Abstract
Free full text

Identification of residues in the human DNA repair enzyme HAP1 (Ref-1) that are essential for redox regulation of Jun DNA binding.
Abstract
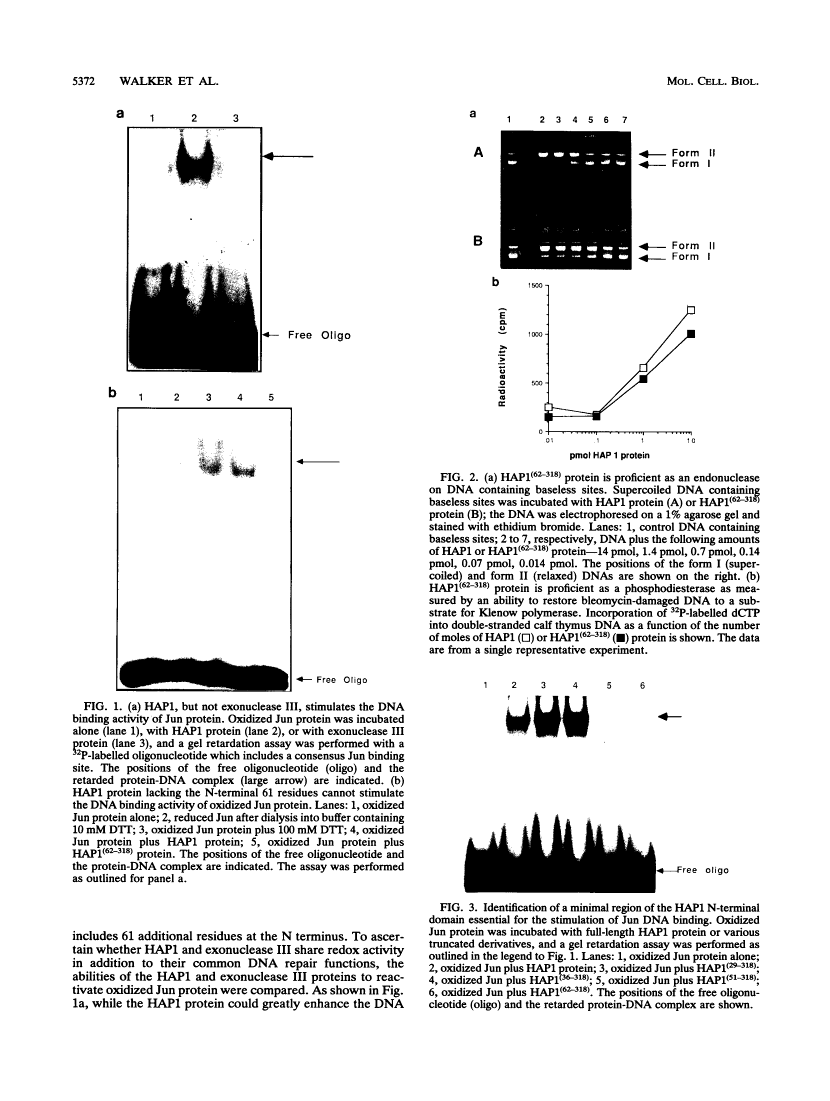
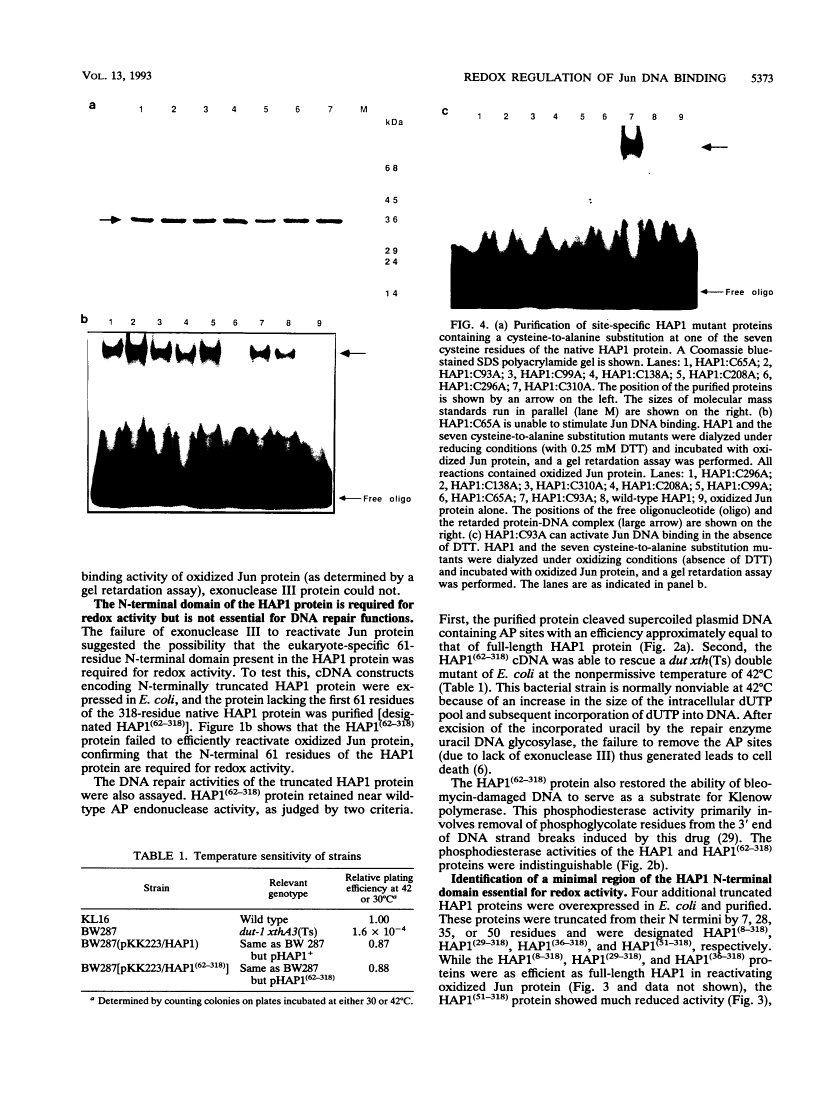
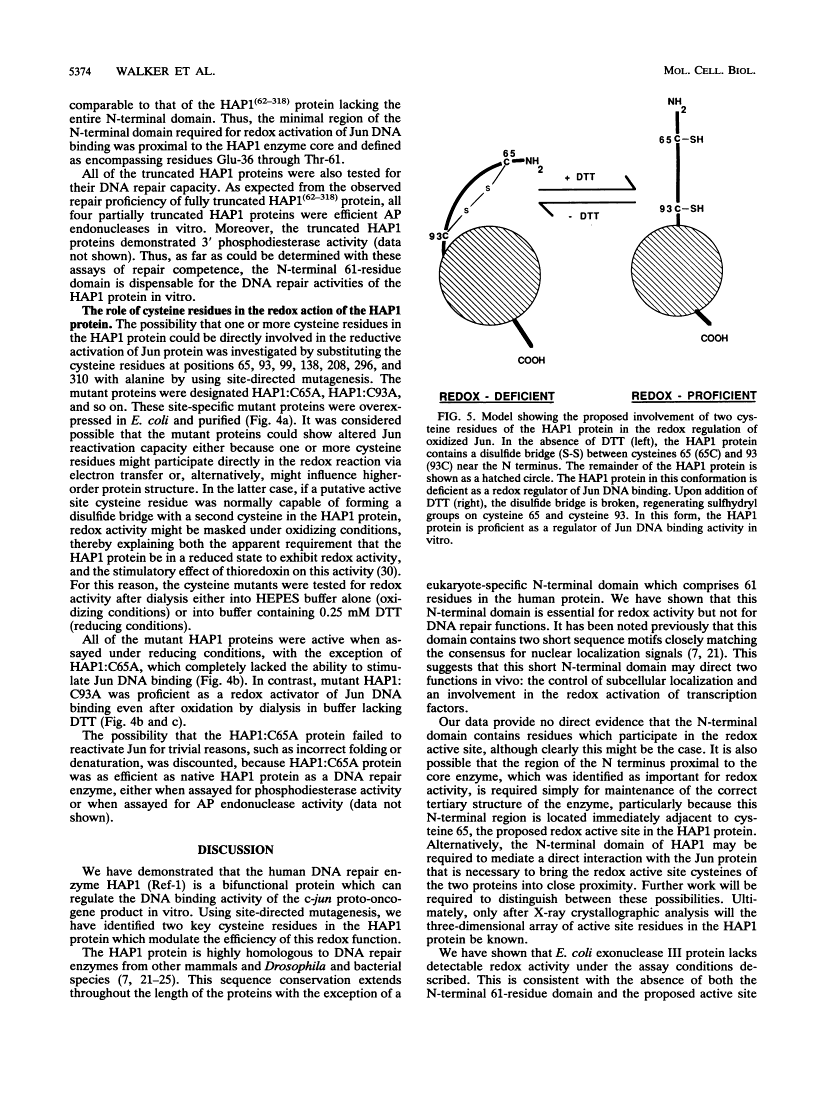
The DNA binding activity of the c-jun proto-oncogene product is inhibited by oxidation of a specific cysteine residue (Cys-252) in the DNA binding domain. Jun protein inactivated by oxidation of this residue can be efficiently reactivated by a factor from human cell nuclei, recently identified as a DNA repair enzyme (termed HAP1 or Ref-1). The HAP1 protein consists of a core domain, which is highly conserved in a family of prokaryotic and eukaryotic DNA repair enzymes, and a 61-amino-acid N-terminal domain absent from bacterial homologs such as Escherichia coli exonuclease III. The eukaryote-specific N-terminal domain was dispensable for the DNA repair functions of the HAP1 protein but was essential for reactivation of the DNA binding activity of oxidized Jun protein. Consistent with this finding, exonuclease III protein could not reactive Jun. A minimal 26-residue region of the N-terminal domain proximal to the core of the HAP1 enzyme was required for redox activity. By site-directed mutagenesis, cysteine 65 was identified as the redox active site in the HAP1 enzyme. In addition, it is proposed that cysteine 93 interacts with the redox active site, probably via disulfide bridge formation. It is concluded that the HAP1 protein has evolved a novel redox activation domain capable of regulating the DNA binding activity of a proto-oncogene product which is not essential for its DNA repair functions. Identification of a putative active site cysteine residue should facilitate analysis of the mechanism by which the HAP1 protein may alter the redox state of a wide range of transcription factors.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (1.6M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Click on the image to see a larger version.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abate C, Luk D, Curran T. A ubiquitous nuclear protein stimulates the DNA-binding activity of fos and jun indirectly. Cell Growth Differ. 1990 Oct;1(10):455–462. [Abstract] [Google Scholar]
