Abstract
Free full text

A Quorum-Sensing-Induced Bacteriophage Defense Mechanism
Associated Data
ABSTRACT
One of the key determinants of the size, composition, structure, and development of a microbial community is the predation pressure by bacteriophages. Accordingly, bacteria have evolved a battery of antiphage defense strategies. Since maintaining constantly elevated defenses is costly, we hypothesize that some bacteria have additionally evolved the abilities to estimate the risk of phage infection and to adjust their strategies accordingly. One risk parameter is the density of the bacterial population. Hence, quorum sensing, i.e., the ability to regulate gene expression according to population density, may be an important determinant of phage-host interactions. This hypothesis was investigated in the model system of Escherichia coli and phage λ. We found that, indeed, quorum sensing constitutes a significant, but so far overlooked, determinant of host susceptibility to phage attack. Specifically, E. coli reduces the numbers of λ receptors on the cell surface in response to N-acyl-l-homoserine lactone (AHL) quorum-sensing signals, causing a 2-fold reduction in the phage adsorption rate. The modest reduction in phage adsorption rate leads to a dramatic increase in the frequency of uninfected survivor cells after a potent attack by virulent phages. Notably, this mechanism may apply to a broader range of phages, as AHLs also reduce the risk of χ phage infection through a different receptor.
IMPORTANCE
To enable the successful manipulation of bacterial populations, a comprehensive understanding of the factors that naturally shape microbial communities is required. One of the key factors in this context is the interactions between bacteria and the most abundant biological entities on Earth, namely, the bacteriophages that prey on bacteria. This proof-of-principle study shows that quorum sensing plays an important role in determining the susceptibility of E. coli to infection by bacteriophages λ and χ. On the basis of our findings in the classical Escherichia coli-λ model system, we suggest that quorum sensing may serve as a general strategy to protect bacteria specifically under conditions of high risk of infection.
Introduction
Many bacterial species possess the ability to regulate gene expression according to the cell density of the population. They do so by releasing signaling molecules called autoinducers into the environment. As the population density increases, autoinducers accumulate extracellularly, and this can be detected by the bacteria—a phenomenon known as quorum sensing. Since quorum-sensing signals vary between different bacterial species, they can be utilized to distinguish groups of bacteria. Hence, quorum sensing enables sensing of local population density as well as species complexity and makes it possible for microorganisms to switch between different gene expression patterns depending on these parameters. Accordingly, quorum sensing is often used to coordinate social behaviors such as virulence and biofilm formation across the population (reviewed in reference 1). This study was motivated by the hypothesis that quorum sensing could additionally be used as a means of regulating phage-bacterium interactions.
Bacteriophages are viruses that attack bacteria. The predation pressure by phages is substantial, as they outnumber bacterial cells by an estimated 10-fold in many natural environments (2). Consequently, host bacteria have evolved a wide range of antiphage mechanisms, including ways of blocking the initial attachment of phages, degradation of the phage genome, or abortive infection by host suicide, preventing the spread of phage progeny in the population (reviewed in reference 3). As phages require a bacterial host to proliferate, phages are expected to be more abundant and diverse in densely populated mixed-species environments than in sparsely populated environments. Therefore, the risk of suffering phage attacks is generally elevated at high microbial cell densities. The costs associated with general phage resistance mechanisms are substantial and serve as a key factor in shaping the evolutionary dynamics between the phage and host (4, 5). We speculate that if bacteria used quorum sensing to regulate their antiphage activities, they could specifically upregulate their defense mechanisms to avoid infection during growth under high-risk conditions, while saving the metabolic burden of maintaining constantly elevated antiphage strategies.
As a first approach to testing whether quorum sensing is used to regulate phage-bacterium interactions, we investigated the role of quorum sensing in the classical model system of phage λ and its host Escherichia coli K-12. Since the discovery of phage λ more than 60 years ago (6), it has been intensively studied and likely represents the most completely understood biological entity. The investigations of λ and its interactions with E. coli have served as a paradigm for molecular biology and paved the way for our understanding of key biological mechanisms, including gene regulation, recombination, molecular cloning, protein folding, and virion assembly (reviewed in reference 7).
Gram-negative bacteria typically quorum sense through the production and detection of N-acyl-l-homoserine lactone (AHL) quorum-sensing signals. AHLs are produced by synthases of the LuxI family and typically detected by receptors of the LuxR family (reviewed in reference 1). E. coli can detect AHL quorum-sensing signals through SdiA, a LuxR-type transcriptional regulator (8, 9). SdiA is able to bind and be activated by a broad range of AHLs (10, 11). Curiously, E. coli and other enterobacteria are not able to produce AHLs, as they lack a LuxI-type AHL synthase, but they can detect AHLs emitted by other bacterial species (10). Only a few gene groups have reproducibly been shown to be regulated by AHLs in E. coli. These include flagellar genes, acid resistance genes, and virulence genes in the LEE pathogenicity island of enterohemorrhagic E. coli (EHEC) (12–14).
