Abstract
Free full text

Antigenicity and Immunogenicity of Transmitted/Founder, Consensus, and Chronic Envelope Glycoproteins of Human Immunodeficiency Virus Type 1
Associated Data
Abstract
Human immunodeficiency virus type 1 (HIV-1) vaccine development requires selection of appropriate envelope (Env) immunogens. Twenty HIV-1 Env glycoproteins were examined for their ability to bind human anti-HIV-1 monoclonal antibodies (MAbs) and then used as immunogens in guinea pigs to identify promising immunogens. These included five Envs derived from chronically infected individuals, each representing one of five common clades and eight consensus Envs based on these five clades, as well as the consensus of the entire HIV-1 M group, and seven transmitted/founder (T/F) Envs from clades B and C. Sera from immunized guinea pigs were tested for neutralizing activity using 36 HIV-1 Env-pseudotyped viruses. All Envs bound to CD4 binding site, membrane-proximal, and V1/V2 MAbs with similar apparent affinities, although the T/F Envs bound with higher affinity to the MAb 17b, a CCR5 coreceptor binding site antibody. However, the various Envs differed in their ability to induce neutralizing antibodies. Consensus Envs elicited the most potent responses, but neutralized only a subset of viruses, including mostly easy-to-neutralize tier 1 and some more-difficult-to-neutralize tier 2 viruses. T/F Envs elicited fewer potent neutralizing antibodies but exhibited greater breadth than chronic or consensus Envs. Finally, chronic Envs elicited the lowest level and most limited breadth of neutralizing antibodies overall. Thus, each group of Env immunogens elicited a different antibody response profile. The complementary benefits of consensus and T/F Env immunogens raise the possibility that vaccines utilizing a combination of consensus and T/F Envs may be able to induce neutralizing responses with greater breadth and potency than single Env immunogens.
INTRODUCTION
Amajor challenge for human immunodeficiency virus type 1 (HIV-1) vaccine development is to design immunogens that can overcome HIV-1 diversity and induce T and B cell responses that cross-react with a majority of HIV-1 transmitted/founder virus strains (1, 2). A number of strategies have aimed to induce broad T cell responses, including mosaic (3), ancestral or consensus (1), and conserved region (4, 5) immunogen designs. Both consensus and mosaic immunogens have been shown to induce superior T cell responses in nonhuman primates compared to wild-type HIV-1 immunogens (6–8). Consensus sequences induced T cell responses that had greater cross-reactivity in terms of breadth and depth than wild-type strains (7, 9, 10), and polyvalent mosaic vaccine designs were able improve the cross-reactive potential even more (6, 8, 11), raising the possibility that global coverage of the diverse forms of the HIV M group might be achievable for a T cell vaccine.
In contrast, induction of broadly neutralizing antibodies (bNAbs) to HIV-1 Env (Env) has been more problematic, with no vaccine designs to date inducing high levels of bNAbs at mucosal surfaces (12). The inability of HIV-1 Env constructs to induce bNAbs is likely to be due to multiple factors acting in concert, including occlusion of bNAbs epitopes in the native trimer (13–15), strain-specific differences in epitopes, the inability to mimic tertiary and quaternary epitopes with monomeric gp120 (16, 17), and host immune controls that prevent bNAbs maturation and expression (18, 19). Nonetheless, Env immunogens must be antigenic with expression of conserved neutralizing determinants to have a chance for induction of neutralizing antibodies with any degree of breadth, where breadth is the extent to which antibodies can neutralize diverse natural HIV isolates.
While transmitted/founder (T/F) viruses have subtle traits that distinguish them from chronic viruses (20, 21), the advantage of the selection of T/F viruses for candidate Env immunogens remains unclear (22, 23). T/F Envs tend to have shorter variable loops and/or fewer N-linked glycosylation sites; this pattern is clearly evident in clades A, C, and D (24–26) but marginal in clade B (24, 27, 28). Protein sequence signatures have been identified that are associated with T/F viruses (27); these signatures may be associated with Env expression levels (29). A critical question for HIV-1 vaccine development is to determine whether T/F HIV-1 Envs differ from chronic and consensus Envs in their ability to induce antibody responses.
To address the need for developing criteria for the choice of candidate Env immunogens, we have compared the antigenicity and immunogenicity of T/F, chronic and in silico-designed consensus recombinant Env proteins. We have produced a panel of 7 T/F Envs from clades B and C; 5 chronic Envs from clades A, B, C, AE_01, and G; and 8 consensus Envs selected from clades A, B, C, AE_01, and G, as well as group M global consensus Envs (1). We then compared Env antigenicity by enzyme-linked immunosorbent assay (ELISA) and surface plasmon resonance (SPR) and determined Env immunogenicity in guinea pigs.
We found chronic Envs induced low or moderate levels of primarily tier 1 neutralizing antibodies, and scattered low-level tier 2 antibodies. Consensus Env immunogens induced higher levels of tier 1 neutralizing antibodies (NAbs) than chronic Envs and moderate levels of NAbs against subsets of tier 2 Envs. T/F Envs immunogens induced low but measurable levels of broad NAbs, neutralizing both tier 1 and most tier 2 HIV-1 strains. We have identified a subset of T/F and consensus Env candidates that may have promise as immunogens alone or that have complementary neutralization profiles that may be more immunogenic as polyvalent mixtures.
MATERIALS AND METHODS
Recombinant HIV-1 Env proteins.
Table S1 in the supplemental material lists all of the recombinant Envs produced for the present study. HIV-1 Env consensus subtype sequences of clades A, B, C, G, and CRF01_AE Envs were derived by aligning complete Env sequences, including one randomly selected Env per sampled infected individual, of each HIV-1 subtype in the 2003 database as described at http://hiv-web.lanl.gov/content/hiv-db/CONSENSUS/M_GROUP/Consensus.html (see Table S1 in the supplemental material) (1, 30). CRF01 is a circulating recombinant form (CRF) that is a common lineage in Asia (31, 32) and is predominantly clade E in Env. Subtype G is not very common globally; however, CRF02 is another major epidemic lineage in west Africa with a recombinant founder, and CRF02 is subtype G in Env (33). (http://www.hiv.lanl.gov/content/sequence/HIV/CRFs/CRFs.html). These clade consensus sequences are relatively stable over time, although they can shift slightly from year to year based on new sample acquisition at the database; obviously, the most variable positions are particularly subject to change, although such shifts are minor relative to between-patient sequence differences. We included two consensus sequences from different years, B clade and M group consensus sequences from 2001 and 2003, to see how clade consensus sequences derived from two different years compared as immunogens. Group M is the “main” group of HIV-1 and refers to a set including all circulating HIV-1 clades and recombinants. Group M consensus Env sequences (CON-S and CON-T) were derived by aligning consensus sequences from HIV-1 subtypes A (including A, A1, and A2), B, C, D, F (including F1 and F2), and G from the 2001 (CON-S) and 2003 (CON-T) HIV-1 sequence database and forming a consensus of the subtype consensus sequences (see Table S1 in the supplemental material) (1, 30). The wild-type subtype A, B, C, and CRF01_AE gp160 Env sequences from HIV-1 primary isolates from chronically infected subjects were selected from HIV-1 sequence database to be representative of their clades; specifically, they were not outliers within a given clade (http://hiv-web.lanl.gov) (Table S1). Finally, seven T/F HIV-1 Env genes were chosen for expression, including five subtype B Envs (B.040, B.65321, B.6240, B.62357, and B.9021) and two subtype C Envs (C.1086 and C.089). T/F Envs were obtained from acutely infected HIV-1 subjects by single genome amplification (22). In one previous study, we found no difference between gp140 and gp120 for the induction of neutralizing antibodies in guinea pigs (9); in contrast, in another study, we found minor augmentation of both binding and neutralizing antibody induction with immunization with gp140 relative to gp120 (34). Env gp140 as immunogens may also have advantages in that they contain membrane-proximal external region (MPER) neutralization epitopes (35) and have more relatively conserved T cell epitopes (2) than do gp120 Envs. Thus, for these immunogenicity studies, we used gp140 oligomeric constructs.
All HIV-1 Env genes were codon optimized by converting amino acid sequences to nucleotide sequences employing the codon usage of highly expressed human housekeeping genes (36) and de novo-synthesized as gp140 (see Table S1 in the supplemental material). HIV-1 gp140 Envs with deletion of the cleavage (C) site and fusion (F) domain are termed HIV-1. gp140CF; HIV-1 gp140 Envs with CF deletions and as well deletions in the immunodominant (I) region in gp41 are termed gp140CFI (9), and HIV-1 gp140 Envs with the deletion of only the cleavage (C) are termed gp140C (Fig. 1A; see Table S1 in the supplemental material). All group M and subtype A, B, C, G, and AE01 consensus and wild-type HIV-1 gp140s were expressed as recombinant vaccinia viruses (rVVs) as described previously (9). All T/F HIV-1 gp140C Env genes were cloned into a mammalian expression plasmid pcDNA3.1/hygromycin (Invitrogen, Grand Island, NY). Recombinant Env glycoproteins were purified using Galanthus nivalis lectin-agarose (Vector Laboratories, Burlingame, CA) column chromatography from supernatants of 293T cell cultures either infected with rVVs or transfected with the HIV-1 gp140C-expressing pcDNA3.1 plasmids and stored at −80°C until use.