- Abate C, Luk D, Gentz R, Rauscher FJ, 3rd, Curran T. Expression and purification of the leucine zipper and DNA-binding domains of Fos and Jun: both Fos and Jun contact DNA directly. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1032–1036. [Europe PMC free article] [Abstract] [Google Scholar]
- Abate C, Patel L, Rauscher FJ, 3rd, Curran T. Redox regulation of fos and jun DNA-binding activity in vitro. Science. 1990 Sep 7;249(4973):1157–1161. [Abstract] [Google Scholar]
- Boyle WJ, Smeal T, Defize LH, Angel P, Woodgett JR, Karin M, Hunter T. Activation of protein kinase C decreases phosphorylation of c-Jun at sites that negatively regulate its DNA-binding activity. Cell. 1991 Feb 8;64(3):573–584. [Abstract] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. [Abstract] [Google Scholar]
- Cunningham RP, Saporito SM, Spitzer SG, Weiss B. Endonuclease IV (nfo) mutant of Escherichia coli. J Bacteriol. 1986 Dec;168(3):1120–1127. [Europe PMC free article] [Abstract] [Google Scholar]
- Demple B, Herman T, Chen DS. Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11450–11454. [Europe PMC free article] [Abstract] [Google Scholar]
- Doetsch PW, Cunningham RP. The enzymology of apurinic/apyrimidinic endonucleases. Mutat Res. 1990 Sep-Nov;236(2-3):173–201. [Abstract] [Google Scholar]
- Frame MC, Wilkie NM, Darling AJ, Chudleigh A, Pintzas A, Lang JC, Gillespie DA. Regulation of AP-1/DNA complex formation in vitro. Oncogene. 1991 Feb;6(2):205–209. [Abstract] [Google Scholar]
- Guehmann S, Vorbrueggen G, Kalkbrenner F, Moelling K. Reduction of a conserved Cys is essential for Myb DNA-binding. Nucleic Acids Res. 1992 May 11;20(9):2279–2286. [Europe PMC free article] [Abstract] [Google Scholar]
- Hunter T, Karin M. The regulation of transcription by phosphorylation. Cell. 1992 Aug 7;70(3):375–387. [Abstract] [Google Scholar]
