Abstract
Free full text

Potential of ancestral sylvatic dengue-2 viruses to re-emerge
Abstract
Dengue viruses (DENV) are the most important arboviral pathogens in tropical and subtropical regions throughout the world. DENV transmission includes both a sylvatic, enzootic cycle between nonhuman primates and arboreal mosquitoes of the genus Aedes, and an urban, endemic/epidemic cycle between Aedes aegypti, a mosquito with larval development in peridomestic water containers, and human reservoir hosts. All 4 serotypes of endemic DENV evolved independently from ancestral sylvatic viruses and have become both ecologically and evolutionarily distinct; this process may have involved adaptation to (i) peridomestic mosquito vectors and/or (ii) human reservoir hosts. To test the latter hypothesis, we assessed the ability of sylvatic and endemic DENV-2 strains, representing major genotypes from Southeast Asia, West Africa and the Americas, to replicate in two surrogate human model hosts: monocyte-derived, human dendritic cells (moDCs), and mice engrafted with human hepatoma cells. Although the various DENV-2 strains showed significant inter-strain variation in mean replication titers in both models, no overall difference between sylvatic and endemic strains was detected in either model. Our findings suggest that emergence of endemic DENV strains from ancestral sylvatic strains may not have required adaptation to replicate more efficiently in human reservoir hosts, implying that the potential for re-emergence of sylvatic dengue strains into the endemic cycle is high. The shared replication profiles of the American endemic and sylvatic strains suggest that American strains have maintained or regained the ancestral phenotype.
Introduction
The four serotypes of dengue virus (DENV) are transmitted among humans by Aedes aegypti and Aedes albopictus mosquitoes (Gubler, 1998), causing approximately 100 million infections leading to dengue fever (DF) annually, and up to 500,000 cases of dengue hemorrhagic fever (DHF) with a case fatality rate of up to 5% (Halstead, 1997). DENV is believed to have evolved as a non-human primate virus transmitted by canopy-dwelling arboreal mosquitoes before diverging into the 4 antigenically distinct DENV serotypes (DENV-1 to DENV-4), each of which later emerged into the human population (Cheong, 1986; Smith, 1956). Wang et al. (2000) tested this hypothesis using phylogenetic analyses to compare envelope protein gene sequences of the of endemic (human and Ae. aegypti) isolates to sequences of sylvatic (nonhuman primate and arboreal Aedes spp.) DENV-1, -2 and -4 strains of Southeast Asian origin, as well as DENV-2 sylvatic strains from West Africa. These analyses indicated that endemic DENV-1, -2 and -4 emerged independently from sylvatic progenitors at a time consistent with the establishment of urban populations in the Asia-Oceania region large enough to support a continuous human transmission cycle (Kuno, 1995). The Asiatic origin of DENV sylvatic progenitors is further supported by serological surveys of ecologically diverse rural habitats in Southeast Asia (Smith, 1956). More recently, the global spread of dengue has given rise to distinct endemic genotypes within each serotype (Gubler, 1997).
Currently, transmission of DENV occurs in 2 distinct cycles: a sylvatic, enzootic cycle between nonhuman primates and treehole mosquitoes of the genus Aedes (Diallo et al., 2003; Rudnick et al., 1967), and an endemic cycle between humans and the peridomestic mosquitoes Ae. aegypti and Ae. albopictus. In rural areas of Africa and Asia (also known as the ‘zone of emergence’) where mosquito vectors often reach high densities, DENV strains could potentially transfer between the two cycles, thus setting the stage for a re-emergence of the sylvatic strains (Gubler, 1988). To anticipate and potentially prevent re-emergence, it is critical to characterize the adaptations that would enable sylvatic DENV to transmit efficiently among humans.
The present study focused on the DENV-2 serotype, which has been grouped into five genotypes (Shurtleff et al., 2001) based on phylogenetic relationships among the envelope protein (E) gene sequences (Chang et al., 1994; Rico-Hesse, 1990; Wang et al., 2000). Sylvatic DENV-2, which circulates in both West Africa and Southeast Asia, shows a broader geographic range than the remaining sylvatic serotypes, which have only been detected in Southeast Asia. There is no evidence that the sylvatic cycles are involved in outbreaks of human dengue, suggesting that these strains are confined to forest habitats and/or produce relatively mild human disease. While transmission of sylvatic DENV-2 strains to humans has been documented in several West African cases of dengue fever (Saluzzo et al., 1986; Zeller et al., 1992), there is no evidence that sylvatic DENV strains of any serotype have subsequently been transmitted among humans to generate outbreaks (Diallo et al., 2003; Rico-Hesse, 1990). Moreover, sylvatic DENV strains have not been detected in the peridomestic vectors of endemic dengue, suggesting that these strains are confined to forest habitats.
We hypothesized that emergence of sylvatic DENV strains into the endemic cycle may require adaptation to peridomestic mosquito vectors and/or human hosts. Previous comparisons of the infectivity of sylvatic and endemic DENV-2 strains for urban anthropophilic Ae. aegypti and Ae. albopictus mosquito vectors demonstrated an advantage for the endemic strain, suggesting that adaptation to these mosquito species had accompanied emergence (Moncayo et al., 2004). Moreover, a cluster of amino acid changes in domain 3 of the viral envelope protein, which is believed to interact with cellular receptors, accompanied DENV emergence (Rico-Hesse, 1990; Wittke et al., 2002), and amino acid substitutions in the envelope glycoprotein of other arboviruses have been implicated in adaptation to novel mosquito vectors (Brault et al., 2004).
In the current study, we compared the human infection phenotypes of endemic and sylvatic DENV-2 strains using in vitro and in vivo models to determine whether adaptation to humans is a necessary component of dengue emergence. We selected virus strains of relatively low passage histories that represent all major DENV-2 genotypes, including both African and Asian sylvatic strains, and Asian, African and American endemic strains (Table 1). Both Asian and American endemic strains were tested because they are reported to differ in their human virulence (Cologna and Rico-Hesse, 2003; Vaughn et al., 2000).
Table 1
History of DENV-2 strains utilized in this study
| Strain | Epidemiological type a | Host | Passage history b | Location | Year |
|---|---|---|---|---|---|
| 1349 | Endemic | Human | SM2, C6/36-2 | Burkina Faso | 1982 |
| 16681 | Endemic | Human | BSC-1–x, LLC-MK2-6, Rh. Macaque-1, Tx. Amboinensis −2, C6/36-4, LLC-MK2-1, C6/36-1 | Thailand | 1964 |
| 1328 | Endemic | Human | Mosq.-2, C6/36-1 | Puerto Rico | 1977 |
| IQT-1950 | Endemic | Human | C6/36-2 | Peru | 1995 |
| PM33974 | Sylvatic | Ae. africanus | Tx. Amboinensis −1, C6/36-2 | Guinea | 1981 |
| P8-1407 | Sylvatic | Sentinel monkey | SM3, C6/36-2 | Malaysia | 1970 |
| A510 | Sylvatic | Ae. taylori | SM4, C6/36-2 | Ivory Coast | 1980 |
| A1247 | Sylvatic | Ae. taylori | SM5, C6/36-1 | Ivory Coast | 1980 |
| 2022 | Sylvatic | Ae. africanus | SM6, C6/36-1 | Burkina Faso | 1980 |
| 2039 | Sylvatic | Human | SM6, C6/36-1 | Burkina Faso | 1980 |
Because no animal model has been identified that recapitulates human infection and disease, we utilized two surrogate human models to compare the viremia and replication profiles of sylvatic versus endemic strains: (1) monocyte-derived dendritic cells (moDCs) from healthy volunteers (Wu et al., 2000), and (2) the severe combined immune deficiency (SCID) mouse xenografted with human hepatoma cells (An et al., 1999). Although the DENV-2 strains showed significant variation in mean replication titers, no overall difference between sylvatic and endemic strains was detected in either model. These findings suggest that emergence of endemic DENV strains from ancestral sylvatic strains may not have required adaptation to replicate more efficiently in humans as reservoir hosts, implying that the probability of re-emergence of human-to-human transmission is high.