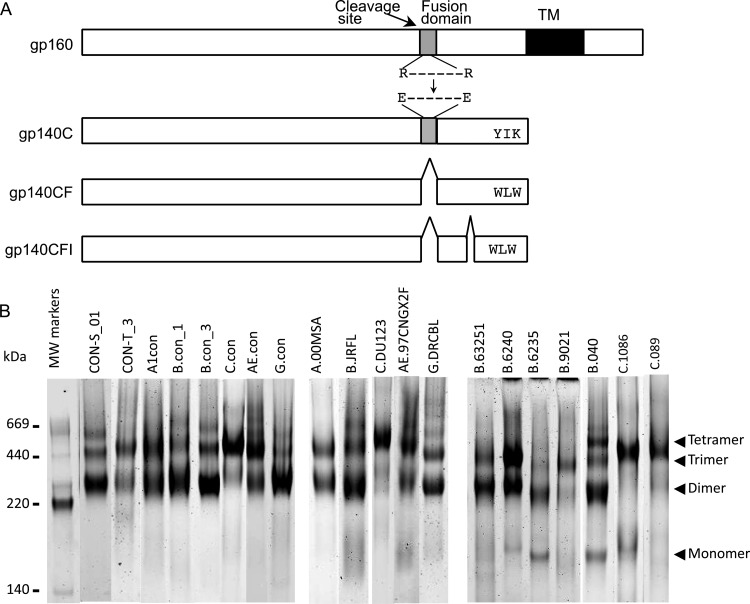
HIV-1 Env designs used in the study. (A) Schematic representation of HIV-1 gp160, as well as recombinant HIV-1 gp140C with a deletion of the cleavage site (C) in the junction of gp120 and gp41 by the mutation of two Arg residues to Glu residues as indicated; gp140CF with deletion of the cleavage site and the fusion domain (CF); and gp140 CFI with deletions of the cleavage site, fusion domain, and immunodominant domain (CFI). (B) Blue native PAGE analysis of HIV-1 gp140 Envs. Names of individual Env proteins are shown on the top of the lane. Arrowheads indicate the monomeric, dimeric, trimeric, and tetrameric forms of gp140.
The gel patterns of HIV-1 Envs proteins analyzed in blue native gels are reproducible. For CON-S gp140, we have produced more than 50 individual batches of proteins, and CON-S gp140 has always had the same gel pattern with predominant dimers and trimers, while clade C Envs, such as 1086C, DU123, and C.con, from different batches consistently migrate as trimers in blue native gels.
MAbs and soluble CD4.
Monoclonal antibody (MAb) b12 was produced by Quality Biological, Inc. (Gaithersburg, MD). MAbs 2G12, 2F5, and 4E10 were produced by Polymun Scientific (Vienna, Austria). MAbs PG9 and PG16 were gifts from D. Burton (Scripps Research Institute, La Jolla, CA); MAbs VRC01, VRC02, and VRC03 were gifts from J. Mascola (National Institutes of Health [NIH], Vaccine Research Center, Bethesda, MD); and MAbs to the C1 region of HIV-1 Env (A32) (37) and the CCR5 bs (17b) (38) were kindly provided by James Robinson (Tulane Medical School, New Orleans, LA). MAbs to the V2 loop of HIV-1, 697D (39) and 2158, were kindly provided by Susan Zolla-Pazner. Soluble CD4 (sCD4) was obtained from the NIH AIDS Research and Reference Reagent Program (Bethesda, MD). T8 is a murine MAb that binds to the gp120 C1 region (a gift from P. Earl, NIH).
Blue native polyacrylamide gel electrophoresis (BN-PAGE) and SDS-PAGE analysis of recombinant HIV-1 Envs.
BN-PAGE analysis of recombinant gp140 Env proteins was carried out as described previously (9, 40–42). Briefly, purified HIV-1 Env glycoproteins were diluted in a buffer containing 50 mM MOPS (morpholinepropanesulfonic acid), 50 mM Tris-HCl (pH 7.7), 20% glycerol, and 0.05% Coomassie blue. Protein samples were loaded onto a 3 to 8% Tris-acetate NuPAGE gel (Invitrogen, Carlsbad, CA), and electrophoresis was carried out for 1.5 h at 150 V with 50 mM MOPS–50 mM Tris-HCl (pH 7.7)–0.03% Coomassie blue as the cathode running buffer and 50 mM MOPS–50 mM Tris-HCl (pH 7.7) as the anode buffer. To determine what forms of oligomers were recognized by select HIV-1 MAbs, after BN-PAGE analysis and transfer onto nitrocellulose filters, HIV-1 gp140 proteins were probed with MAbs 2F5, PG9, CH31, and 17B and detected by with goat anti-human whole IgG specific (heavy- and light-chain)-AP (1:3,000 dilution) (Sigma, St. Louis, MO). The immunoblots were developed with Western-blue substrate (Promega, Madison, WI). HIV-1 gp140 proteins were also analyzed in SDS-PAGE under nonreducing and reducing conditions to ensure no major degradations of the HIV-1 gp140 preparations used in the study.
ELISA.
The reactivities of HIV-1 Envs with select HIV-1 MAbs and serum antibodies of immunized guinea pigs with autologous HIV-1 Env and select HIV-1 V3 peptides were tested in ELISA as described previously (43). To probe the antibody specificity induced by HIV-1 Env proteins, sCD4 protein, MAbs 2F5, PG9, CH31, and 17B were biotinylated and used as targets. Blocking the binding of biotinylated sCD4 protein and MAbs to HIV-1 Envs by immunized guinea pigs serum antibodies was also determined by ELISA. Briefly, purified HIV-1 Env proteins (1 μg/ml) in coating buffer (0.1 M NaHCO3 [pH 9.6]) were used to coat 384-well high binding ELISA plates at 15 μl/well at 4°C overnight. The plates were washed with superwash (1% Tween 20, phosphate-buffered saline [PBS]) and blocked with 40 μl of superblock (4% wheat protein, 1.5% goat serum, 0.5% Tween 20, and 0.05% sodium azide in PBS)/well at room temperature for 1 h. The blocked plates were washed with superwash and then incubated with 10 μl/well antibody in superblock at room temperature for 1.5 h. All MAbs were tested in serial dilution with a typical starting concentration of 50 μg/ml and no less than 20 μg/ml. After incubation, these plates were washed twice with PBS and then incubated with horseradish peroxidase (HRP)-conjugated goat anti-human IgG (1:5,000 dilution in 5% goat serum-PBS) at room temperature for 1 h. These plates were then washed four times with PBS and developed with 30 μl of TMB substrate (SureBlue Reserve, Gaithersburg, MD)/well. The HRP reaction was stopped with 30 μl of 1 M HCl/well, and the well optical density was determined at 450 nm. To determine antibody titers, pre- and postimmunization serum samples of guinea pigs were tested in 3-fold serial dilutions (from 1:30 to 1:5,314,410). Half-maximal effective concentrations (EC50) of antibody binding titers were determined as described previously (43).
SPR.
Surface plasmon resonance (SPR) assays were performed on a BIAcore 3000 instrument and analyzed with BIAevaluation 3.0 software (BIAcore, Inc., Uppsala, Sweden) as described previously (9, 44). Briefly, anti-human IgG Fc antibody (Sigma Chemicals) was immobilized on a CM5 sensor chip to about 12,000 to 14,000 response units (RU), and each antibody was then captured to about 1,000 to 2,000 RU on individual flow cells of the same sensor chip. Env proteins were injected in the solution phase at 100 μg/ml for 1 min over each of the flow cells. Nonspecific binding of Env proteins to the captured control Synagis (anti-RSV) MAb was subtracted from each binding curve (9, 44).
Animals and immunizations.
Outbred guinea pigs were purchased from Jackson Laboratory and housed in the vivarium at Duke University Animal Research Facility. Animals were studied under a protocol approved by the Animal Use and Care Committee of Duke University. Guinea pigs (four animals per immunogen group) were immunized intramuscularly on day 0 and weeks 3, 6, 9, and 12 with 100 μg/injection of recombinant HIV-1 Env gp140 adjuvanted with type B oCpG in Emulsigen (MVP Technologies, Omaha, NE). Serum samples were collected 10 days after each immunization and stored at −30°C until use.
Neutralization assays.