- Kumar S, Rabson AB, Gélinas C. The RxxRxRxxC motif conserved in all Rel/kappa B proteins is essential for the DNA-binding activity and redox regulation of the v-Rel oncoprotein. Mol Cell Biol. 1992 Jul;12(7):3094–3106. [Europe PMC free article] [Abstract] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. [Abstract] [Google Scholar]
- Landt O, Grunert HP, Hahn U. A general method for rapid site-directed mutagenesis using the polymerase chain reaction. Gene. 1990 Nov 30;96(1):125–128. [Abstract] [Google Scholar]
- Maki Y, Bos TJ, Davis C, Starbuck M, Vogt PK. Avian sarcoma virus 17 carries the jun oncogene. Proc Natl Acad Sci U S A. 1987 May;84(9):2848–2852. [Europe PMC free article] [Abstract] [Google Scholar]
- Matthews JR, Wakasugi N, Virelizier JL, Yodoi J, Hay RT. Thioredoxin regulates the DNA binding activity of NF-kappa B by reduction of a disulphide bond involving cysteine 62. Nucleic Acids Res. 1992 Aug 11;20(15):3821–3830. [Europe PMC free article] [Abstract] [Google Scholar]
- McBride AA, Klausner RD, Howley PM. Conserved cysteine residue in the DNA-binding domain of the bovine papillomavirus type 1 E2 protein confers redox regulation of the DNA-binding activity in vitro. Proc Natl Acad Sci U S A. 1992 Aug 15;89(16):7531–7535. [Europe PMC free article] [Abstract] [Google Scholar]
- Oehler T, Pintzas A, Stumm S, Darling A, Gillespie D, Angel P. Mutation of a phosphorylation site in the DNA-binding domain is required for redox-independent transactivation of AP1-dependent genes by v-Jun. Oncogene. 1993 May;8(5):1141–1147. [Abstract] [Google Scholar]
- Okuno H, Akahori A, Sato H, Xanthoudakis S, Curran T, Iba H. Escape from redox regulation enhances the transforming activity of Fos. Oncogene. 1993 Mar;8(3):695–701. [Abstract] [Google Scholar]
- Robson CN, Hickson ID. Isolation of cDNA clones encoding a human apurinic/apyrimidinic endonuclease that corrects DNA repair and mutagenesis defects in E. coli xth (exonuclease III) mutants. Nucleic Acids Res. 1991 Oct 25;19(20):5519–5523. [Europe PMC free article] [Abstract] [Google Scholar]
- Robson CN, Milne AM, Pappin DJ, Hickson ID. Isolation of cDNA clones encoding an enzyme from bovine cells that repairs oxidative DNA damage in vitro: homology with bacterial repair enzymes. Nucleic Acids Res. 1991 Mar 11;19(5):1087–1092. [Europe PMC free article] [Abstract] [Google Scholar]
