and Supplementary Fig. 8c). This suggests that the reduction in T and B cell reconstitution from the bone marrow of Col2.3-cre; Cxcl12fl/fl mice reflected the depletion of early lymphoid progenitors.
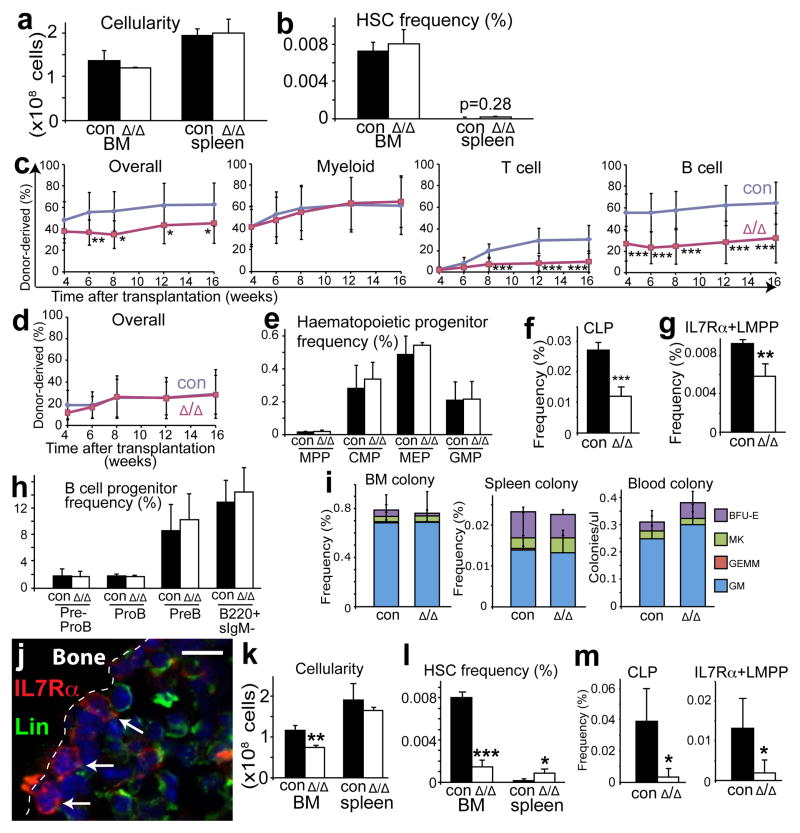
a, b, Cellularity (a, n=4) and HSC frequency (b, n=4) in the bone marrow and spleen of Col2.3-cre; Cxcl12fl/fl mice and littermate controls. c, 3×105 bone marrow cells from Col2.3-cre; Cxcl12fl/fl mice gave significantly lower levels of donor cell reconstitution in the T and B cell lineages but not in the myeloid lineage relative to control bone marrow cells (three experiments with a total of 13–14 recipients per genotype). d, 20 CD150+CD48−Lineage−Sca1+cKit+ HSCs from Col2.3-cre; Cxcl12fl/fl mice gave normal donor cell reconstitution (three experiments with a total of 14–15 recipients per genotype), including normal levels of myeloid, B, and T cells (Supplementary Fig. 8c). e–h, Col2.3-cre; Cxcl12fl/fl bone marrow had normal frequencies of MPPs, CMPs, MEPs, GMPs (e), and committed B lineage progenitors (h) but significantly reduced frequencies of CLPs (f) and IL7Rα+LMPPs (g) (n=3–5). i, Col2.3-cre; Cxcl12fl/fl mice had normal frequencies of myeloerythroid colony-forming progenitors in the bone marrow, spleen, and blood (n=3–6). j, Some Lin−IL7Rα+ early lymphoid progenitors were adjacent to the endosteum. k–m, Prx1-cre; Cxcl12fl/fl mice exhibited significant reductions in bone marrow cellularity (k, n=3–4) and the frequencies of HSCs (l), CLPs, and IL7Rα+LMPPs (m, n=4–5).Δ, recombined Cxcl12fl allele; con, control mice. *P<0.05, **P<0.01, ***P<0.001.