Serum neutralizing antibody levels were measured against a panel of pseudotyped HIV-1 viruses containing Envs from subtype B (B.QH0692, B.SF162, B.SS1196, B.BaL, B.BG1168, B.3988 B.WITO [from acute HIV-1 infection], and B.6101 [from acute HIV-1 infection]), subtype C (C.TV-1, C.DU123, C.DU172, C.92ZM233M, C.92BR025, C.92ZM197M, C.96ZM651, C.DU151, C.97ZA012, C.DU422, C.DU156, C.02ZM233M, and C.02ZM197M), subtype A (A.92RW010, A.92UG37, A.93TH976.01, A.Q23, A.Q168, A.Q259, A.Q461, A.Q769, and A.Q842), and a single circulating recombinant form CRF01_AE Env, AE_93TH976 (45). Neutralization assays were based on reductions in luciferase (Luc) reporter gene expression after a single round of virus infection with pseudotyped HIV-1 viruses in TZM-bl cells (25, 46–48). For each assay, serum samples were collected prior to vaccination (prebleed) and postvaccination (postbleed) from guinea pigs and assayed against individual pseudotyped HIV-1 viruses and also tested against murine leukemia virus (MuLV) as a negative control.
We performed parallel analyses of neutralization data using two categorizations of neutralization data. The first was a traditional strategy, typical of what we and others have used in the past. Here, we considered a neutralization value positive only if the neutralization titer was 3-fold over both the prebleed level and the level seen with the MuLV control, and the value of the experimental neutralization titer was ≥30. Samples that did not meet these criteria were considered equivalent to below the threshold of detection (<20). We refer to this as the “traditional approach.” When applying this method, we noted that we were frequently disregarding low levels of postbleed activity that were reproducibly above background in all four animals in a group, but not 3-fold above background, and it is not known whether such low-level responses may be useful in a vaccine context. To have greater confidence in measures of low-level activity, for every guinea pig that had a higher than background prevaccination spurious NAb activity in its sera, IgG was purified from both pre- and postbleed, and purified IgG was used instead of sera from those animals to test the NAb response to the immunogen. The IgG levels were brought to a concentration equivalent to the IgG level in serum, so the titers were equivalent. A high prebleed background was essentially removed by this step. Thus, we also analyzed the data considering all 50% infective doses (ID50) and values above the background, including the IgG purification step for all animals with high prebleed values. We refer to this as the “inclusive approach.” Using this approach, low-level responses against the panel of Envs were more reproducible within a group of animals that received the same immunogen than when using the traditional approach (see Results). We included both the traditional approach, because it is the way such data have been analyzed in the past, and the inclusive approach, because the results were more consistent between animals and the increased sensitivity might be relevant to human studies, in parallel analyses presented throughout the present study. In our inclusive analysis, values that were recorded as “20” and so were below the threshold of detection were replaced with “10,” as were any values less than the controls. Left censored data points were replaced with their lower limits (i.e., 540 and 43,740, respectively). Our “traditional analysis” was similar except that any postimmunization data point that was <3 times the preimmunization bleed was also replaced with a 10.
Analysis and statistical methods. (i) Hierarchical clustering.
Neutralization data were organized by hierarchical clustering so that similar patterns of NAb susceptibility among pseudovirions and similar patterns of NAb response among immunogens could be grouped and displayed using a heat map and dendrogram clustered according to distances between vectors of log ID50 NAb scores using the Los Alamos HIV database tool heat map (http://www.hiv.lanl.gov/content/sequence/HEATMAP/heatmap.html). Euclidean distances and the average clustering method were used to create the dendrograms. This tool uses the statistical package R (49).
(ii) Nonparametric statistics used to compare distributions of values among consensus, T/F, and chronic immunogen sets.
The analysis was conducted using a Kruskal-Wallis test for omnibus effects and Wilcoxon tests for pairwise effects. We used these nonparametric methods to obviate any potential violations of normality assumptions. The z approximation was used to determine significance when there were ties in the data. These standard tests were performed using either the R package (49) or SAS v9.2. Benjamini-Hochberg (50) false discovery rates (FDR) to adjust for multiple tests were computed using PROC MULTTEST in SAS v9.2.
(iii) GLM analysis.
Because the data were heavily censored, they did not have a log-normal distribution but were markedly skewed with a bolus of points recorded as <20 or below the threshold of detection. (There were 3,015 tests performed, and 1,209 of 3,015 (40%) were recorded as <20; a few other measurements lay above the threshold of the assay, with 36 of 3,015 recorded as >540 [1.2%] and 5 of 3,015 recorded as >43,740 [0.17%]). We compared a Gaussian model, an inverse Gaussian model, and several models designed to address censored data that are discussed below. All modeling and model comparisons were done using the R package (49). For these data, using both the conserved and inclusive cutoffs, the inverse Gaussian model was significantly better by Akaike's information criterion; thus, it was used for the generalized linear model (GLM) comparison. The two approaches we tested designed to handle censoring were the “lmec” (linear-mixed-effects model for left censored data implemented in the R lmec package) and the MCMCglmm algorithm (for fitting generalized linear mixed effects models for left, right, or interval censored data in the R MCMCglmm package). The results obtained in each censored data model were consistent with those obtained in the normal fit and the inverse Gaussian mode, but the latter provided a better fit.
For the model, let yi be the log10 titer observed for guinea pig i. Also, let the regression coefficient βk,0 represent the effect of the kth immunogen relative to a specified reference immunogen, where βk,0 = 0 represents no difference between the kth immunogen and the reference immunogen, βk,0 > 0 indicates that a stronger immune response was elicited by the kth immunogen relative to the reference, and βk,0 < 0 indicates a weaker elicited response. Finally, let Gi be an intercept, which augments the immunogen effect for a particular guinea pig. The model of the mean log10 titer response is: E[yi] = β0 + β1,0 Immunogen1 +[r] β2,0 Immunogen2 + … +[r] β19,0 Immunogen19 + Gi, where Immunogenk is an indicator variable, so that Immunogenk = 1 if guinea pig i received the reference immunogen and is zero otherwise. Further, Immunogenk = 0 for all k if guinea pig i received the reference immunogen. In the following development, we consider models whose reference immunogens were those that produced the strongest responses. CON-S gave the highest overall levels of response, so all other vaccines are measured relative to CON-S.
(iv) Model-fitting using the censored data strategy that estimated the probability of a response.
Model fitting and figure preparation used (i) the statistics package R (49), supplemented with the packages GAMLSS (51), used to fit Gaussian models to the censored titer data, (ii) multcomp (52), used to make multiple comparisons, and (iii) the suite of utilities called BSagri (53).
RESULTS
Production of HIV-1 Env gp140 oligomers.
To examine and compare the antigenicity of consensus, chronic, and T/F HIV-1 gp140 Envs, the HIV-1 gp140s in Table S1 in the supplemental material were expressed and purified. Purified HIV-1 Env gp140C oligomeric proteins migrated in blue native gels as dimers, trimers, and tetramers (Fig. 1B) with no major degradation in SDS-PAGE under reducing and nonreducing conditions (see Fig. S1 in the supplemental material). In contrast, subtype C HIV-1 gp140 Envs of consensus, chronic, and T/F Envs migrated predominantly as trimers (Fig. 1B). The glycosylation status of several HIV-1 Env proteins included here has been studied in detailed by mass spectrometry (40, 54–56). The carbohydrate composition of the same HIV-1 gp140 construct produced in 293T cells by either recombinant vaccinia virus or by transfection was analyzed, and we found essentially no change in the glycosylation between the two samples (E. Go, H.-X. Liao, B. Haynes, and H. Desaire, unpublished data).
Binding reactivity of HIV-1 MAbs to Env gp140 proteins.
The antigenicity of the three groups of HIV-1 gp140 Envs were tested and compared using ELISA (Table 1) and SPR (Table 2) for their ability to bind a panel of HIV-1 Env antibodies including CD4 binding site antibodies, b12, and VRC01, VRC02, and VRC03 from a related lineage; CCR5 coreceptor binding site (CCR5 bs) MAb 17b; glycan antibody 2G12; quaternary antibodies PG9 and PG16, a pair from a related lineage; and MPER MAbs 2F5 and 4E10. The relative binding affinity of each of the individual HIV-1 Envs to specific antibodies measured by ELISA (EC50, nM) and SPR (response unit) are summarized in Tables 1 and and2.2. Of note, consensus proteins are artificial constructs, a concatenation of the most common form of each amino acid in each position in an alignment. Thus, HIV-1 population consensus sequences contained combinations of amino acids that are not found in nature, and there is always a concern that they may not fold appropriately. Therefore, the consistency of binding of antibodies with discontinuous epitopes to the consensus sequences was reassuring. CON-S gp140 Env bound with the highest affinity among the consensus sequences, and the only MAb tested that did not bind to CON-S was VRC03 (Tables 1 and and2).2). VRC01 and VRC02 are derived from the same clonal lineage as VRC03 (57), and these two MAbs did bind to CON-S with high affinity. The only consensus Env that did not bind to most MAbs tested was A1.con.