Results
Phylogenetic analyses
We expanded previous phylogenetic analyses (Holmes and Twiddy, 2003; Rico-Hesse, 1990; Wang et al., 2000) of the evolutionary relationships among endemic serotypes and their sylvatic progenitors by using additional endemic and sylvatic strains in the phylogeny. The sylvatic strains were isolated between 1970 and 1981 in West Africa and Southeast Asia, whereas the endemic strains were isolated between 1964 and 1995, in West Africa, South America and Southeast Asia (Table 1). Analysis of 1485 nucleotides encoding the envelope protein (E) gene from 55 DENV-2 isolates, representing strains from diverse localities throughout the tropics and neotropics, included homologous DENV-1 and DENV-3 sequences as an outgroup to root the DENV-2 tree. All phylogenetic methods revealed 5 major DENV-2 lineages or genotypes, as has been demonstrated in previous studies (Holmes and Twiddy, 2003; Twiddy et al., 2002; Wang et al., 2000). The trees also revealed considerable genetic diversity within the DENV-2 genotypes, reflecting their continual divergence and diverse geographic distribution. Representative members of the major genotypes (Fig. 1) were selected for experimental studies.

Phylogenetic analysis of DENV isolates. Phylogenetic tree derived from the envelope protein gene nucleotide sequences of sylvatic and representative endemic DENV-2 strains using Bayesian analysis or maximum likelihood (ML) (PAUP, version 4.10) and drawn using branch lengths obtained using the Rogers–Swofford approximation method. The following ML parameters corresponding to the GTR+G+I model were used: empirical values for nucleotide frequencies (A = 0.32989, C=0.19737, G = 0.25924 and T = 0.21350); and among-site rate variation as: at invariable sites as estimated and γ distribution (discrete approximation) of rates at variable sites. The scale shows a genetic distance of 0.01 or 1% nucleotide sequence divergence. Homologous sequences from dengue sister serotypes 1 and 3 were used as an outgroup to root the DENV-2 tree. Numbers indicate bootstrap values for groups to the right. Asterisks indicate the strains used for this study.
All sylvatic DENV-2 isolates from Malaysia and West Africa were genetically distinct from endemic DENV-2 isolates. The endemic DENV-2 strains showed further subdivisions between the American and Asian lineages, with high bootstrap support using Bayesian and/or maximum parsimony methods, consistent with previous reports (Holmes and Twiddy, 2003; Twiddy et al., 2002; Wang et al., 2000). The Asian lineage included isolates clustered in 3 groups, Asian, American/Asian and Cosmopolitan, with bootstrap support of 71%, 100% and 99% respectively (Fig. 1).
Replication of sylvatic and endemic DENV-2 strains in vivo
A major limitation in dengue research is the lack of an inexpensive laboratory animal model that recapitulates human disease and viremia. A large number of nonhuman primates support DENV replication without developing clinical signs of illness (Halstead et al., 1973; Rosen, 1958). Nevertheless, because the duration and magnitude of virus replication in non-human primates often correlates with patterns of replication in humans, primates have become the gold standard for evaluation of live attenuated dengue vaccine candidates (Angsubhakorn et al., 1988; Blaney et al., 2005; Edelman et al., 1994; Hanley et al., 2004; Markoff et al., 2002; Men et al., 1996; Robert Putnak et al., 2005). However, economic and animal facility limitations preclude widespread utility of nonhuman primates as a useful model for DENV replication. Several murine models (Boonpucknavig et al., 1981; Chaturvedi et al., 1991; Cole and Wisseman, 1969; Hotta et al., 1981) developed to evaluate protective immune responses, have proven to be ineffective due to the absence of DENV replication and illness or the requirement for murine-adapted DENV (Cole and Wisseman, 1969; Sabin, 1952). Human peripheral blood lymphocytes (hu-PBL) engrafted in SCID mice (Wu et al., 1995) infected with DENV produce high viremia and virus loads in the spleen and lymph nodes. Limitations of this model include extensive labor requirements and variable reconstitution (successful engraftment) rates.
Observations documenting hepatic lesions (Couvelard et al., 1999; Lum et al., 1993), as well as detection of DENV antigen in hepatocytes (Kuo et al., 1992) during human infections formed the rationale for development of the SCID-xenograft model, where human hepatoma cells (HepG2 or Huh-7) are grafted intraperitoneally (i.p.) into SCID mice and DENV is injected directly into the i.p. tumor (An et al., 1999; Blaney et al., 2002). Mice develop gradual illness with peak viremia on day 7, whereas peak viral replication in the liver is detected by day 5 with gradual declines, and by day 11 virus has reached peak titers in the brain. This pattern of replication is similar to that which accompanies human infection, where DENV initially replicates at the site of infection and gradually disseminates to other organs and to the brain (although cerebral involvement is not believed to be common in human infections) via the circulation (Rothman, 1997). These are the first animal models where DHF/DSS-like manifestations (gastrointestinal bleeding and small focal hemorrhages in the livers of some mice) similar to those seen in human infections (Burke, 1968; Rosen et al., 1989) occur, and have proved useful for virulence testing of DENV vaccine candidates (Blaney et al., 2002; Whitehead et al., 2003). More recently, a humanized mouse model based on the grafting of human CD34+ cells in nonobese diabetic/severely compromised immunodeficient mice (NOD/SCID), has also been proposed as a model for studying the pathogenicity of DENV infection (Bente et al., 2005). However, based on the consistency of the SCID-Huh-7 xenograft model in evaluating the virulence of DENV vaccine candidates, we selected this model for the in vivo evaluation of our hypothesis.
To assess the human infection phenotypes of sylvatic versus endemic DENV-2 strains and to determine statistically dictated cohort numbers and tumor development efficiency, we conducted a pilot experiment using the Asian endemic 1349 and sylvatic PM33974 strains. The sylvatic strain generated moderate peak viremia levels of 4 log10ffu/ml (N = 3), while the endemic strain generated significantly higher viremia of 6 log10ffu/ml on day 7 (N = 7) (t-test, df = 7, t = 4.49, P<0.002) (data not shown). We also bled the mice at days 2, 4 and 7 post-infection (p.i.), and mean replication titers consistently peaked at day 7 p.i. (data not shown) as described earlier (An et al., 1999; Blaney et al., 2002). Power analyses indicated that a minimum of 6 animals per group were required to detect significant differences (P = 0.861, α = 0.05), with tumor development efficiency at 88%. Therefore, to compare the sylvatic versus endemic human infection phenotypes, six groups of 10 SCID-huh7 mice were infected with one of 2 sylvatic (PM33974, P8-1407) or 4 endemic (Asian 1349 and 16681, and American IQT-1950 and 1328) DENV-2 strains (Fig. 1). A significant overall difference among the mean serum titers of the six strains was detected (one-way ANOVA, P<0.0001) and a Tukey-Kramer post hoc test revealed multiple significant differences among individual pairs of strains (Table 2). However, this complex pattern of differences did not indicate a consistent or overall difference between sylvatic and endemic strains or between Asian endemic and American endemic strains. For example, the titer of Asian endemic strain 16681 was significantly higher than any of the other three endemic strains but not significantly different from sylvatic strain P8-1407.
Table 2
Serum viremia of endemic and sylvatic DENV-2 in vivo
| Virus a | Epidemiological type | No. of mice b | Mean peak virus titer c, d (log10 ffu/ml±SE) | Statistical group e |
|---|---|---|---|---|
| 16681 | Asian endemic | 7 | 5.9±0.2 | A |
| 1349 | Asian endemic | 6 | 4.9±0.3 | B, C |
| 1328 | American endemic | 6 | 2.9±0.3 | D |
| IQT-1950 | American endemic | 6 | 4.0±0.3 | C, E |
| P8-1407 | Sylvatic | 7 | 5.4±0.2 | A, B |
| PM33974 | Sylvatic | 6 | 3.6±0.2 | D, E |
Replication of sylvatic and endemic DENV-2 strains ex vivo
Natural DENV infection involves virus deposition in the skin by the mosquito vector during blood feeding. Blood and resident skin dendritic cells (DC) including Langerhans cells (LC), may be the initial target of viral infection following exposure to DENV during injection of mosquito saliva (Marovich et al., 2001; Wu et al., 2000). DCs efficiently take up pathogens through phagocytocis, pinocytosis and receptor-mediated endocytosis. The latter is mediated through lectins (i.e. CD209, DC-SIGN) (Navarro-Sanchez et al., 2003; Tassaneetrithep et al.,2003), as well asimmunoglobulin Fc receptors. DENV-infected DCs then migrate to the lymphoid tissues through the afferent lymphatic system, where they mature and interact with T-cells (Austyn et al., 1988; Steinman, 1991), and could shed virus, aiding in dissemination.