In this study, we have identified a novel quorum-sensing-regulated antiphage defense mechanism in E. coli K-12. We find that E. coli utilizes AHL quorum-sensing signals to reduce its susceptibility to infection by phage λ as well as the broad-host-range phage χ. This is, to our knowledge, the first example of a quorum-sensing-regulated antiphage defense mechanism. We propose that this mechanism serves to protect E. coli under conditions of high risk of infection, namely, during growth in high-cell-density, mixed-species environments, where the quorum-sensing signals would accumulate. Quorum-sensing control of phage susceptibility may be a general phenomenon in microbial communities.
RESULTS
λ phages accumulate in AHL-treated cultures due to reduced superinfection of lysogenic cells.
Many known prophages can be induced to follow the lytic pathway and kill the host cell to release phage progeny. These prophages have evolved to incorporate sensory inputs into the genetic switches that govern this developmental decision. Known induction signals generally provide the prophage with information on the metabolic state and stress level of the host cell (15–17). As a culture of λ lysogens grows, a small fraction (10−5) of the resident λ prophages will induce and lyse the host cell to release progeny phages into the medium. To assess the effect of quorum sensing on the interaction between E. coli and phage λ, cultures of lysogenic E. coli BW25113 λi434 were grown in the presence or absence of a cocktail of synthetic AHL autoinducer molecules, and free λ phages were enumerated as PFUs on a lawn of λ-sensitive bacteria. The cultures were grown at 30°C, where SdiA has been shown to be most active (18, 19).
Figure 1 shows the concentrations of free λ phages in AHL-treated or control lysogenic cultures. In wild-type lysogens, AHL treatment leads to a 2- to 3-fold increase in free phage levels (blue bars). Thus, quorum-sensing signals do indeed influence phage-bacterium interactions in the classic host-phage pair of E. coli and λ. Deletion of the sdiA gene encoding the AHL receptor abolishes the AHL-mediated increase in free phage (green bars), suggesting that the AHL effect is mediated by the AHL receptor SdiA. Similar observations have been reported previously (20). However, the mechanism underpinning phage accumulation has not been determined and is the focus of the present study.
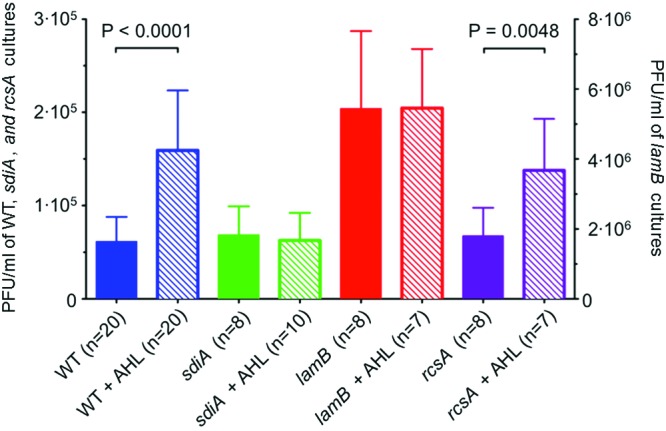
AHL quorum-sensing signals reduce λ phage superinfection. The concentrations of free phage from lysogenic cultures with or without AHLs are shown for E. coli BW25113 (wild type [WT]), AHL receptor mutant (sdiA mutant), λ receptor mutant (lamB mutant), and a mutant of a transcriptional regulator for exopolysaccharide (rcsA mutant). The cultures were grown to an OD600 of 1.2 in the presence or absence of 5 µM AHLs. Free phage concentration is indicated as PFU per ml of culture. The number of independent cultures tested (n) is indicated. Each error bar indicates 1 standard deviation from the mean. Reported differences were evaluated using a Student’s t test.
The concentration of free phages in a culture of lysogens is a result of two opposing factors, production and loss. First, it depends on the rate of production and release of progeny phages into the medium by host cell lysis. Second, it depends on the rate of loss of free phages due to superinfection of the remaining cells. Specifically, a free λ phage can initiate infection of a lysogenic cell by injecting its DNA into the host cytoplasm, but the superinfection immunity system of the resident prophage will prevent expression of the infecting DNA, leading to loss of the superinfecting phage (21). Hence, the observed increase in free phage in the presence of quorum-sensing signals could be a result of increased prophage induction or decreased superinfection of the lysogenic cells or both. In order to distinguish between these possibilities, we measured the effects of AHLs on prophage induction in bacteria where superinfection is not possible. Specifically, the concentration of free phages in a culture of lysogens immune to superinfection due to deletion of the λ receptor LamB was used as a direct measure of prophage induction. Since there is no loss of free phage due to superinfection, cultures of lamB lysogens show higher concentrations of free phages than do wild-type cultures (compare the solid red bar to the solid blue bar in Fig. 1). Importantly, growth in the presence of AHLs does not stimulate additional phage accumulation in the lamB lysogen (red bars). This observation proves that AHLs do not stimulate induction of the λ prophage as suggested previously (20). Rather, the observed AHL effect on free phage concentration must be due to a decreased loss of free phage, most likely due to a reduction in the rate of superinfection of the lysogenic cells.