Table 1
Binding reactivity of HIV-1 Envs to HIV-1-specific MAbs in ELISAa
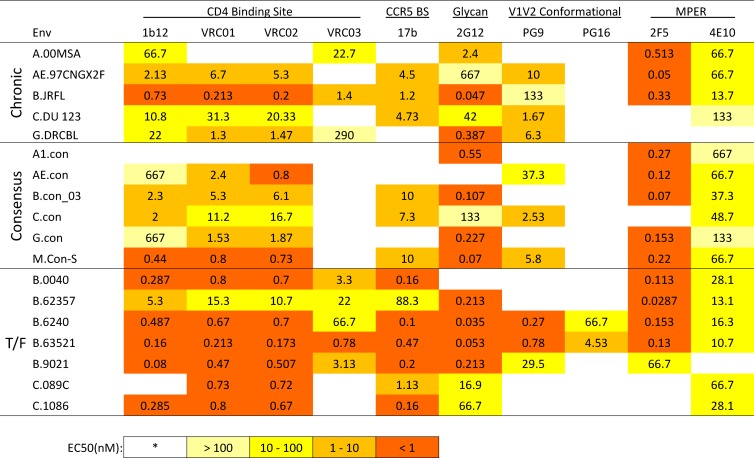
Table 2
Binding reactivity of HIV-1 Envs to neutralizing MAbs in SPR analysisa
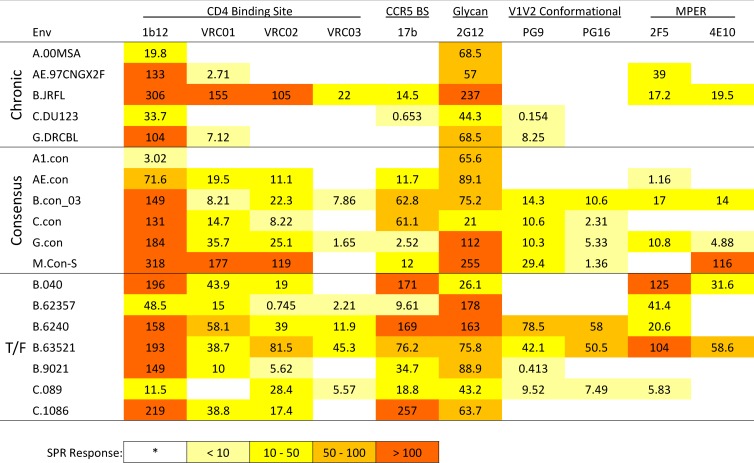
We next compared the overall binding scores of the consensus, chronic, and T/F Envs to the panel of MAbs in shown in Tables and and11 and and2.2. We first used the nonparametric Kruskal-Wallis omnibus test to determine whether, for a given antibody, one of the three groups was distinct from the others, followed by Wilcoxon pairwise comparisons. VRC01, VRC02, and VRC03 were all derived from the same clonal lineage, with VRC01 being most potent neutralizing MAb among them (57). PG9 and PG16 are similarly clonally related, with PG9 being the most potent (17). Thus, only VRC01 and PG9 were used in the statistical comparisons, to represent the two lineages and minimize the number of tests. Because multiple tests were performed, we performed an FDR comparison. The T/F binding tended to be higher in number of epitopes expressed, and the binding of MAb 17b was significantly different between the three groups; 17b had significantly higher binding affinity for T/F viruses (Tables 1 and and22 and see Table S2 in the supplemental material, false discovery adjusted P value = 0.02 for ELISA, 0.03 for SPR). 17b is a CD4-inducible antibody that binds near the CCR5-coreceptor binding site (38) and, interestingly, increased T/F binding affinity was observed even though sCD4 was not present in the binding assay. Similar reactivity patterns of MAbs 2F5, PG9, CH31, and 17b with HIV-1 Envs detected by ELISA and by SPR were also observed in BN-PAGE Western blot analysis. When these MAbs did bind to HIV-1 Envs, MAb 2F5 had the strongest reactivity and recognized all forms of gp140 oligomers, whereas the PG9, CH31, and 17B primarily recognized dimers, trimers, and oligomers but also recognized the gp140 monomer weakly or not at all (see Fig. S2 in the supplemental material).
Immunogenicity of T/F, chronic, and consensus Envs.
Next, the 20 gp140 Envs in Table S1 in the supplemental material were used to immunize guinea pigs. Most consensus gp140 sequences were based on the database from 2003; two B clade consensus Envs (B.con_01 from 2001 and B.con_03 from 2003) and two group M consensus Envs (CON-S from 2001 and CON-T from 2003) were included in the immunogenicity studies to ensure that we were not missing minor Env changes that might lead to induction of the breadth of responses. The immune responses elicited were indeed somewhat different when using immunogens from different years (Fig. 2). The T/F Envs were from 2005 to 2007 isolates (22). NAb responses to these HIV-1 Envs were evaluated using a panel of 36 Env-pseudotyped viruses from clades A, B, C, and CRF01_AE (see Table S3 in the supplemental material).
Neutralization activity of guinea pig groups immunized with HIV-1 Envs. Geometric means of IC50 neutralization titers (y axis) for groups of guinea pig that received the indicated HIV-1 Envs (grouped into consensus, T/F, and chronic families) were determined against a panel of subtype A, B, C, and E pseudoviruses using the traditional approach (A) or the inclusive approach (B). The same color codes were used for each subtype for both Env immunogens, as shown in the lower right portions of panels A and B. Pseudoviruses are ordered the same way in each panel, according to the key at the bottom, to facilitate comparisons between vaccine responses.
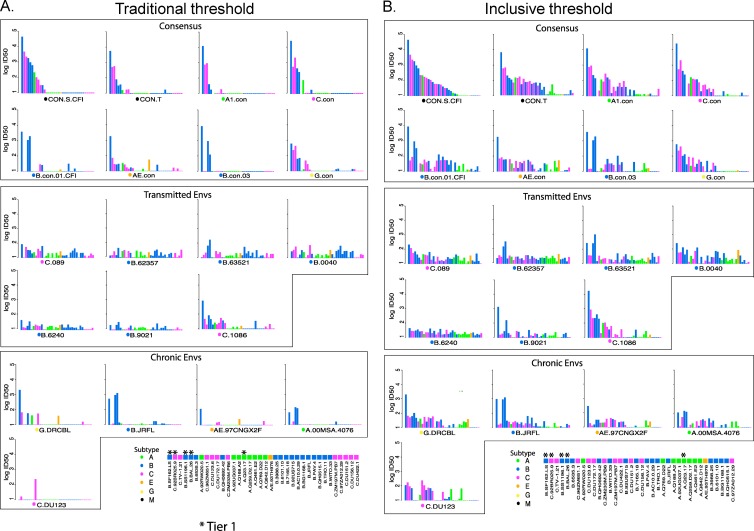
Bar plots illustrating the relative potency of ID50 NAb responses elicited by each immunogen against each of the pseudovirus Envs are shown in Fig. 2 and Fig. S1 in the supplemental material (Fig. 2 shows the geometric mean response for each immunized group of four animals to each Env; Fig. S3 in the supplemental material illustrates the same data, but the results for each animal are shown individually). As described in Materials and Methods, we analyzed the data using two strategies: a traditional approach (Fig. 2A), in which a positive neutralization value of immune guinea pig serum had to be at least three times higher than both the prebleed serum and the negative control virus MuLV-SVA to be considered positive, and an inclusive approach where IgG was purified to minimize background, and then any responses that were above both negative controls were considered positive (Fig. 2B). By definition, more reactions were deemed positive when using the inclusive method (Fig. 2A compared to Fig. 2B). Using both data interpretation strategies, a pattern is evident that is associated with each of the classes of immunogens. Consensus sequence immunogens tended to have the greatest responses to tier 1 Envs (45) and elicited a moderately high level of response to a subset tier 2 Envs. T/F immunogens gave low levels of response across most Envs tested, but with greater breadth than consensus immunogens. Chronic Env immunogens showed moderate to low-level responses to some tier 1 Envs and sporadic low responses to tier 2 Envs (45). Within-clade matched responses are indicated by the colors in Fig. 2. Little clade specificity was manifested in these data, although occasionally it is evident. For example, the B.con.03 responses favored B clade pseudoviruses. Of note, the B and C clade T/F viruses did not elicit markedly stronger responses within clades than between clades (Fig. 2).
Heat maps, organized by two-dimensional hierarchical clustering, further illustrate the over distinctive patterns between consensus T/F and chronic immunogens. The traditional threshold data heat map is shown in Fig. 3A, with the inclusive method of analysis in Fig. 3B. In these heat maps, pseudotyped Envs with similar susceptibility patterns are clustered together and, likewise, immunogens that elicited antibodies that had similar neutralizing susceptibility are clustered together, regardless of their classification as T/F, consensus, or chronic. Between three and seven animals per immunogen group were tested. The three patterns associated with chronic, T/F, and consensus immunogens are clearly discernible (Fig. 3). The chance that the animals who received the same type of immunogen would cluster according to reactivity pattern to the extent observed in the heat maps is very small if you consider each animal separately (P < 2 × 10−16, the Fisher exact test for a 3×3 table for the traditional method, and P = 0.0000002 for the inclusive method), and still low when animals given the same immunogen are considered as a group rather than single animals, and the group is assigned to the cluster that most of the animals in the group fall into (P = 0.003 for the conserved approach, P = 0.01 for the inclusive). Although the distinct patterns were statistically significant, there were a few exceptions to this overall pattern. The T/F founder virus, C.1086, behaved more like a consensus immunogen than the other T/F strains, in that C.1086 did not have great breadth, and it generates more intense tier 1 than tier 2 responses (for example, the C.1086-vaccinated animals have a NAb profile very similar to that of the G consensus-vaccinated animals). Also, the two subtype B consensus sequences from different years were subtype specific and elicited responses similar to the chronic B clade isolate B.JRFL.