The bone marrow of Col2.3-cre; Cxcl12fl/fl mice had normal frequencies of MPPs, CMPs, MEPs, and GMPs (Fig. 4e) but significantly fewer CLPs and IL7Rα+LMPPs – cells thought to be specified for lymphoid differentiation24,25 (Fig. 4f, g). The thymus in Col2.3-cre; Cxcl12fl/fl mice had a normal frequency of early thymic progenitors (B220−Gr1−Ter119−CD4−CD8−CD44+cKit+CD25− ETPs; Supplementary Fig. 8d, e) and the bone marrow had a normal frequency of committed B lineage progenitors (Fig. 4h). This suggests that homeostatic mechanisms can restore normal frequencies of downstream lymphoid progenitors despite depletion of primitive lymphoid progenitors. CLPs from Col2.3-cre; Cxcl12fl/fl mice had normal frequencies of BrdU+ and Annexin V+ cells, suggesting that these cells might be depleted by premature differentiation (Supplementary Fig. 8f, g).
Col2.3-cre; Cxcl12fl/fl mice had normal frequencies of myeloerythroid colony-forming progenitors in the bone marrow, spleen, and blood (Fig. 4i) and we were unable to detect HSCs in the blood of these mice in competitive reconstitution assays (data not shown). CXCL12 from osteoblasts is thus required for the maintenance of certain early lymphoid progenitors but not for the maintenance or bone marrow retention of HSCs or myeloerythroid progenitors.
In stained bone marrow sections we found 30% of Lineage−IL7Rα+ cells at the endosteum, adjacent to bone-lining cells (Fig. 4j and Supplementary Fig. 9). Only 5% of all bone marrow cells were adjacent to the endosteum in the sections we examined, so these early lymphoid progenitors were 6 times more likely than the average bone marrow cell to be adjacent to endosteum. Both functional genetic data and localization data therefore suggest there is an endosteal niche for a subset of early lymphoid progenitors.
To test whether both HSCs and early lymphoid progenitors would be depleted by deleting Cxcl12 in perivascular stromal cells and in osteoblasts we used Prx1-cre26. Greenbaum et al. discovered that Prx1-Cre deletes broadly in perivascular stromal cells as well as in osteoblasts27. Consistent with this, Prx1-Cre recombined a conditional reporter in osteoblasts as well as in 95±4% CD45/Ter119−PDGFRα+ perivascular stromal cells (Supplementary Fig. 10a–c) but not in bone marrow endothelial cells (Supplementary Fig. 10d). Thus, Prx1-Cre recombined more broadly in perivascular stromal cells than Lepr-Cre, which recombined a conditional reporter in 70±9% of CD45/Ter119−PDGFRα+ cells (Supplementary Fig. 1f). Consistent with our results with other Cre alleles, Prx1-cre; Cxcl12fl/fl mice exhibited significant reductions in bone marrow cellularity (Fig. 4k) and in the frequencies of HSCs (Fig. 4l), CLPs, and IL7Rα+LMPPs (Fig. 4m). The bone marrow of Prx1-cre; Cxcl12fl/fl mice also had fewer committed B lineage progenitors, but CMPs, MEPs, and GMPs were normal (Supplementary Fig. 10e, f).
Our data indicate that HSCs depend upon a perivascular niche created by endothelial cells and Lepr-cre/Prx1-cre-expressing perivascular stromal cells, while some early lymphoid progenitors depend upon an endosteal niche created by osteoblasts, and committed B lineage progenitors depend upon a distinct perivascular niche created by Prx1-cre-expressing stromal cells but not endothelial cells. Greenbaum et al.27 come to similar conclusions. Osteoblasts promote the proliferation and differentiation of lymphoid progenitors in culture and conditional ablation of osteoblasts in vivo acutely depletes lymphoid progenitors but not HSCs28,29. Conditional deletion of G protein alpha from osteoblasts depletes B lineage progenitors but not other haematopoietic progenitors30. Our data are thus consistent with a number of prior studies, including our recent work10, in suggesting that osteoblasts create a niche for certain early lymphoid progenitors but not for HSCs.