- Sander M, Lowenhaupt K, Rich A. Drosophila Rrp1 protein: an apurinic endonuclease with homologous recombination activities. Proc Natl Acad Sci U S A. 1991 Aug 1;88(15):6780–6784. [Europe PMC free article] [Abstract] [Google Scholar]
- Saporito SM, Smith-White BJ, Cunningham RP. Nucleotide sequence of the xth gene of Escherichia coli K-12. J Bacteriol. 1988 Oct;170(10):4542–4547. [Europe PMC free article] [Abstract] [Google Scholar]
- Seki S, Akiyama K, Watanabe S, Hatsushika M, Ikeda S, Tsutsui K. cDNA and deduced amino acid sequence of a mouse DNA repair enzyme (APEX nuclease) with significant homology to Escherichia coli exonuclease III. J Biol Chem. 1991 Nov 5;266(31):20797–20802. [Abstract] [Google Scholar]
- Seki S, Hatsushika M, Watanabe S, Akiyama K, Nagao K, Tsutsui K. cDNA cloning, sequencing, expression and possible domain structure of human APEX nuclease homologous to Escherichia coli exonuclease III. Biochim Biophys Acta. 1992 Jul 15;1131(3):287–299. [Abstract] [Google Scholar]
- Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. [Abstract] [Google Scholar]
- Winters TA, Weinfeld M, Jorgensen TJ. Human HeLa cell enzymes that remove phosphoglycolate 3'-end groups from DNA. Nucleic Acids Res. 1992 May 25;20(10):2573–2580. [Europe PMC free article] [Abstract] [Google Scholar]
- Xanthoudakis S, Curran T. Identification and characterization of Ref-1, a nuclear protein that facilitates AP-1 DNA-binding activity. EMBO J. 1992 Feb;11(2):653–665. [Europe PMC free article] [Abstract] [Google Scholar]
- Xanthoudakis S, Miao G, Wang F, Pan YC, Curran T. Redox activation of Fos-Jun DNA binding activity is mediated by a DNA repair enzyme. EMBO J. 1992 Sep;11(9):3323–3335. [Europe PMC free article] [Abstract] [Google Scholar]
Associated Data
Articles from Molecular and Cellular Biology are provided here courtesy of Taylor & Francis
Full text links
Read article at publisher's site: https://doi.org/10.1128/mcb.13.9.5370-5376.1993
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc360239?pdf=render