Because of the great genetic diversity of the human population at risk as well as epidemiologic evidence suggesting the influence of host genetics in the development of DENV viremia and disease (Bravo et al., 1987), we utilized moDC from anonymous, healthy donors of Caucasian, West African and Southeast Asian origin (3 donors per ethnic group) to assess the human infection phenotypes of 2 sylvatic (PM33974, P8-1407) and 4 endemic (Asian 1349 and 16681, and American IQT-1950 and 1328) DENV-2 strains. Prior to infection, the immature moDC phenotype was confirmed with a panel of fluorescein-isothiocyanate (FITC)-conjugated monoclonal antibodies against common DC markers, CD40, CD80, CD83, CD86 and DC-specific ICAM-3 grabbing non-integrin (DC-SIGN) using a FACScan flow cytometer (Fig. 2A). DC-SIGN is not a marker of the moDC phenotype, but mediates the infection of human DCs by DENV (Tassaneetrithep et al., 2003). Immature moDCs are susceptible to DENV infection, whereas mature moDCs are refractory (Wu et al., 2000). The moDC infection rates determined by FACS analysis varied between 6.4% and 18% (Fig. 2B). We also observed interdonor variation among the levels of moDC infections (data not shown), which was within the range of variation described in previous reports of DENV infection of human moDCs (Cologna et al., 2005; Sanchez et al., 2006). The cell-free supernatants of the infected moDCs were collected at 24 and 48 h post-infection and viral output was evaluated by focus forming assay (FFA) on C6/36 cells. Mean replication titers peaked consistently at 48 h post-infection (Tables 3 and and4),4), as observed in previous reports (Palmer et al., 2005; Wu et al., 2000). As illustrated in Table 3, the 6 DENV-2 isolates showed significant differences in levels of replication (df = 5, F = 69.9, P<0.0001); endemic Asian strains consistently generated higher virus outputs than the American DENV-2 strains, as demonstrated previously (Cologna et al., 2005), as well as the sylvatic DENV-2 isolates (Tukey-Kramer post hoc test, P < 0.05). However, no consistent difference between endemic American and sylvatic DENV-2 replication was detected. One of the 2 sylvatic strains, African PM33974, failed to replicate in the moDCs of all but one donor (the limit of detection of FFA assay was 0.5 log10 ffu/ml) (data not shown). To eliminate the possibility that this strain was an abberant outlier, possibly due to artificial selection during its passage history, 4 additional African sylvatic strains (Table 1) were evaluated in moDCs from 2 human volunteers. The poor replication profile of sylvatic PM33974 was shared by the sylvatic A1247 strain, but not by the other 3 sylvatic strains analyzed (Table 4). As in the previous analyses, all of these sylvatic strains replicated to lower titers than the endemic Asian strains, but did not differ consistently from the endemic American strains (Tukey-Kramer post hoc test, Table 4). A comparison of the mean titers of endemic (N = 4) and sylvatic (N = 6) strains showed no consistent difference in the mean level of replication of this expanded group of sylvatic DENV-2 strains compared to the endemic isolates (Student’s t-test: df = 8; t = 1.35; P = 0.21).
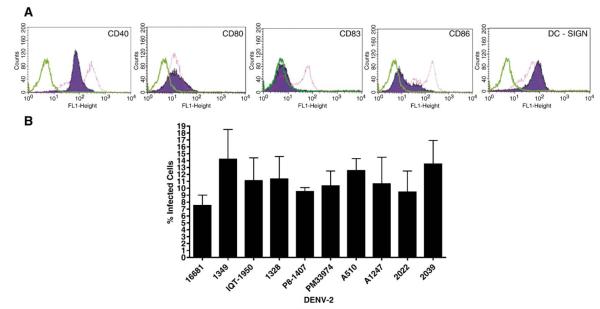
FACS analysis of blood-derived DCs. (A) Cell surface staining of immature blood-derived moDCs at day 6 post stimulation with IL-4 and GM-CSF cytokine cocktail prior to DENV-1349 infection and infected blood-derived DCs at day 2 post-infection with DENV-1349. Surface expression of CD40, CD80, CD83, CD86 and DC-SIGN was evaluated to determine the maturity of the DCs immediately prior to DENV infection. DC-SIGN is not a marker of moDC phenotype but mediates the infection of human DCs by DENV. Green peaks, isotype control; purple-shaded peaks, non-infected immature DCs; pink peaks, infected mature DCs. Data are representative of three independent experiments. (B) Infected DCs were examined by FACS analysis 48 h post-infection with an α-dengue virus-specific antibody, followed by an FITC-conjugated secondary antibody to determine the percentage of infected cells. Graph represents pooled data generated from 2 individual donors and error bars represent the standard errors of the means.
Table 3
Replication profile of endemic and sylvatic DENV-2 ex vivo
| Virus a | Epidemiological type | Mean peak virus titer b, c, d (log10 ffu/ml±SE) | Statistical group e |
|---|---|---|---|
| 16681 | Asian endemic | 4.8±0.1 | A |
| 1349 | Asian endemic | 5.3±0.2 | A |
| 1328 | American endemic | 2.7±0.3 | B |
| IQT-1950 | American endemic | 3.3±0.4 | B |
| P8-1407 | Sylvatic | 3.1±0.2 | B |
| PM33974 | Sylvatic | 0.6±0.1 | C |
Table 4
Replication profile of expanded endemic and sylvatic DENV-2 ex vivo
| Virus a | Epidemiological type | Mean peak virus titer b, c, d (log10 ffu/ml±SE) | Statistical group e |
|---|---|---|---|
| 16681 | Asian endemic | 5.4±0.1 | A |
| 1349 | Asian endemic | 5.0±0.3 | A |
| 1328 | American endemic | 3.1±0.6 | B |
| IQT-1950 | American endemic | 4.3±0.2 | B |
| P8-1407 | Sylvatic | 3.2±0.0 | B |
| PM33974 | Sylvatic | 0.5±0.0 | C |
| A510 | Sylvatic | 4.4±0.2 | B |
| A1247 | Sylvatic | 0.5±0.0 | C |
| 2022 | Sylvatic | 2.6±0.1 | B |
| 2039 | Sylvatic | 4.6±0.2 | B |
Discussion
The presence of neutralizing antibodies in canopy-dwelling nonhuman primates (Rodhain, 1991; Rudnick, 1986; Smith, 1956; Wolfe et al., 2001), the relatively recent development of large, concentrated human populations in the zone of emergence and phylogenetic (Wang et al., 2000) evidence suggest that sylvatic DENV strains are the ancestors of the existing endemic DENV strains that circulate in most of the tropics, putting a third of the global human population at risk and causing a huge burden of morbidity and mortality. These sylvatic DENV are maintained in enzootic cycles between forest-dwelling Aedes spp. mosquito vectors (not Ae. aegypti or Ae. albopictus) and probably nonhuman primate reservoir hosts in the sylvan ecosystems of Southeast Asia (Rudnick, 1986) and West Africa (Saluzzo et al., 1986; Traore-Lamizana et al., 1994).