SUPPLEMENTARY METHODS
Mice
Targeting vectors for making Cxcl12DsRed and Cxcl12fl mice were generated by recombineering31. Linearized targeting vector was electroporated into C57BL-derived Bruce4 ES cells. Correctly targeted clones were identified by Southern blotting. Following expansion, these ES cell clones were injected into C57BL/6-Tyrc-2J blastocysts. Chimeric mice were bred with C57BL/6-Tyrc-2J mice to obtain germline transmission. The Frt-flanked Neo selection cassette was removed by subsequent mating with Flpe mice32. The resulting mice were backcrossed for at least three generations onto a C57BL/Ka background. Mice used in this study included Ubc-creER42, CMV-cre43, Vav1-cre44, Nestin-cre45, Tie2-cre46, Lepr-cre47, Prx1-cre26, and LoxpEYFP48 (all from Jackson Laboratory). Scfgfp/+ mice were described previously10. Col2.3-cre mice23 were obtained from Drs. F. Liu and B. Kream (University of Connecticut). Col2.3-GFP mice49 were obtained from Dr. D. Rowe. Tamoxifen chow (Harlan) containing tamoxifen citrate (Spectrum Chemical) at 400mg/kg, with 5% sucrose added, was administrated to mice for 2–4 months to induce recombination by CreER.
All mice were housed in the Unit for Laboratory Animal Medicine at the University of Michigan or in the Animal Resource Center at the University of Texas Southwestern Medical Center. All protocols were approved by the University of Michigan Committee on the Use and Care of Animals and by the UT Southwestern Institutional Animal Care and Use Committee.
PCR genotyping
Primers for genotyping of the Cxcl12DsRed allele were: OLD322, 5′-GATGCGCGTCAGAGACCCC-3′; OLD355, 5′-GCGCAGAGCTGCGAGCCTTTC-3′; and OLD 415, 5′-CTTGAAGCGCATGAAGGGCTTG-3′. Primers for genotyping of the Cxcl12fl allele were: OLD312, 5′-GAGCCCAGAACTCTGCCACC-3′; and OLD313, 5′-TCTTGCAAAGACCATCCCCTC-3′. Primers for genotyping of the Cxcl12− allele were: OLD312, 5′-GAGCCCAGAACTCTGCCACC-3′, and OLD353, 5′-ACTCTGAGTGAGATATTCACATC-3′.
Long-term competitive reconstitution assay
Adult mice were administered a minimum lethal dose of radiation using a Cesium 137 GammaCell40 Irradiator (MDS) or an XRAD 320 x-ray irradiator (Precision X-Ray Inc.) to deliver two doses of 540 rads (total 1080 rads) at least 2 hours apart. C57BL/6-SJL (CD45.1) mice were used as recipient mice in transplantation experiments. Cells were injected into the retro-orbital venus sinus of anaesthetized mice. Blood cells were subjected to Ficoll centrifugation prior to transplantation to isolate mononucleated cells. Mononucleated cells were counted prior to transplantation either using a CBC machine or a hemocytometer with Turks solution. In most experiments recipient mice were maintained on antibiotic water (neomycin sulfate 1.11g/l and polymixin B 0.121g/l) for 14 days after transplantation and then switched to regular water. Recipient mice were bled from 4 to 16 weeks after transplantation to examine the levels of donor-derived myeloid, B and T cells in their blood. Red blood cells were lysed with ammonium chloride potassium buffer before antibody staining. The antibodies used to analyze donor chimerism in the blood were anti-CD45.1 (A20), anti-CD45.2 (104), anti-Gr1 (8C5), anti-Mac1 (M1/70), anti-B220 (6B2) and anti-CD3 (KT31.1).