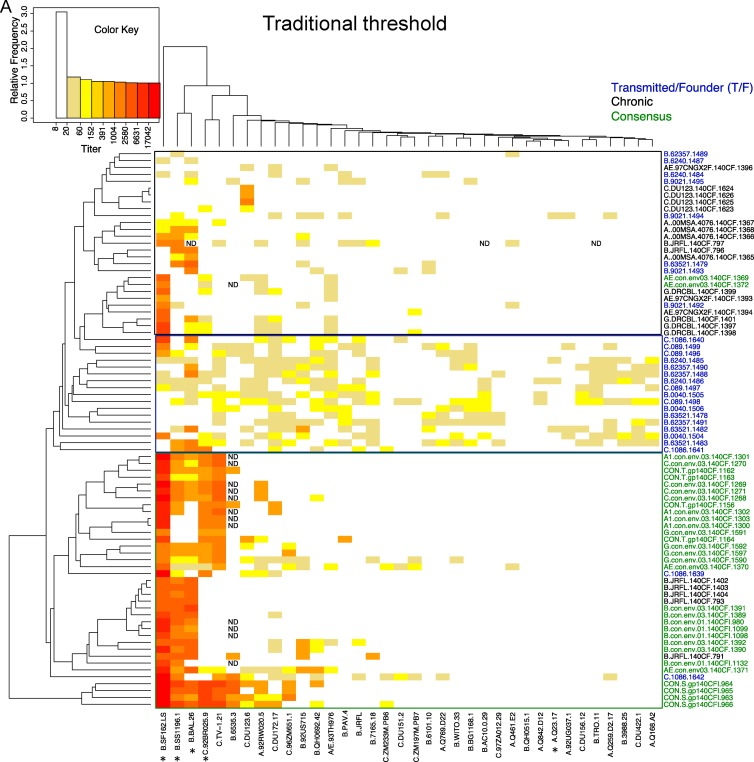
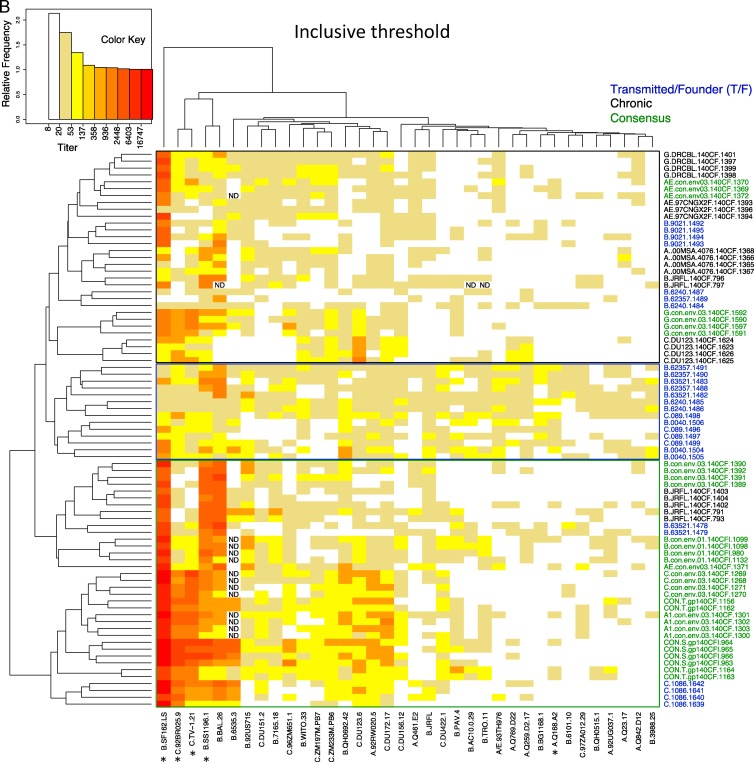
Heat maps reflecting HIV-1 neutralization activity of each individual guinea pigs immunized with HIV-1 gp140 Envs. The neutralizing activity of individual guinea pigs immunized with a particular HIV-1 Env is presented as a heat map, with stronger neutralizing activity indicated by color, as shown in the key at the upper left. Serum samples from individual guinea pigs are identified by a string beginning with the name of the HIV-1 Env used to immunize the animal, followed by a unique guinea pig ID number. The panel of 36 HIV-1 pseudoviruses is listed at the bottom of the heat map, arranged in order of decreasing susceptibility to neutralization. The neutralizing activity of individual guinea pig sera was analyzed using both the traditional approach (A) and the inclusive approach (B).
Hierarchical clustering strategy used to create the heat map brought the replicate animals in 12 of the 20 immunogen groups that received the same immunogen most closely together, when the inclusive strategy was used (see brackets in Fig. 3B). This compares with only 2 of 20 immunogen group clusters using the traditional method (see brackets in Fig. 3A) (P = 0.002, calculated using a Fisher exact test comparing inclusive versus traditional strategies). This reflects the greater consistency of the response data between animals that received the same immunogen when the inclusive cutoff was used and is an indication that the inclusion of low-level responses does not just add noise but rather captures consistent reproducible patterns in the Env immunogen-induced neutralizing responses.
Comparison of neutralizing breadth induced by T/F, chronic, and consensus Env immunogens.
For an initial comparison of neutralizing breadth responses induced by the three classes of Env immunogens, we counted the number of times an immunogen group of animals had at least two responding animals that had detectable neutralizing titer for each of the 36 Envs and compared the distribution of counts for groups of T/F, consensus, and chronic immunogens (Fig. 4A and andBB illustrate the breadth of Env responses among the 36 Env pseudovirions for each immunogen using the traditional and inclusive strategies, respectively). The omnibus Kruskal-Wallis test indicated that there was a significant difference among the distributions using the traditional approach (P = 0.0027), and T/F Envs ranked significantly higher than consensus Envs, which in turn were significantly higher than chronic Envs (all P values of each pairwise comparison are provided in Fig. 4). Using the less stringent inclusive strategy (Fig. 4B), many more low responses against a spectrum of Envs were observed within all groups, and so the groups were not statistically distinguishable, although there was a trend again suggesting that T/F Envs induced a greater breadth of response than did chronic Envs (P = 0.061).
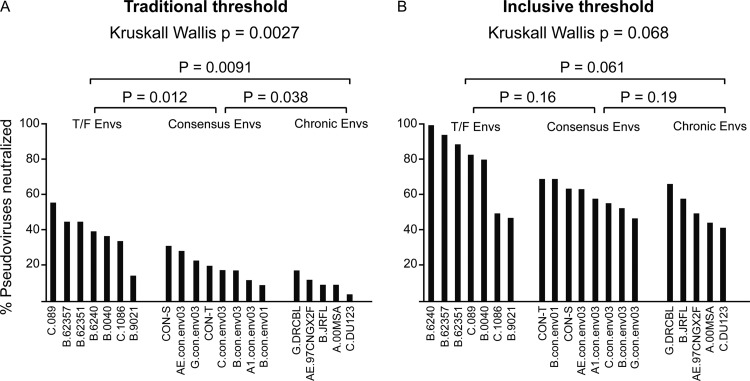
Comparison of breadth of antibody responses induced by HIV-1 T/F, consensus, and chronic Env immunogens. The breadth (the vertical axis shows the percentage of the panel of 36 pseudoviruses neutralized, averaged across all immunized animals) for each of the individual HIV-1 Env immunogens (x axis) using the inclusive approach (A) and the traditional approach (B). The immunogens were classed as T/F, consensus, or chronic Envs, and the P values are for comparisons between these classes.
Comparison of the magnitude of responses to single immunogens.
We initially used a generalized linear model to compare the potency of the neutralizing antibody responses using the different immunogens. The model selection is described in Materials and Methods; the model which fit the data the best was an inverse Gaussian model, and animal-to-animal immune system differences were accounted for by treating the interanimal variation as a random effect. The analysis requires as input both the neutralization titers (one for each measured combination of animal and isolate) and a separate list of responses that should be regarded as definitely positive, based on whether the traditional or the inclusive approach described above is used. The CON-S immunogen showed the highest overall levels of response using both traditional (Table 3) and inclusive (Table 4) threshold approaches and so was used as the reference strain, and all other immunogens were measured relative to CON-S. Using the traditional approach, CON-S Env induced significantly greater antibody responses than all other HIV-1 Env immunogens, and chronic responses, along with T/F B.9201, ranked the lowest (Table 3). Using the inclusive approach, CON-S again ranked the highest but was not statistically distinguishable from other consensus immunogens, including CON-T, A1-con, C-con, and B-con_01; the CON-S response was significantly higher than all T/F and chronic Env immunogens.