Endemic dengue viruses are believed to have originated from ancestral sylvatic strains that switched from arboreal to urban mosquito vectors and from nonhuman primate to human reservoir hosts. Assessing the likelihood of current sylvatic DENV strains to undergo a similar emergence is important for public health. Specifically, programs to reduce or eradicate dengue from human populations by vector control or vaccination might be short-lived if sylvatic DENV strains can readily re-emerge from sylvatic cycles not amenable to intervention. The success of these programs could hinge on sustainable vaccination coverage of the susceptible population at risk and/or virtual elimination of endemic vector mosquitoes. Active employment of these methods should prevent the successful introduction of sylvatic DENV into the urban cycle and thus eliminate the resurgence of a major public health problem. Nevertheless, if the endemic DENV transmission cycle is eradicated as the result of public health measures, cessation of the anti-dengue campaigns (vaccination and/or vector control) for a sufficiently long time will lead to the rise of susceptible human populations, as well as the establishment of adequate populations of endemic vector mosquitoes. Under such conditions, the re-emergence of human replication competent disease-producing sylvatic DENV could pose a problem to human health. Information critical to assessing the potential for re-emergence includes: (1) the ability of humans to serve as reservoir hosts for sylvatic DENV strains with or without adaptation; (2) the degree of ecological contact between humans and sylvatic DENV strains; (3) ability of peridomestic mosquitoes to serve as vectors for sylvatic DENV transmission; and (4) the ability of the sylvatic strains to evolve to efficiently complete a transmission cycle in human hosts and peridomestic vectors.
Collectively, our findings do not support the hypothesis that emergence of endemic DENV strains involved adaptation to human reservoir hosts. The endemic strains we tested did not produce higher viremia in the mouse model or higher levels of DENV-2 replication in dendritic cells, compared to sylvatic strains. Assuming that the phenotype of the extant sylvatic strains is ancestral, this suggests that emergence of endemic DENV-2 strains from sylvatic strains did not require adaptation to replicate more efficiently in human reservoir hosts. Our data therefore imply that re-emergence of human-to-human transmission may occur readily. Comparable studies with DENV-1 and -4 strains [sylvatic DENV-3 strains have not been isolated but are believed to exist in Malaysia based on the seroconversion of sentinel monkeys (Rudnick, 1978)] are needed to determine if the other sylvatic DENV strains are also capable of efficient human infection.
Data in the literature on disease severity in humans with different racial backgrounds suggest that host genetics may influence the outcome of DENV infection (Bravo et al., 1987; Kouri et al., 1987). The utilization of the human surrogate animal and human dendritic cell models allowed us to control for host genetics to evaluate putative differences in the DENV-2 replication phenotypes among sylvatic and endemic strains. The significant differences in the replication profiles of Southeast Asian and American DENV strains observed in both models support previous evidence for the higher pathogenic potential of Southeast Asian strains (Cologna et al., 2005; Leitmeyer et al., 1999;Watts et al., 1999). Although higher replication profiles in vivo have been associated with severe disease (Vaughn et al., 2000; Wang et al., 2003), other factors such as immune enhancement (Halstead, 2003), genetic predisposition (Bravo et al., 1987), gender and age (Guzman et al., 1984) and nutrition (Thisyakorn and Nimmannitya, 1993) may influence the pathogenic outcome to DENV infections. Moreover, although our moDCs were obtained from a small number of human donors, they represented different ethnicity and no differences were observed in their ability to support DENV-2 replication. Lastly, without coming to conclusions about which was the first to evolve, the shared replication profiles of the American endemic and sylvatic strains suggest that American strains may have “relaxed” into the lower replication phenotype after evolution in the Americas.
Twiddy et al. (2002) attempted to identify evidence of positive selective pressure during the evolution of sylvatic and human DENV strains, but found no evidence of adaptive evolution in the E gene that coincided with the emergence of endemic strains. Although the computational methods used to identify positive selection have limitations, the lack of evidence for selective pressure during endemic emergence would imply that currently circulating sylvatic strains may not have difficulty spreading into the human reservoir host should the appropriate conditions occur. However, there is no evidence for the recent, direct involvement of these enzootic strains in DENV epidemics, which always involve genetically distinct endemic strains. Nevertheless, in rural areas of Africa and in Asia where the peridomestic Ae. albopictus mosquito vectors often reach high densities, DENV may be transmitted at low frequency from its sylvatic reservoir and humans. Additional ecological studies are necessary to identify and assess the role of vertebrate hosts in the maintenance and amplification of sylvatic DENV, which may provide valuable insight to the degree of the ecological contact between humans and sylvatic DENV. Ideally infectivity and disease expression of these viruses should be evaluated in human volunteers, but ethical or legal considerations may preclude their evaluation in human volunteers. However a comparison of the pattern of replication of sylvatic and endemic viruses in nonhuman primates is certainly warranted at this juncture. This information is needed to predict the risk of sylvatic DENV emergence and establishment of an epidemic in humans.
Materials and methods
Cell cultures and viruses
C6/36 (mosquito, Ae. albopictus) cells were maintained in Eagle’s minimal essential medium (MEM) supplemented with 5% fetal bovine serum (FBS), 50 mg/ml penicillin/streptomycin, 2 mM l-glutamine and non-essential amino acids (NEAA) (Invitrogen, Carlsbad) at 28 °C. Human hepatoma Huh-7 cells (clone JTC-39) were obtained from the Japanese Health Sciences Foundation, Osaka and were maintained in Dulbecco’s MEM supplemented with 10% FBS, 50 mg/ml penicillin/streptomycin and 2 mM l-glutamine. DENV-2 isolates with the lowest in vitro passage histories (Table 1) were selected from the World Reference Center for Emerging Viruses and Arboviruses, University of Texas Medical Branch to reduce the chance of cell culture or mouse adaptive mutations. Viral isolates were passaged twice in C6/36 cultures to obtain high titer stocks. Supernatants were clarified from cellular debris by low-spin centrifugation (630 × g, 20 min, 4 °C) stabilized with the addition of 10 × SPG (2.18 M Sucrose, 0.038 M KH2PO4, 0.072 M K2HPO4 and 0.054 M l-glutamate), and stored at −80 °C.
Focus forming assays (FFA) and immunostaining
Ten-fold serial dilutions of virus were added to confluent C6/36 cell monolayers. The virus inoculum was removed 1 h later, cell monolayers were washed and overlayed with 0.8% methylcellulose (Sigma-Aldrich, St. Louis) diluted in Optimem (Invitrogen, Carlsbad) supplemented with 2% FBS, antibiotics, 1% l-glutamine and 1% NEAA. Plates were incubated for 4 days at 28 °C, the methylcellulose overlay was removed and plates were rinsed with phosphate-buffered saline (PBS), pH 7.4. Plates were fixed with the addition of ice-cold acetone and methanol (1:1) for 30 min at room temperature (RT). The fixation solution was aspirated and plates were allowed to air dry. Plates were then washed with PBS, followed by blocking (PBS supplemented with 3% FBS) and addition of mouse anti-DENV-2 ascites fluid (1:1000) and incubation for 30 min. The antibody was aspirated and plates washed 3 times in PBS followed by addition of secondary antibody conjugated to horseradish peroxidase (HRP) (KPL, Gaithersburg) (1:1000) and incubation at RT for 30 min. Plates were washed 3 times with PBS and aminoethylcarbazole (AEC) substrate (ENZO Diagnostics, Farmingdale), prepared according to the manufacturer’s instructions, was added and allowed to incubate in the dark for 10 min. Substrate solution was aspirated, washed with water and plates were allowed to air dry before scoring.