Flow cytometry
Bone marrow cells were isolated by flushing the long bones or by crushing the long bones with mortal and pestle in Ca2+ and Mg2+ free HBSS with 2% heat-inactivated bovine serum. Spleen cells were obtained by crushing the spleen between two glass slides. The cells were dissociated to a single cell suspension by gently passing through a 25G needle then filtering through a 70μm nylon mesh. The following antibodies were used to isolate HSCs: anti-CD150 (TC15-12F12.2), anti-CD48 (HM48-1), anti-CD41 (MWReg30), anti-Sca1 (E13-161.7), anti-cKit (2B8) and the following antibodies against lineage markers (anti-Ter119, anti-B220 (6B2), anti-Gr1 (8C5), anti-CD2 (RM2-5), anti-CD3 (17A2), anti-CD5 (53-7.3) and anti-CD8 (53-6.7)). Haematopoietic progenitors were identified by flow cytometry using the following antibodies: anti-Sca1 (E13-161.7), anti-cKit (2B8) and the following antibodies against lineage markers (anti-Ter119, anti-B220 (6B2), anti-Gr1 (8C5), anti-CD2 (RM2-5), anti-CD3 (17A2), anti-CD5 (53-7.3) and anti-CD8 (53-6.7)), anti-CD34 (RAM34), anti-CD135 (Flt3) (A2F10), anti-CD16/32 (FcγR) (93), anti-CD127 (IL7Rα) (A7R34), anti-CD24 (M1/69), anti-CD43 (1B11), anti-B220 (6B2), anti-IgM (II/41), anti-CD3 (17A2), anti-Gr1 (8C5), anti-Mac1 (M1/70), anti-CD41 (MWReg30), anti-CD71 (C2), anti-Ter119, anti-CD44 (IM7) and anti-CD25 (PC61). DAPI was used to exclude dead cells.
For flow cytometric analysis of Scf-GFP+ or Lepr-cre; loxpEYFP+ stromal cells, bone marrow was flushed using HBSS- with 2% bovine serum. Then whole bone marrow was digested with Collagenase IV (200U/ml) and DNase I (200U/ml) at 37°C for 15min. Samples were then stained with antibodies and analyzed by flow cytometry. Anti-CD140a (APA5), anti-CD45 (30F-11) and anti-Ter119 antibodies were used to isolate perivascular stromal cells. For analysis of bone marrow endothelial cells, mice were i.v. injected with 10ug/mouse Alexa Fluor 647 conjugated anti-VE-cadherin antibody (BV13, eBiosciences)51. Ten minutes later, the long bones were removed and bone marrow was flushed, digested and stained as above. Samples were run on FACSAria or FACSCanto II flow cytometers. Data were analyzed by FACSDiva (BD) or FlowJo (Tree Star) software. Osteoblasts were dissociated as described50.
Methylcellulose cultures
Cells were sorted into methylcellulose culture medium (3434, Stemcell Technology) and incubated at 37°C as described9. Blood cells were obtained by Ficoll centrifuge according to manufacture’s recommendation before plating (GE Healthcare).
Immunostaining bone sections
Freshly dissected long bones were fixed in a Formalin-based fixative at 4°C for 3 hours. Then the bones were embedded in 8% gelatin in PBS. Samples were snap frozen with liquid nitrogen and stored at −80°C. Bones were sectioned using CryoJane (Instrumedics). Sections were dried overnight at room temperature (RT) and stored at −80°C. Sections were re-hydrated in PBS for 5 min before immunostaining. 5% goat serum in PBS was used for blocking. Primary antibodies were applied to the slides for 1 hour at RT, followed by secondary antibody incubation for 30min at RT with repeated washes in between. Slides were mounted with anti-fade prolong gold (Invitrogen) and images were acquired on an Olympus IX81 microscope or a Zeiss LSM 780 confocal microscope. Primary antibodies were: chicken-anti-GFP (Aves), rat-anti-mouse pan-endothelial cell antigen (Meca32, Biolegend), rat-anti-endoglin (eBioscience), rat-anti-RFP (AlleleBiotechnology) and goat-anti-osteopontin (R&D). For staining Lin−IL7Rα+ cells in bone marrow sections, mice were intravenously injected with 2ug Alexa Fluor 647 conjugated monoclonal antibody against IL7Rα (A7R34, Biolegend). After 5 minutes, the mice were killed, and long bones were dissected, fixed, sectioned, and stained with antibodies against Lineage markers (CD2, CD3, CD5, CD8, Ter119, Gr1 and B220) as described above. Images were acquired using a Zeiss LSM 780 confocal microscope.