Table 3
Results of a model fit to the data with log10 neutralization titers using traditional strategya
| Vaccine | Estimate | SE | t | P |
|---|---|---|---|---|
| (Intercept)- CON-S | 1.5 | 0.082 | 19 | 9.5 × 10−75 |
| C.con | –0.21 | 0.11 | –2 | 0.047 |
| B.0040 | –0.23 | 0.11 | –2.1 | 0.034 |
| C.089 | –0.22 | 0.1 | –2.1 | 0.033 |
| C.1086 | –0.25 | 0.1 | –2.4 | 0.016 |
| CON-T | –0.27 | 0.1 | –2.6 | 0.0084 |
| B.63521 | –0.28 | 0.1 | –2.8 | 0.0058 |
| B.62357 | –0.29 | 0.1 | –2.8 | 0.0044 |
| B.con_01 | –0.3 | 0.1 | –2.9 | 0.0032 |
| A1.con | –0.3 | 0.1 | –3 | 0.0029 |
| AE.con | –0.3 | 0.1 | –3 | 0.0028 |
| G.con | –0.3 | 0.1 | –3 | 0.0027 |
| B.6240 | –0.35 | 0.099 | –3.6 | 0.00038 |
| B.con_03 | –0.37 | 0.099 | –3.7 | 0.00002 |
| G.DRCBL | –0.38 | 0.098 | –3.8 | 0.00014 |
| B.JRFL | –0.35 | 0.092 | –3.8 | 0.00013 |
| B.9021 | –0.43 | 0.096 | –4.5 | 0.0000062 |
| A.00MSA | –0.44 | 0.096 | –4.6 | 0.0000040 |
| AE.97CNGX2F | –0.48 | 0.098 | –4.8 | 0.0000014 |
| C.DU123 | –0.5 | 0.094 | –5.3 | 0.00000012 |
Table 4
Results of a model fit to the data with log10 neutralization titers using inclusive strategya
| Vaccine | Estimate | SE | t | P |
|---|---|---|---|---|
| (Intercept)- CON-S | 1.8 | 0.11 | 17 | 1.2 × 10−61 |
| CON-T | –0.13 | 0.14 | –0.92 | 0.36 |
| A1.con | –0.17 | 0.14 | –1.2 | 0.24 |
| C.con | –0.21 | 0.14 | –1.5 | 0.13 |
| B.con_01 | –0.22 | 0.14 | –1.6 | 0.12 |
| B.0040 | –0.3 | 0.14 | –2.2 | 0.031 |
| C.1086 | –0.29 | 0.13 | –2.2 | 0.029 |
| B.63521 | –0.33 | 0.13 | –2.5 | 0.012 |
| C.089 | –0.34 | 0.13 | –2.6 | 0.009 |
| AE.con | –0.35 | 0.13 | –2.7 | 0.0073 |
| B.62357 | –0.36 | 0.13 | –2.8 | 0.0051 |
| G.DRCBL | –0.39 | 0.13 | –3 | 0.0024 |
| B.con_03 | –0.4 | 0.13 | –3.1 | 0.0018 |
| G.con | –0.4 | 0.13 | –3.1 | 0.0017 |
| B.JRFL | –0.41 | 0.12 | –3.4 | 0.00063 |
| B.6240 | –0.44 | 0.13 | –3.5 | 0.00052 |
| C.DU123 | –0.44 | 0.13 | –3.5 | 0.00045 |
| A.00MSA | –0.52 | 0.12 | –4.3 | 0.000022 |
| AE.97CNGX2F | –0.55 | 0.13 | –4.3 | 0.000017 |
| B.9021 | –0.55 | 0.12 | –4.5 | 0.0000081 |
Evaluation of neutralizing breadth versus strength of responses induced by consensus, T/F, and chronic Env immunogens.
To reexamine the question of breadth and depth in a probabilistic framework, given our uncertainty about near-threshold responses, we developed a new class of models designed to separate the breadth of the NAb response—i.e., the number and variety of strains to which an immunogen induces a response—from the strength of that response—i.e., the raw neutralizing power of the sera from those animals in which the immunogen induces a positive response. This model also incorporated a self-consistent estimation process for the censored data. The resulting model has two aspects: a binomial regression that produces a probability of positive response for each combination of immunogen and isolate and then, again for each immunogen-isolate combination, a pair of linear regression models—one for the positive responses and another for the negative ones—that predict the logs of the neutralization titer.
The novelty of the approach is that we require internal consistency between the two aspects of the model: if some immunogen-isolate combinations produce only modestly strong positive responses, then it is possible that some near-threshold responses, initially classified as negative, may contribute to a better-fitting model if they are reclassified as weak, but positive responses. After an initial round of parameter estimation, we assign a suitable positive weight to such near-threshold responses and fold them back into the analysis, using the EM-algorithm (58) to repeatedly fit parameters and reweight the data until we obtain a self-consistent model.
As before, we fit models based on the two different positivity criteria, the conserved and inclusive strategies. Using this model, the conserved data fit better (in the sense of having the higher log-likelihood), and so the parameters of conserved model are illustrated in Fig. 5 and and6.6. Figure 5 shows the fitted probability of a positive response as a function of immunogen-isolate combination. Using this measure and modeling the probability of a response being positive (in contrast to our previous measure of breadth in Fig. 4), the broadest neutralizing responses were induced by the consensus Env AE.con, followed closely by the immunogens derived from the T/F strains B.63521, B.0040, and C.089. The reason underlying this reordering compared to Fig. 4 can be understood by looking closely at the data in Fig. S3 in the supplemental material; while AE.con has fewer detected responses, many of the responses to the T/F immunogens are borderline, and when AE.con does have a response, it tends to be a higher value and so is more likely to be rescored as positive in this probabilistic model. Three of the four immunogens with the highest probability of response to the most Envs are T/F viruses, and these three consistently yield great breadth by both measures (Fig. 4 and and55).
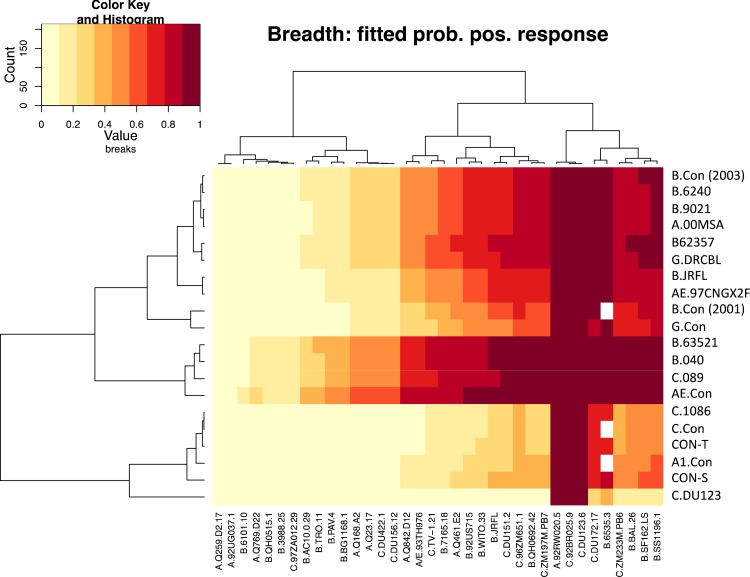
Heat map reflecting breadth of antibody responses induced by HIV-1 consensus, chronic, and T/F Env immunogens. Heat map showing the fitted probability that a guinea pig in each of the individual immunization groups (rows) would have shown neutralizing activity against the panel of 36 HIV-1 pseudoviruses (columns). A higher probability of a positive response (traditional criterion) is indicated by a darker color, as detailed in the key at the upper left.
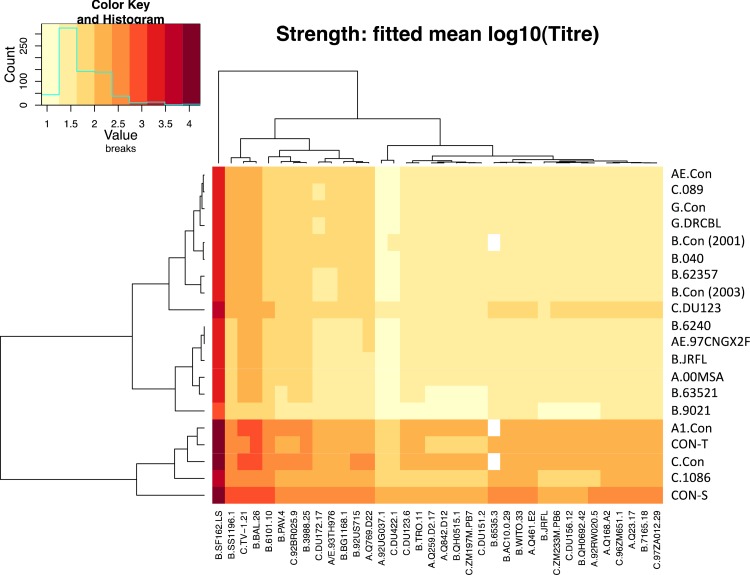
Heat map showing the fitted strength (predicted log10 titer) of positive responses induced by HIV-1 immunogens. As in Fig. 5, rows correspond to immunogens, whereas columns are labeled by the pseudovirus strain. The key at the upper left shows both the color scale (with the strongest predicted responses corresponding to titers of 10,000, as seen for some tier 1 Envs) and the distribution of fitted values: many lie near log10(20) ≈ 1.3, indicating that some responses near the threshold of the assay may be weak but positive.