Isolation, stimulation of peripheral blood mononuclear cells and DENV infections of moDCs
Nine consenting healthy volunteers, with no history of infection to any of DENV serotypes and confirmed as negative by plaque reduction neutralization test (PRNT) assay, were used to obtain approximately 100 ml of blood. Peripheral blood mononuclear cells were then isolated from buffy coats by centrifugation over an Accuprep gradient according to the manufacturer’s protocol (Accurate Chemical Corp, Westbury). CD14+ monocytes were positively selected using a magnetic cell sorting (MACS) isolation column (Miltenyi Biotec, Auburn). Cells were counted and seeded in 6-well plates at a density of 1–2 × 106 cells per well in RPMI 1640 culture medium supplemented with 10% heat-inactivated FBS, 100 U/ml penicillin, 100 μg/ml streptomycin and 2 mM l-glutamine, containing 1000 U/ml recombinant human interleukin 4 (IL-4) (R&D Systems, Minneapolis) and 1400 U/ml granulocyte–monocyte colony-stimulating factor (Immunex, Thousand Oaks). On alternate days, one-half of the volume was removed and replaced with medium containing double (2×) the concentration of fresh cytokines. Cultures were maintained under these conditions for a total of 6 days, then cells were collected with gentle pipetting, washed and the moDC phenotype was confirmed by FACS analysis using a panel of FITC-conjugated monoclonal antibodies against CD40, CD80, CD83, CD86 and DC-SIGN. Subsequently, 1 × 105 cells were infected with the selected DENV at MOI = 2 for 2 h at 37 °C. The virus inoculum was removed and the cells were washed 3 times with PBS to ensure removal of unadsorbed virus. Cells were resuspended in 2 ml of RPMI 1640 culture medium supplemented with cytokines and incubated at 37 °C. Cell-free supernatant aliquots were removed immediately prior and after infection, at day one and two post-infection and were assayed on C6/36 cells to obtain infection and progeny titers respectively. moDC infection rates were established by FACS analysis using a DENV-specific antibody (7E11, courtesy of Dr. Putnak), as well as IHC on cell cytospins.
Flow cytometry
Uninfected and/or infected DCs were washed once in FACS staining buffer (PBS containing 1% FBS) prior to confirmation of their immature phenotype with a panel of FITC-conjugated monoclonal antibodies against CD40, CD80, CD83, CD86, DC-SIGN (Beckman-Coulter), as well as for intracellular DENV antigen. As a control, the appropriate isotype was also utilized. Cells were allowed to incubate on ice for 30 min followed by washing twice with FACS staining buffer to remove unbound antibody. Cells were then fixed in buffered 2% paraformaldehyde (pH 9.5) and analyzed with a FACScan flow cytometer (Becton Dickinson) within 24 h of staining. Prior to fixation, cells that had been probed for DENV antigen were subjected to secondary staining with FITC-conjugated antibody (Molecular Probes) and washed twice with FACS staining buffer.
SCID mice
Ten 5–6-week-old SCID mice (Tac:Icr:Ha[ICR]-Prkdcscid; Taconic Farms) per group were xenografted intraperitoneally (i.p.) with 1 × 107 Huh-7 cells suspended in 0.2 ml phosphate-buffered saline. Tumors were detected 5–6 weeks later by palpation, and mice were infected by direct injection into the tumor mass with 1 × 104 focus-forming units (ffu) of virus diluted in 50 ml of PBS. Seven days post-infection, the mice were sacrificed and blood was isolated by cardiac puncture and the serum stored at −80 °C. Virus titers were determined by FIA in C6/36 cells. Mice that failed to develop viremia were excluded from the experiment. Prior to implantation, Huh-7 cells were certified to be free of mycoplasma, human and mouse viruses using PCR or RT-PCR (Taconic Anmed, Rockville). Animal experimentation was approved by the UTMB IACUC and mice were maintained in gnotobiotic isolators on specific pathogen-free environment.
Statistical analyses
GraphPad Prism version 4 software (GraphPad Software) was used for data analyses. Virus outputs from 11 ex vivo or 2 in vivo experiments were grouped by virus strain and compared by one-way ANOVA. The Tukey–Kramer post hoc test was used to test for differences between pairs of strains. Student’s t-tests were used for two sample comparisons.
RNA extraction and sequencing
Viral RNA was extracted from virus preparations using the QIAamp Viral RNA mini kit (Qiagen). PCR primers designed to amplify the E protein gene (Wang et al., 2000) were used with the Titan one step RT-PCR kit (Roche, Indianapolis). Viral RNA was denatured for 2 min at 70 °C and cDNA was synthesized at 50 °C for 30 min, followed by 35 rounds of amplification in a 50 μl reaction volume. The PCR products were purified from 1% agarose gels and both strands were sequenced directly using an Applied Biosystems (Foster City, California) Prism automated DNA sequencing kit and model 3100 Genetic Analyzer sequencer according to the manufacturer’s protocol.
Phylogenetic analyses
The obtained nucleotide sequences encoding the envelope (E) protein, and representative sequences from the GenBank library, were aligned using the ClustalW multiple sequence alignment program with default gap penalties. Phylogenetic analyses of the aligned nucleotide sequences were performed using Bayesian analysis with 1 million reiterations and/or maximum likelihood, neighbor joining and maximum parsimony methods implemented in the PAUP 4.0 software package (Swofford, 1998). Homologous nucleotide sequences from DENV-1 and -3 were used as an outgroup to root the DENV-2 tree. Bootstrapping with 1000 replicates was used to place confidence values on grouping within the tree (Felsenstein, 1985).
Nucleotide sequence accession numbers
The GenBank accession numbers for the DENV viruses used in the phylogenetic analyses are as follows: for DENV-2 strain 40274, accession number L10041; 124B, AY158340; 49255, AY577430; 6663, AY079424; 360236, AY158331; 360281, AY577431; 102954, AY158330; 19966, AY577433; Mara3, AY158329; Oax468, AY158341; 15957, AY577434; N.1409, M20558; M56309, X15433; b87MS8455, X15434; ThNH-28, U31950; ThNH-52, U31951; ThNH-7, U31959; ThNH-p11, U31952; Puo-218, U87331; 16681, U87411; TH-36, D10514; 1897, L10052; 2088, L10045; NGC, AF038403; #10, L10051; 0190, L10042; 1051, L10044; S-44554, L10048; S-44552, L10047; 1583, L10050; 1592, L10040; 271206, L10049; 206714, L10055; 271235, L10054; P7-863, AF231716; P8-377, AF231715; PR158, L10046; 131, AY158332; 132, AY158333; 328298, AY158338; Ven2, AY158328; IQT2133, AY577439; IQT2913, AY158339; 780477, AY158327; 200787, L04561; P9122, L10043; TR1751, L10053; DAK Ar578, AF231718; PM33974, AF231719; DAK HD10674, AF231720; P8-1407, AF231717; D1/45A25, U88536; D1/836-1, D00503; D1/CV1636, D00501; D1/AHF82, D00502; D1/P72-1244, AF231721; D3/2783, L11438; D3/260698, L11437; D3/5987, L11430; D3/D86-007, L11441; D3/CH3489D73-1, L11620; H87, M93130; 29472, L11422; and 168-Ap-2, L11432. The following sequences were determined in this study: IQT-1950; 1328; A510; A1247; 2039 and 2022. Nucleotide sequences were submitted to GenBank under accession nos. DQ917242 to DQ917247.
Acknowledgments
NV was supported by the Centers for Disease Control and Prevention Fellowship Training Program in Vector-Borne Infectious Diseases, T01/CCT622892. KAH was supported by a NM-INBRE (P20 RR016480-05) grant from the National Institutes of Health. We thank Robert Tesh and Hilda Guzman for kindly providing DENV strains and antisera. Gerald Kovacs and Mark Endsley for their critical comments on the manuscript. Two anonymous referees also provided valuable comments.