Quantitative reverse transcription PCR
Cells were sorted directly into Trizol. Total RNA was extracted according to manufacture’s instructions. Total RNA was subjected to reverse transcription using SuperScript III (Invitrogen). Quantitative real-time PCR was run using SYBR green on a LightCycler 480 (Roche). β-actin was used to normalize the RNA content of samples. Primers used in this study were: Cxcl12: OLD35, 5′-TGCATCAGTGACGGTAAACCA-3′ and OLD36, 5′-GTTGTTCTTCAGCCGTGCAA-3′. β-actin: OLD27, 5′-GCTCTTTTCCAGCCTTCCTT-3′ and OLD28, 5′-CTTCTGCATCCTGTCAGCAA-3′.
Acknowledgments
This work was supported by the Howard Hughes Medical Institute (HHMI) and the National Heart, Lung and Blood Institute (5R01-HL097760). L.D. was supported by a Helen Hay Whitney Foundation Fellowship and by HHMI. We thank M. White and D. Adams for flow cytometry, E. Hughes and T. Saunders at the University of Michigan transgenic core for helping to generate Cxcl12DsRed and Cxcl12fl mice, and S. Grove, R. Coolon, S. Manning, M. Gross and K. Correll for managing the mouse colony.
Footnotes
AUTHOR CONTRIBUTIONS
L.D. performed all of the experiments. L.D. and S.J.M. conceived the project, designed the experiments, interpreted the results, and wrote the manuscript.
AUTHOR INFORMATION
The authors declare no competing financial interests.
References
SUPPLEMENTARY REFERENCES
Full text links
Read article at publisher's site: https://doi.org/10.1038/nature11885
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3600153?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Cells and signals of the leukemic microenvironment that support progression of T-cell acute lymphoblastic leukemia (T-ALL).
Exp Mol Med, 01 Nov 2024
Cited by: 0 articles | PMID: 39482533
Review
The evolving hematopoietic niche during development.
Front Mol Biosci, 11:1488199, 02 Oct 2024
Cited by: 0 articles | PMID: 39417006 | PMCID: PMC11480086
Review Free full text in Europe PMC
The role of the haematopoietic stem cell niche in development and ageing.
Nat Rev Mol Cell Biol, 10 Sep 2024
Cited by: 0 articles | PMID: 39256623
Review
Influences on the Hematopoietic Stem Cell Niche.
SciBase Hematol Blood Disord, 1(1):1002, 06 Sep 2024
Cited by: 0 articles | PMID: 39429505 | PMCID: PMC11486556
Bone marrow niches for hematopoietic stem cells.
Hemasphere, 8(8):e133, 31 Jul 2024
Cited by: 1 article | PMID: 39086665 | PMCID: PMC11289431
Review Free full text in Europe PMC
Go to all (765) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance.
Nature, 495(7440):227-230, 24 Feb 2013
Cited by: 840 articles | PMID: 23434756 | PMCID: PMC3600148
Endothelial and perivascular cells maintain haematopoietic stem cells.
Nature, 481(7382):457-462, 25 Jan 2012
Cited by: 1192 articles | PMID: 22281595 | PMCID: PMC3270376
Mesenchymal and haematopoietic stem cells form a unique bone marrow niche.
Nature, 466(7308):829-834, 01 Aug 2010
Cited by: 2080 articles | PMID: 20703299 | PMCID: PMC3146551
Regulation of hematopoietic stem cells by bone marrow stromal cells.
Trends Immunol, 35(1):32-37, 05 Nov 2013
Cited by: 162 articles | PMID: 24210164 | PMCID: PMC3947365
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Howard Hughes Medical Institute
NCI NIH HHS (1)
Grant ID: P30 CA142543
NHLBI NIH HHS (2)
Grant ID: R01 HL097760
Grant ID: 5R01-HL097760