The strengths of positive responses induced by different Envs were also compared using this model and were consistent with the findings reported above based on an inverse Gaussian GLM. The consensus Envs as a class were by far the most effective at eliciting potent responses (Fig. 6). If we again make suitably adjusted multiple comparisons between the two best immunogens from each category, the best consensus Env, CON.S, produced significantly stronger responses than any of the others (P = 0.0233 for C.con, the second best consensus, while P < 0.001 for all T/F and chronic Envs).
Serological analysis of serum antibodies induced by HIV-1 Envs.
To determine whether differences in serum binding antibodies induced by individual HIV-1 Env immunogens could contribute to differences observed in neutralizing antibody responses, we evaluated pre- and postimmunization sera in multiple epitope-binding ELISAs. We found chronic and consensus Envs induced comparable binding antibody responses to autologous Envs (Fig. 7A), while significantly lower binding antibody titers were induced by T/F Envs (P = 0.005 compared to chronic Envs and P = 0.001 compared to consensus Envs [two-sided Wilcoxon rank sum test]). Next, we probed the serum antibody specificities by serum inhibition assays of biotinylated MAbs or sCD4 using the MAbs 1b12, PG9, and 2G12. Serum antibodies blocked sCD4 and MAb 1b12 binding to HIV-1 subtype B.JRFL gp140 and MAb PG9 binding to AE.A244 gp120 (Fig. 7B to toD),D), while no induced 2G12 serum blocking antibodies were detected (Fig. 7E). Thus, CD4bs and V2 region antibodies contribute to the immune responses in consensus, chronic, and T/F immunizations. Serum antibodies from animals immunized with T/F Envs tended to have the highest inhibition of sCD4 and 1b12 binding to JRFL gp140, followed by consensus Envs and then chronic Envs (Fig. 7B and andC),C), but the distinction between classes of immunogen was not significant. In contrast, chronic and consensus Envs induced antibodies that had significantly higher inhibition of MAb PG9 binding to AE.A244 gp120 than antibodies induced by T/F Envs (P = 0.03 and P = 0.017, respectively) (Fig. 7D).

Levels and specificities of serum antibodies of immunized guinea pigs to HIV-1 Envs. Serum samples of immunized guinea pigs grouped by immunogens of consensus (red), chronic (blue), and T/F (green) were tested in ELISA for binding to the autologous Env in serial 1:3 dilutions (1:30 to 1:5,314,410) to compare the levels of antibody responses for the different classes of immunogens. Prebleed serum was used as a negative control and had negligible binding to HIV-1 Env. Serum binding antibody titers were expressed as the EC50 (reciprocal serum dilutions) on the y axis (A). The open circles in each column represent the EC50 for each member of a group of animals immunized with the same immunogen; the filled circles represent the geometric mean of the group. This geometric mean score was used to represent each of the immunogens and to statistically compare the classes of vaccines. The median value of the geometric means is indicated by the horizontal bar. Vaccine classes (consensus, chronic, and T/F) were compared by using a Kruskal-Wallis omnibus test; if it was significant (P = 0.003), pairwise comparisons were then made between the vaccine classes, and consensus and chronic vaccines were shown to show higher-magnitude autologous responses than T/F vaccines. The immune sera were also tested for blocking the binding of sCD4 to HIV-1 Env JRFL gp140 (B), MAbs 1b12 to HIV-1 Env JRFL (C), PG9 to HIV-1 Env A244 (D), and 2G12 to HIV-1 Env JRFL (E) at a 1:50 serum dilution. The percentage of inhibition of the binding by guinea pig sera of each group was plotted and statistically compared in a in a manner analogous to the strategy described in panel A, except that for panels B to E, the filled circles represent the mean value for each vaccine group and not the geometric mean. Among the inhibition assays (B-E), only the inhibition of PG9 binding was statistically significant (D), and the T/F vaccines had lower levels of PG9 inhibition than either consensus or chronic.
We detected V3 peptide binding antibodies in all immunized guinea pig groups with robust reactivity to consensus subtypes B and C and group M CON-S V3 peptides (see Fig. S4 and S5 in the supplemental material). There was a within-clade preference in the V3 responses, such that vaccines using an Env of a particular clade tended to react most intensely with the consensus peptide of that same clade, most likely due to higher degrees of sequence similarity (see Fig. S4 and S5A to C in the supplemental material). Consensus Envs induced the highest measured levels of V3 antibodies relative to chronic or T/F Envs (Wilcoxon P = 0.048 and P = 0.037, respectively) (see Fig. S5D in the supplemental material), but consensus peptides were used to assess V3 reactivity, so the consensus immunogens were either a perfect match or a closer match to the test peptides than were natural strain immunogens, which each had strain-specific differences in V3 (see Fig. S4 in the supplemental material). The V3 regions were quite diverse among the T/F and chronic immunogen sequences (see Fig. S4 in the supplemental material). Thus, we could not resolve whether the higher V3 peptide antibody responses observed among the consensus immunized animals were due to a more intense V3 antibody response or simply the result of greater similarity with the V3 peptides used for testing. We had previously observed that the CON-S neutralization of tier 1 Envs could be blocked by CON-S V3 peptides (9). Consistent with this, the tier 1 neutralization potency of consensus immunogens was highly correlated within-clade V3 peptide binding (see Fig. S5E and F); i.e., the responses to the V3 peptide based on the B.con sequence were highly correlated with neutralization of the tier 1 virus SF162 (P = 0.008), and the responses to the V3 peptide based on the C.con sequence were highly correlated with neutralization of the tier 1 virus C.BR025 (P = 0.008) (see Fig. S5D and E in the supplemental material). Neutralization of tier 2 viruses was not correlated V3 peptide reactivity.
DISCUSSION
In this study, we produced a panel of 20 recombinant T/F, chronic, and consensus gp140 Envs derived from 293T cells either by transfection or recombinant vaccinia virus and then compared them as Env immunogen candidates by antigenicity and immunogenicity studies. First, we found that T/F, chronic, and consensus Envs bound comparably to a panel of Env antibodies, with the exception of increased CCR5 binding site antibody 17b to T/F viruses. The CCR5 binding site region has been identified as a focus of Env signature mutations associated with developing bNAbs during chronic infection (59), and CD4-independent exposure of this region is related to the intrinsic reactivity of an Env protein (60), suggesting that epitope exposure of this region may be an important aspect of the bNAb potential of an immunogen. Second, we found that of the Envs, the best inducer of potency of the NAb responses was the group M consensus Env, CON-S, which had the highest magnitude of response to tier 1 and a subset of tier 2 pseudotyped virions (45). In contrast, the T/F recombinant Envs consistently outperformed both chronic and consensus Envs in terms of the induction of antibodies with neutralization breadth across all Envs tested, although these tended to be low level, even against tier 1 viruses. One might expect the results with chronic and consensus Envs to be the same since the consensus Env sequences are primarily derived from chronic-Env sequences. However, because the consensus sequences are central, they are likely to capture common recurring aspects of the virus. Thus, while chronic infections after exposure to autologous NAbs may regularly accrue characteristics that tend to make them more resistant, such changes may also have fitness costs that are selected against at transmission. Particularly if there are multiple pathways to such chronic escape phenotypes, a population consensus would not necessarily reflect them. Thus far, we have only tested five B and two C clade T/F immunogens, and whether the observed patterns will hold for T/F viruses of other clades remains to be determined. An alternative to T/F Envs being more broadly immunogenic than chronic Envs is that there may be clade-specific effects, and the single B Env in the chronic HIV-1 group may not have been comparable to the majority of clade B Envs in the T/F group.
The recombinant HIV-1 Envs in the present study all induced antibodies that had high binding antibody titers to the autologous Env and to V3 peptides and were capable of blocking the binding of sCD4 and MAbs 1b12 and PG9 to HIV-1 Env proteins but were not able to block MAb 2G12 binding to JRFL gp140. Despite the complexity of the polyclonal immune responses, anti-V3 peptide responses correlate strongly with potent tier 1 neutralization.