References
- An J, Kimura-Kuroda J, Hirabayashi Y, Yasui K. Development of a novel mouse model for dengue virus infection. Virology. 1999;263(1):70–77. [Abstract] [Google Scholar]
- Angsubhakorn S, Yoksan S, Bhamarapravati N, Moe JB, Marchette NJ, Pradermwong A, Sahaphong S. Dengue-4 vaccine: neurovirulence, viraemia and immune responses in rhesus and cynomolgus monkeys. Trans. R Soc. Trop. Med. Hyg. 1988;82(5):746–749. [Abstract] [Google Scholar]
- Austyn JM, Kupiec-Weglinski JW, Hankins DF, Morris PJ. Migration patterns of dendritic cells in the mouse. Homing to T cell-dependent areas of spleen, and binding within marginal zone. J. Exp. Med. 1988;167(2):646–651. [Europe PMC free article] [Abstract] [Google Scholar]
- Bente DA, Melkus MW, Garcia JV, Rico-Hesse R. Dengue fever in humanized NOD/SCID mice. J. Virol. 2005;79(21):13797–13799. [Europe PMC free article] [Abstract] [Google Scholar]
- Blaney JE, Jr., Johnson DH, Manipon GG, Firestone CY, Hanson CT, Murphy BR, Whitehead SS. Genetic basis of attenuation of dengue virus type 4 small plaque mutants with restricted replication in suckling mice and in SCID mice transplanted with human liver cells. Virology. 2002;300(1):125–139. [Abstract] [Google Scholar]
- Blaney JE, Jr., Matro JM, Murphy BR, Whitehead SS. Recombinant, live-attenuated tetravalent dengue virus vaccine formulations induce a balanced, broad, and protective neutralizing antibody response against each of the four serotypes in rhesus monkeys. J. Virol. 2005;79(9):5516–5528. [Europe PMC free article] [Abstract] [Google Scholar]
- Boonpucknavig S, Vuttiviroj O, Boonpucknavig V. Infection of young adult mice with dengue virus type 2. Trans. R Soc. Trop. Med. Hyg. 1981;75(5):647–653. [Abstract] [Google Scholar]
- Brault AC, Powers AM, Ortiz D, Estrada-Franco JG, Navarro-Lopez R, Weaver SC. Venezuelan equine encephalitis emergence: enhanced vector infection from a single amino acid substitution in the envelope glycoprotein. Proc. Natl. Acad Sci. U.S.A. 2004;101(31):11344–11349. [Europe PMC free article] [Abstract] [Google Scholar]
- Bravo JR, Guzman MG, Kouri GP. Why dengue haemorrhagic fever in Cuba? 1. Individual risk factors for dengue haemorrhagic fever/dengue shock syndrome (DHF/DSS) Trans. R Soc. Trop. Med. Hyg. 1987;81(5):816–820. [Abstract] [Google Scholar]
- Burke T. Dengue haemorrhagic fever: a pathological study. Trans. R Soc. Trop. Med. Hyg. 1968;62(5):682–692. [Abstract] [Google Scholar]
- Chang GJ, Trent DW, Vorndam AV, Vergne E, Kinney RM, Mitchell CJ. An integrated target sequence and signal amplification assay, reverse transcriptase-PCR-enzyme-linked immunosorbent assay, to detect and characterize flaviviruses. J. Clin. Microbiol. 1994;32(2):477–483. [Europe PMC free article] [Abstract] [Google Scholar]
- Chaturvedi UC, Dhawan R, Khanna M, Mathur A. Breakdown of the blood–brain barrier during dengue virus infection of mice. J. Gen. Virol. 1991;72(Pt. 4):859–866. [Abstract] [Google Scholar]
- Cheong WH. The vectors of dengue and dengue hemorrhagic fevers in Malaysia. In: Rudnick A, Lim TW, editors. Dengue fever studies in Malaysia. The Institute for Medical Research Jalan Pahang; Kuala Lampour, Malaysia: 1986. Bulletin No. 23. [Google Scholar]
- Cole GA, Wisseman CL., Jr. Pathogenesis of type 1 dengue virus infection in suckling, weanling and adult mice: 1. The relation of virus replication to interferon and antibody formation. Am. J. Epidemiol. 1969;89(6):669–680. [Abstract] [Google Scholar]
- Cologna R, Rico-Hesse R. American genotype structures decrease dengue virus output from human monocytes and dendritic cells. J. Virol. 2003;77(7):3929–3938. [Europe PMC free article] [Abstract] [Google Scholar]
- Cologna R, Armstrong PM, Rico-Hesse R. Selection for virulent dengue viruses occurs in humans and mosquitoes. J. Virol. 2005;79(2):853–859. [Europe PMC free article] [Abstract] [Google Scholar]
- Couvelard A, Marianneau P, Bedel C, Drouet MT, Vachon F, Henin D, Deubel V. Report of a fatal case of dengue infection with hepatitis: demonstration of dengue antigens in hepatocytes and liver apoptosis. Hum. Pathol. 1999;30(9):1106–1110. [Abstract] [Google Scholar]
- Diallo M, Ba Y, Sall AA, Diop OM, Ndione JA, Mondo M, Girault L, Mathiot C. Amplification of the sylvatic cycle of dengue virus type 2, Senegal, 1999–2000: entomologic findings and epidemiologic considerations. Emerg. Infect. Dis. 2003;9(3):362–367. [Europe PMC free article] [Abstract] [Google Scholar]
- Edelman R, Tacket CO, Wasserman SS, Vaughn DW, Eckels KH, Dubois DR, Summers PL, Hoke CH. A live attenuated dengue-1 vaccine candidate (45AZ5) passaged in primary dog kidney cell culture is attenuated and immunogenic for humans. J. Infect. Dis. 1994;170(6):1448–1455. [Abstract] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39(4):783–791. [Abstract] [Google Scholar]
- Gubler DJ. Dengue. In: Monath TP, editor. The Arboviruses: Epidemiology and Ecology. vol. II. CRC Press; Boca Raton, FL: 1988. pp. 223–260. 5 vols. [Google Scholar]
- Gubler DJ. Dengue and dengue hemorrhagic fever: its history and resurgence as a global public health problem. In: Gubler DJ, Kuno G, editors. Dengue and Dengue Hemorrhagic Fever. CABI Publishing; Oxon: 1997. pp. 1–22. [Google Scholar]
- Gubler DJ. Dengue and dengue hemorrhagic fever. Clin. Microbiol. Rev. 1998;11(3):480–496. [Europe PMC free article] [Abstract] [Google Scholar]
- Guzman MG, Kouri G, Morier L, Soler M, Fernandez A. A study of fatal hemorrhagic dengue cases in Cuba, 1981. Bull. Pan. Am. Health Organ. 1984;18(3):213–220. [Abstract] [Google Scholar]
- Halstead SB. Epidemiology of dengue and dengue hemorrhagic fever. In: Gubler DJ, Kuno G, editors. Dengue and Dengue Hemorrhagic Fever. CBA international; Oxon, UK: 1997. [Google Scholar]
- Halstead SB. Neutralization and antibody-dependent enhancement of dengue viruses. Adv. Virus Res. 2003;60:421–467. [Abstract] [Google Scholar]
- Halstead SB, Shotwell H, Casals J. Studies on the pathogenesis of dengue infection in monkeys: I. Clinical laboratory responses to primary infection. J. Infect. Dis. 1973;128(1):7–14. [Abstract] [Google Scholar]
- Hanley KA, Manlucu LR, Manipon GG, Hanson CT, Whitehead SS, Murphy BR, Blaney JE., Jr. Introduction of mutations into the nonstructural genes or 3′ untranslated region of an attenuated dengue virus type 4 vaccine candidate further decreases replication in rhesus monkeys while retaining protective immunity. Vaccine. 2004;22(25–26):3440–3448. [Abstract] [Google Scholar]
- Holmes EC, Twiddy SS. The origin, emergence and evolutionary genetics of dengue virus. Infect. Genet. Evol. 2003;3(1):19–28. [Abstract] [Google Scholar]