Free after 4 months at mcb.asm.org
http://mcb.asm.org/cgi/reprint/13/9/5370
Free to read at mcb.asm.org
http://mcb.asm.org/cgi/content/abstract/13/9/5370
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/mcb.13.9.5370-5376.1993
Article citations
APE1/Ref-1 as a Therapeutic Target for Inflammatory Bowel Disease.
Biomolecules, 13(11):1569, 24 Oct 2023
Cited by: 3 articles | PMID: 38002251 | PMCID: PMC10669584
Review Free full text in Europe PMC
Knockout and Inhibition of Ape1: Roles of Ape1 in Base Excision DNA Repair and Modulation of Gene Expression.
Antioxidants (Basel), 11(9):1817, 15 Sep 2022
Cited by: 16 articles | PMID: 36139891 | PMCID: PMC9495735
APE1 assembles biomolecular condensates to promote the ATR-Chk1 DNA damage response in nucleolus.
Nucleic Acids Res, 50(18):10503-10525, 01 Oct 2022
Cited by: 14 articles | PMID: 36200829 | PMCID: PMC9561277
Cysteine Oxidation to Sulfenic Acid in APE1 Aids G-Quadruplex Binding While Compromising DNA Repair.
ACS Chem Biol, 17(9):2583-2594, 29 Aug 2022
Cited by: 3 articles | PMID: 36037088 | PMCID: PMC9931449
The human AP-endonuclease 1 (APE1) is a DNA G-quadruplex structure binding protein and regulates KRAS expression in pancreatic ductal adenocarcinoma cells.
Nucleic Acids Res, 50(6):3394-3412, 01 Apr 2022
Cited by: 13 articles | PMID: 35286386 | PMCID: PMC8990529
Go to all (172) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Redox activation of Fos-Jun DNA binding activity is mediated by a DNA repair enzyme.
EMBO J, 11(9):3323-3335, 01 Sep 1992
Cited by: 497 articles | PMID: 1380454 | PMCID: PMC556867
The redox and DNA-repair activities of Ref-1 are encoded by nonoverlapping domains.
Proc Natl Acad Sci U S A, 91(1):23-27, 01 Jan 1994
Cited by: 217 articles | PMID: 7506414 | PMCID: PMC42878
Identification and characterization of Ref-1, a nuclear protein that facilitates AP-1 DNA-binding activity.
EMBO J, 11(2):653-665, 01 Feb 1992
Cited by: 395 articles | PMID: 1537340 | PMCID: PMC556497
The structure and functions of the HAP1/Ref-1 protein.
Oncol Res, 9(6-7):275-280, 01 Jan 1997
Cited by: 15 articles | PMID: 9406232
Review