We recently demonstrated that the Thai RV144 HIV-1 vaccine that resulted in an estimated vaccine efficacy of 31% in a phase III trial correlated directly with antibodies to HIV-1 envelope variable regions 1 and 2 (V1 and V2) (61, 62). We have also recently demonstrated that MAbs CH58, CH59, HG107, and HG120 isolated from RV144 vaccinees share the same V2 epitope region with the broad neutralizing antibody PG9, have neutralizing activity against tier 1 isolates, and mediate the killing of field isolate HIV-1-infected CD4+ T cells (63). PG9 bNAb binds both to amino acids K168 and K169 (positions 164 and 172 in HXB2 numbering) of the V2 region and binds to glycans at N156 and N160 in that same region (64). The inhibitory binding of PG9 binding to Env by our guinea pig sera most likely reflects the induction of V2 antibodies targeted at or near this region. This region contributes to an epitope recognized by V2 anti-peptide antibodies that are frequently induced with Env immunization as well as by PG9 bNAb, but the PG9 inhibition we observed in this study does not necessarily reflect the induction of anti-glycan PG9-like bNAbs.
It is well known that the antigenicity of an Env does not predict its capacity to induce an immune response to an expressed epitope. Nonetheless, we reasoned it is important for candidate Env immunogens to be as antigenic as possible in terms of bNAb binding. Thus, of the 18 T/F, chronic, and consensus Envs, we found that B.63521 and B.6240 Env gp140s expressed all of the Env epitopes studied. In addition, despite being artificial constructs, several of the consensus Envs also expressed each of these epitopes, in particular B.con_03 and G.con. CON-S had particularly high levels of reactivity to most of the panel of MAbs, comparable to the best T/F strains (Tables 1 and and2).2). These epitopes included the epitopes recognized by the bNAbs 2F5 (65), 4E10 (66), PG9, PG16, and VRC01 (57) and the glycan bNAb, 2G12 (67), in addition to the PG9 and PG16 (17) V2V3 conformational epitope bNAbs (64), as well as CH01-CH04 MAbs against the same V1/V2 bNAb epitope (68, 69). Thus, choosing candidate recombinant Env immunogens that react with V1/V2 conformational epitope bNAbs, as well as other bNAbs, seems prudent. In this regard, the C.1086 gp120 and a chronic C.TV-1 gp120 Env pair have been selected for production as a bivalent clade C gp120 boost for a poxvirus prime-gp120 protein boost clinical trial. The antigenicity of these two gp120 Envs are complementary with C.1086, binding better to sCD4, 17b (CCR5 bs), A32 (C1, ADCC), VRC01 (CD4bs), 19b (V3), and various V2 MAbs (e.g., 2158), whereas C.TV-1 gp120 binds better to other V2 antibodies (830A, 697-D), and in particular to PG9 and PG16 V1V2 bNAbs (S. A. Alam, H.-X. Liao, S. Barnett, and B. F. Haynes, unpublished data). CON-S Env will be tested in a phase I human trial, and its immunogenicity in terms of NAb responses in humans will be explored. In a recent immunogenicity test in macaques of the vaccine constructs going into phase I human trials, CON-S was shown to be highly immunogenic and to elicit potent tier 1 and some moderate tier 2 NAb responses, in good accord with the results of the comparative vaccination study in guinea pigs presented here (B. F. Haynes, H.-X. Liao, B. Korber, S. Santra, and N. Letvin, unpublished data).
The findings we present here are based on using gp140 Envs that may have incremental immunological advantages over gp120 (2, 34, 70), although gp120 is easier to produce and so historically has been used in human trials (62, 71, 72). In several studies, stable soluble trimeric HIV-1 Envs have elicited more potent neutralizing antibodies compared to gp120s, but still of limited breadth (70, 73). Stable, rigorously homogeneous trimers that have antigenic properties markedly different from those of monomeric gp120 induced more potent neutralizing antibodies than those elicited by the corresponding gp120 monomers; however, antibodies induced by such stable trimers also have very limited neutralizing potency and breadth (73, 74). It remains to be definitively determined whether gp140 Env constructs are indeed superior and worth the production cost for use in clinical trials.
The key criterion for Env immunogen selection should be Env immunogenicity. We found three distinct patterns of Env immunogenicity in isolates from acute infection, chronic infection, and in silico-designed consensus Envs. Although these patterns were statistically significant, our sample size for each group was limited, and the clade compositions of the groups were biased because of reagent availability when the study was initiated. Further studies to confirm these findings are warranted. Chronic Envs gave the poorest tier 1 and only weak sporadic tier 2 neutralization responses. Consensus Envs had a pattern of high NAb titers against tier 1 Envs, and moderately high titer responses (higher than T/F immunogens) to a subset of tier 2 Envs. The group M CON-S Env is the optimal Env tested among the consensus group and yielded the highest responses overall. In contrast, the T/F Envs induced tier 1 responses of lower magnitude, but which consistently had greater neutralization breadth, inducing low titer neutralizing responses to most tier 2 HIV-1 strains. The levels of antibody needed for protection in the SHIV/macaque model were initially thought to be quite high and difficult to achieve in vivo (100-fold over the in vitro 50% infectious concentration [IC50] levels) (75–78). However, recently lower levels of neutralizing antibodies were shown to be needed for protection against mucosal challenge in this model (79), and very low levels of neutralizing antibodies have been shown in humans in vivo to induce autologous T/F virus escape mutants (22), providing hope that low levels of NAbs may be able to prevent HIV-1 acquisition at mucosal sites. The distinctive patterns suggest different epitopes may be targeted in responses to consensus and T/F immunogens, and both classes of response may be beneficial in different ways in a vaccine setting. If the NAb effects were additive, as some studies suggest (69, 80), and both types of response can be triggered simultaneously, a polyvalent combination of complementary immunogens may prove a useful vaccine strategy. Analysis of these and similar data sets should be useful both for the choice of individual candidate Env immunogens and for identifying the most promising polyvalent Env mixtures for immunogenicity testing in nonhuman primates and humans.
ACKNOWLEDGMENTS
This study was supported by grant U19AI067854 from the Center for HIV/AIDS Vaccine Immunology, by grant UM1AI100645 from the Center for HIV/AIDS Vaccine Immunology-Immunogen Discovery of the Division of AIDS, NIAID, NIH, and by Vaccine Discovery Center of the Collaboration for AIDS Vaccine Development Program grant OPP1033098s from the Bill and Melinda Gates Foundation.
Footnotes
Published ahead of print 30 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02297-12.
REFERENCES
Articles from Journal of Virology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jvi.02297-12
Read article for free, from open access legal sources, via Unpaywall:
https://jvi.asm.org/content/jvi/87/8/4185.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/jvi.02297-12
Article citations
Hepatitis C Virus E1E2 Structure, Diversity, and Implications for Vaccine Development.
Viruses, 16(5):803, 18 May 2024
Cited by: 0 articles | PMID: 38793684 | PMCID: PMC11125608
Review Free full text in Europe PMC
Spatial-temporal transmission dynamics of HIV-1 CRF01_AE in Indonesia.
Sci Rep, 14(1):9917, 10 May 2024
Cited by: 0 articles | PMID: 38730038 | PMCID: PMC11087524
Two-photon fluorescence lifetime imaging microscopy of NADH metabolism in HIV-1 infected cells and tissues.
Front Immunol, 14:1213180, 16 Aug 2023
Cited by: 1 article | PMID: 37662898 | PMCID: PMC10468605
New SHIVs and Improved Design Strategy for Modeling HIV-1 Transmission, Immunopathogenesis, Prevention and Cure.
J Virol, 95(11):JVI.00071-21, 03 Mar 2021
Cited by: 22 articles | PMID: 33658341 | PMCID: PMC8139694
Recombinant MVA-prime elicits neutralizing antibody responses by inducing antigen-specific B cells in the germinal center.
NPJ Vaccines, 6(1):15, 25 Jan 2021
Cited by: 4 articles | PMID: 33495459 | PMCID: PMC7835239
Go to all (72) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Comparison of Immunogenicity in Rhesus Macaques of Transmitted-Founder, HIV-1 Group M Consensus, and Trivalent Mosaic Envelope Vaccines Formulated as a DNA Prime, NYVAC, and Envelope Protein Boost.
J Virol, 89(12):6462-6480, 08 Apr 2015
Cited by: 35 articles | PMID: 25855741 | PMCID: PMC4474309
Characterization of neutralizing antibody responses elicited by clade A envelope immunogens derived from early transmitted viruses.
J Virol, 82(12):5912-5921, 09 Apr 2008
Cited by: 17 articles | PMID: 18400850 | PMCID: PMC2395128
Antigenicity and immunogenicity of HIV-1 consensus subtype B envelope glycoproteins.
Virology, 360(1):218-234, 13 Nov 2006
Cited by: 54 articles | PMID: 17097711 | PMCID: PMC1945152
Centralized HIV-1 envelope immunogens and neutralizing antibodies.
Curr HIV Res, 5(6):572-577, 01 Nov 2007
Cited by: 17 articles | PMID: 18045113
Review
Funding
Funders who supported this work.
NIAID NIH HHS (5)
Grant ID: R01 AI094604
Grant ID: UM1 AI100645
Grant ID: U19 AI067854
Grant ID: P30 AI050410
Grant ID: U19AI067854
 a,b
a,b 