- Hotta H, Murakami I, Miyasaki K, Takeda Y, Shirane H, Hotta S. Localization of dengue virus in nude mice. Microbiol. Immunol. 1981;25(1):89–93. [Abstract] [Google Scholar]
- Kouri GP, Guzman MG, Bravo JR. Why dengue haemorrhagic fever in Cuba? 2. An integral analysis. Trans. R. Soc. Trop. Med. Hyg. 1987;81(5):821–823. [Abstract] [Google Scholar]
- Kuno G. Review of the factors modulating dengue transmission. Epidemiol. Rev. 1995;17(2):321–335. [Abstract] [Google Scholar]
- Kuo CH, Tai DI, Chang-Chien CS, Lan CK, Chiou SS, Liaw YF. Liver biochemical tests and dengue fever. Am. J. Trop. Med. Hyg. 1992;47(3):265–270. [Abstract] [Google Scholar]
- Leitmeyer KC, Vaughn DW, Watts DM, Salas R, Villalobos I, de C, Ramos C, Rico-Hesse R. Dengue virus structural differences that correlate with pathogenesis. J. Virol. 1999;73(6):4738–4747. [Europe PMC free article] [Abstract] [Google Scholar]
- Lum LC, Lam SK, George R, Devi S. Fulminant hepatitis in dengue infection. Southeast Asian J. Trop. Med. Public Health. 1993;24(3):467–471. [Abstract] [Google Scholar]
- Markoff L, Pang X, Houng Hs HS, Falgout B, Olsen R, Jones E, Polo S. Derivation and characterization of a dengue type 1 host range-restricted mutant virus that is attenuated and highly immunogenic in monkeys. J. Virol. 2002;76(7):3318–3328. [Europe PMC free article] [Abstract] [Google Scholar]
- Marovich M, Grouard-Vogel G, Louder M, Eller M, Sun W, Wu SJ, Putvatana R, Murphy G, Tassaneetrithep B, Burgess T, Birx D, Hayes C, Schlesinger-Frankel S, Mascola J. Human dendritic cells as targets of dengue virus infection. J. Investig. Dermatol. Symp. Proc. 2001;6(3):219–224. [Abstract] [Google Scholar]
- Men R, Bray M, Clark D, Chanock RM, Lai CJ. Dengue type 4 virus mutants containing deletions in the 3′ noncoding region of the RNA genome: analysis of growth restriction in cell culture and altered viremia pattern and immunogenicity in rhesus monkeys. J. Virol. 1996;70(6):3930–3937. [Europe PMC free article] [Abstract] [Google Scholar]
- Moncayo AC, Fernandez Z, Ortiz D, Diallo M, Sall A, Hartman S, Davis CT, Coffey L, Mathiot CC, Tesh RB, Weaver SC. Dengue emergence and adaptation to peridomestic mosquitoes. Emerg. Infect Dis. 2004;10(10):1790–1796. [Europe PMC free article] [Abstract] [Google Scholar]
- Navarro-Sanchez E, Altmeyer R, Amara A, Schwartz O, Fieschi F, Virelizier JL, Arenzana-Seisdedos F, Despres P. Dendritic-cell-specific ICAM3-grabbing non-integrin is essential for the productive infection of human dendritic cells by mosquito-cell-derived dengue viruses. EMBO Rep. 2003;4(7):723–738. [Europe PMC free article] [Abstract] [Google Scholar]
- Palmer DR, Sun P, Celluzzi C, Bisbing J, Pang S, Sun W, Marovich MA, Burgess T. Differential effects of dengue virus on infected and bystander dendritic cells. J. Virol. 2005;79(4):2432–2439. [Europe PMC free article] [Abstract] [Google Scholar]
- Rico-Hesse R. Molecular evolution and distribution of dengue viruses type 1 and 2 in nature. Virology. 1990;174(2):479–493. [Abstract] [Google Scholar]
- Robert Putnak J, Coller BA, Voss G, Vaughn DW, Clements D, Peters I, Bignami G, Houng HS, Chen RC, Barvir DA, Seriwatana J, Cayphas S, Garcon N, Gheysen D, Kanesa-Thasan N, McDonell M, Humphreys T, Eckels KH, Prieels JP, Innis BL. An evaluation of dengue type-2 inactivated, recombinant subunit, and live-attenuated vaccine candidates in the rhesus macaque model. Vaccine. 2005;23(35):4442–4452. [Abstract] [Google Scholar]
- Rodhain F. The role of monkeys in the biology of dengue and yellow fever. Comp. Immunol. Microbiol. Infect. Dis. 1991;14(1):9–19. [Abstract] [Google Scholar]
- Rosen L. Experimental infection of New World monkeys with dengue and yellow fever viruses. Am. J. Trop. Med. Hyg. 1958;7(4):406–410. [Abstract] [Google Scholar]
- Rosen L, Khin MM, U. T. Recovery of virus from the liver of children with fatal dengue: reflections on the pathogenesis of the disease and its possible analogy with that of yellow fever. Res. Virol. 1989;140(4):351–360. [Abstract] [Google Scholar]
- Rothman A. Viral pathogenesis of dengue infections. In: Gubler DJ, Kuno G, editors. Dengue and Dengue Hemorrhagic Fever. CAB International; Oxon: 1997. pp. 245–271. [Google Scholar]
- Rudnick A. Ecology of dengue virus. Asian J. Infect. Dis. 1978;2:156–160. [Google Scholar]
- Rudnick A, Marchette NJ, Garcia R. Possible jungle dengue—Recent studies and hypotheses. Jpn. J. Med. Sci. Biol. 1967;20:69–74. [Abstract] [Google Scholar]
- Rudnick A.a.L., T. W. Dengue fever studies in Malaysia. Inst. Med. Res. Malays. Bull. 1986;23:51–152. [Google Scholar]
- Sabin AB. Research on dengue during World War II. Am. J. Trop. Med. Hyg. 1952;1(1):30–50. [Abstract] [Google Scholar]
- Saluzzo JF, Cornet M, Adam C, Eyraud M, Digoutte JP. Dengue 2 in eastern Senegal: serologic survey in simian and human populations. 1974–85. Bull. Soc. Pathol. Exot. Filiales. 1986;79(3):313–322. [Abstract] [Google Scholar]
- Sanchez V, Hessler C, DeMonfort A, Lang J, Guy B. Comparison by flow cytometry of immune changes induced in human monocyte-derived dendritic cells upon infection with dengue 2 live-attenuated vaccine or 16681 parental strain. FEMS Immunol. Med. Microbiol. 2006;46(1):113–123. [Abstract] [Google Scholar]
- Shurtleff AC, Beasley DW, Chen JJ, Ni H, Suderman MT, Wang H, Xu R, Wang E, Weaver SC, Watts DM, Russell KL, Barrett AD. Genetic variation in the 3′ non-coding region of dengue viruses. Virology. 2001;281(1):75–87. [Abstract] [Google Scholar]
- Smith CE. The history of dengue in tropical Asia and its probable relationship to the mosquito Aedes aegypti. J. Trop. Med. Hyg. 1956;59(10):243–251. [Abstract] [Google Scholar]
- Steinman RM. The dendritic cell system and its role in immunogenicity. Annu. Rev. Immunol. 1991;9:271–296. [Abstract] [Google Scholar]
- Swofford DL. Phylogenetic Analysis Using Parsimony (*and Other Methods) Version 4 Sinauer Associates; Sunderland, Massachusetts: 1998. PAUP*. [Google Scholar]
- Tassaneetrithep B, Burgess TH, Granelli-Piperno A, Trumpfheller C, Finke J, Sun W, Eller MA, Pattanapanyasat K, Sarasombath S, Birx DL, Steinman RM, Schlesinger S, Marovich MA. DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. J. Exp. Med. 2003;197(7):823–829. [Europe PMC free article] [Abstract] [Google Scholar]
- Thisyakorn U, Nimmannitya S. Nutritional status of children with dengue hemorrhagic fever. Clin. Infect. Dis. 1993;16(2):295–297. [Abstract] [Google Scholar]
- Traore-Lamizana M, Zeller H, Monlun E, Mondo M, Hervy JP, Adam F, Digoutte JP. Dengue 2 outbreak in southeastern Senegal during 1990: virus isolations from mosquitoes (Diptera: Culicidae) J. Med. Entomol. 1994;31(4):623–637. [Abstract] [Google Scholar]
- Twiddy SS, Farrar JJ, Vinh Chau N, Wills B, Gould EA, Gritsun T, Lloyd G, Holmes EC. Phylogenetic relationships and differential selection pressures among genotypes of dengue-2 virus. Virology. 2002;298(1):63–72. [Abstract] [Google Scholar]
- Vaughn DW, Green S, Kalayanarooj S, Innis BL, Nimmannitya S, Suntayakorn S, Endy TP, Raengsakulrach B, Rothman AL, Ennis FA, Nisalak A. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J. Infect. Dis. 2000;181(1):2–9. [Abstract] [Google Scholar]
- Wang E, Ni H, Xu R, Barrett AD, Watowich SJ, Gubler DJ, Weaver SC. Evolutionary relationships of endemic/epidemic and sylvatic dengue viruses. J. Virol. 2000;74(7):3227–3234. [Europe PMC free article] [Abstract] [Google Scholar]
- Wang WK, Chao DY, Kao CL, Wu HC, Liu YC, Li CM, Lin SC, Ho ST, Huang JH, King CC. High levels of plasma dengue viral load during defervescence in patients with dengue hemorrhagic fever: implications for pathogenesis. Virology. 2003;305(2):330–338. [Abstract] [Google Scholar]
- Watts DM, Porter KR, Putvatana P, Vasquez B, Calampa C, Hayes CG, Halstead SB. Failure of secondary infection with American genotype dengue 2 to cause dengue haemorrhagic fever. Lancet. 1999;354(9188):1431–1434. [Abstract] [Google Scholar]
- Whitehead SS, Hanley KA, Blaney JE, Jr., Gilmore LE, Elkins WR, Murphy BR. Substitution of the structural genes of dengue virus type 4 with those of type 2 results in chimeric vaccine candidates which are attenuated for mosquitoes, mice, and rhesus monkeys. Vaccine. 2003;21(27–30):4307–4316. [Abstract] [Google Scholar]
- Wittke V, Robb TE, Thu HM, Nisalak A, Nimmannitya S, Kalayanrooj S, Vaughn DW, Endy TP, Holmes EC, Aaskov JG. Extinction and rapid emergence of strains of dengue 3 virus during an interepidemic period. Virology. 2002;301(1):148–156. [Abstract] [Google Scholar]
- Wolfe ND, Kilbourn AM, Karesh WB, Rahman HA, Bosi EJ, Cropp BC, Andau M, Spielman A, Gubler DJ. Sylvatic transmission of arboviruses among Bornean orangutans. Am. J. Trop. Med. Hyg. 2001;64(5–6):310–316. [Abstract] [Google Scholar]
- Wu SJ, Grouard-Vogel G, Sun W, Mascola JR, Brachtel E, Putvatana R, Louder MK, Filgueira L, Marovich MA, Wong HK, Blauvelt A, Murphy GS, Robb ML, Innes BL, Birx DL, Hayes CG, Frankel SS. Human skin Langerhans cells are targets of dengue virus infection. Nat. Med. 2000;6(7):816–820. [Abstract] [Google Scholar]
- Wu SJ, Hayes CG, Dubois DR, Windheuser MG, Kang YH, Watts DM, Sieckmann DG. Evaluation of the severe combined immunodeficient (SCID) mouse as an animal model for dengue viral infection. Am. J. Trop. Med. Hyg. 1995;52(5):468–476. [Abstract] [Google Scholar]
- Zeller HG, Traore-Lamizana M, Monlun E, Hervy JP, Mondo M, Digoutte JP. Dengue-2 virus isolation from humans during an epizootic in southeastern Senegal in November, 1990. Res. Virol. 1992;143(2):101–102. [Abstract] [Google Scholar]
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Genomic surveillance reveals a dengue 2 virus epidemic lineage with a marked decrease in sensitivity to Mosnodenvir.
Nat Commun, 15(1):8667, 09 Oct 2024
Cited by: 1 article | PMID: 39384752 | PMCID: PMC11464713
Does the Presence or a High Titer of Yellow Fever Virus Antibodies Interfere with Pregnancy Outcomes in Women with Zika Virus Infection?
Viruses, 15(11):2244, 11 Nov 2023
Cited by: 1 article | PMID: 38005922 | PMCID: PMC10675107
Contribution of phylogenetics to understanding the evolution and epidemiology of dengue virus.
Animal Model Exp Med, 5(5):410-417, 17 Oct 2022
Cited by: 8 articles | PMID: 36245335 | PMCID: PMC9610151
Review Free full text in Europe PMC
Viral Coinfections.
Viruses, 14(12):2645, 26 Nov 2022
Cited by: 8 articles | PMID: 36560647 | PMCID: PMC9784482
Review Free full text in Europe PMC
Why Did ZIKV Perinatal Outcomes Differ in Distinct Regions of Brazil? An Exploratory Study of Two Cohorts.
Viruses, 13(5):736, 23 Apr 2021
Cited by: 4 articles | PMID: 33922578 | PMCID: PMC8146858
Go to all (62) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (Showing 66 of 66)
- (1 citation) ENA - AY158328
- (1 citation) ENA - AY158327
- (1 citation) ENA - AY158329
- (1 citation) ENA - U87331
- (1 citation) ENA - D10514
- (1 citation) ENA - L04561
- (1 citation) ENA - AY577434
- (1 citation) ENA - L10048
- (1 citation) ENA - L10047
- (1 citation) ENA - L10049
- (1 citation) ENA - AF038403
- (1 citation) ENA - AY577439
- (1 citation) ENA - L10040
- (1 citation) ENA - U88536
- (1 citation) ENA - L10042
- (1 citation) ENA - L10041
- (1 citation) ENA - AY577430
- (1 citation) ENA - L10044
- (1 citation) ENA - AY079424
- (1 citation) ENA - AY577431
- (1 citation) ENA - L10043
- (1 citation) ENA - L10046
- (1 citation) ENA - AY577433
- (1 citation) ENA - L10045
- (1 citation) ENA - AY158339
- (1 citation) ENA - AY158338
- (1 citation) ENA - AF231715
- (1 citation) ENA - AF231716
- (1 citation) ENA - AF231717
- (1 citation) ENA - AF231718
- (1 citation) ENA - AY158331
- (1 citation) ENA - AF231719
- (1 citation) ENA - AY158330
- (1 citation) ENA - AY158333
- (1 citation) ENA - AY158332
- (1 citation) ENA - U31950
- (1 citation) ENA - U31952
- (1 citation) ENA - U31951
- (1 citation) ENA - DQ917247
- (1 citation) ENA - U31959
- (1 citation) ENA - L11441
- (1 citation) ENA - DQ917242
- (1 citation) ENA - AY158340
- (1 citation) ENA - AY158341
- (1 citation) ENA - M20558
- (1 citation) ENA - AF231720
- (1 citation) ENA - AF231721
- (1 citation) ENA - L11438
- (1 citation) ENA - L11437
- (1 citation) ENA - L11430
- (1 citation) ENA - L11432
- (1 citation) ENA - M93130
- (1 citation) ENA - X15434
- (1 citation) ENA - X15433
- (1 citation) ENA - D00503
- (1 citation) ENA - L11620
- (1 citation) ENA - L11422
- (1 citation) ENA - D00501
- (1 citation) ENA - D00502
- (1 citation) ENA - L10051
- (1 citation) ENA - L10050
- (1 citation) ENA - U87411
- (1 citation) ENA - L10053
- (1 citation) ENA - L10052
- (1 citation) ENA - L10055
- (1 citation) ENA - L10054
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The history and evolution of human dengue emergence.
Adv Virus Res, 72:1-76, 01 Jan 2008
Cited by: 108 articles | PMID: 19081488
Review
Dengue emergence and adaptation to peridomestic mosquitoes.
Emerg Infect Dis, 10(10):1790-1796, 01 Oct 2004
Cited by: 59 articles | PMID: 15504265 | PMCID: PMC3323252
Potential role of sylvatic and domestic African mosquito species in dengue emergence.
Am J Trop Med Hyg, 73(2):445-449, 01 Aug 2005
Cited by: 59 articles | PMID: 16103619
Genetic and phenotypic characterization of sylvatic dengue virus type 2 strains.
Virology, 377(2):296-307, 01 Aug 2008
Cited by: 38 articles | PMID: 18570968 | PMCID: PMC3612928
Funding
Funders who supported this work.
NCRR NIH HHS (2)
Grant ID: P20 RR016480
Grant ID: P20 RR016480-05