A positive regulator of exopolysaccharide synthesis, RcsA (22), was previously reported to be required for the AHL-mediated increase in free λ phage accumulation (20). Contrary to these results, we do not observe a difference between the wild type and the rcsA mutant with respect to the AHL effect on free phage concentrations (Fig. 1, purple bars).
λ phage adsorption rate is reduced when E. coli is grown in the presence of AHL signals.
The results displayed in Fig. 1 show that superinfection of λ lysogens is reduced when the cells are grown in the presence of AHL autoinducers. We hypothesized that AHLs may act to reduce the rate of phage adsorption to the cell surface. To test this hypothesis directly, we subjected wild-type, nonlysogenic E. coli cells grown with or without AHLs to primary infection by radioactively labeled λ phages. After the addition of phages to a cell culture, unadsorbed free phages were removed by filtration of an aliquot of the cells every 2 min, and the radioactivity of the adsorbed phages retained on the filters was quantified (Fig. 2).
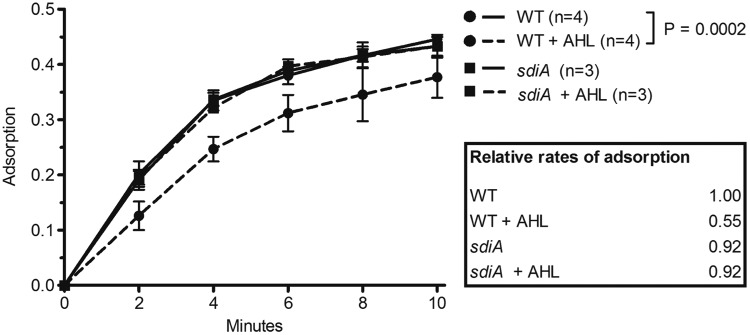
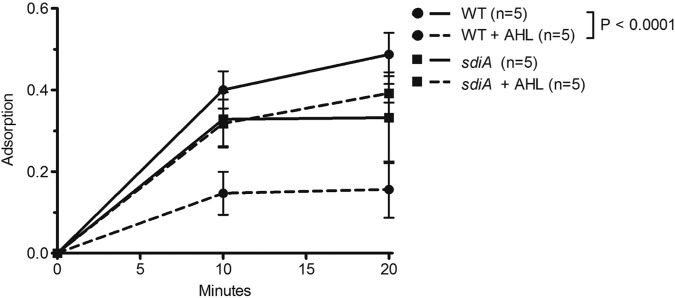
Growth in the presence of AHL signals reduces the rate of phage adsorption. 35S-labeled λ phages were added to a shaking culture of wild-type (WT) or AHL receptor mutant (sdiA mutant) cells grown in the presence or absence of 5 µM AHLs. Prior to the addition of phage, chloramphenicol was added to cells to arrest growth at an OD600 of 1.0 and prevent phage multiplication. At 2-min intervals, an aliquot of the cell-phage mixture was filtered through a 0.45-µm filter, and the radioactivity of the adsorbed phages retained on the filter was measured. The radioactivity of filters subjected to the identical treatment using lamB mutant cells has been subtracted as background, and each sample has been normalized to the total radioactivity of an unfiltered sample aliquot. The number of independent cultures tested (n) is indicated. Each error bar indicates 1 standard deviation from the mean. Reported differences were evaluated using a two-way analysis of variance (ANOVA). The experiment was repeated on three separate days with similar results. The relative rates of adsorption were calculated as shown in the symbol key to Fig. S1 in the supplemental material and are shown in the figure.
Figure 2 shows that AHL treatment of wild-type cells significantly reduces the rate of λ adsorption. The rates of adsorption were calculated under the assumption that the decrease in free phage over time follows an exponential decay law (see Fig. S1 in the supplemental material). The rate of adsorption of λ phages to AHL-treated wild-type cells is 2.7 × 10−10 ml min−1 phage−1 cell−1, about half that of untreated cells (4.9 × 10−10 ml min−1 phage−1 cell−1). In contrast, AHL treatment has no effect on the λ adsorption rate in the sdiA mutant, demonstrating that the AHL signals must be transduced through the SdiA receptor to decrease phage adsorption rates. Thus, AHL quorum sensing leads to a reduction in the rate of adsorption of phage λ to E. coli cells.
AHL-induced downregulation of the λ receptor LamB.
The molecular mechanism underlying the reduced infection rate caused by growth in the presence of AHLs was investigated. As phage λ infects E. coli through the outer membrane maltoporin LamB, the adsorption rate depends on the concentration of LamB receptors on the cell surface. We therefore investigated whether AHL autoinducers directly affect LamB protein levels in the outer membrane. Wild-type and sdiA and lamB mutant cells were grown in the presence or absence of AHLs, and outer membrane proteins were purified and separated by SDS-PAGE (Fig. 3A). A maltose-induced culture was used as a positive control for identifying LamB on the gel. We find that stimulation of wild-type cells by AHLs lead to a reduction in LamB protein levels by 40% (Fig. 3B). AHL treatment does not affect the LamB levels in the sdiA mutant. This finding clearly shows that AHL signaling through SdiA leads to a decrease in the levels of LamB in the outer membrane of E. coli BW25113.
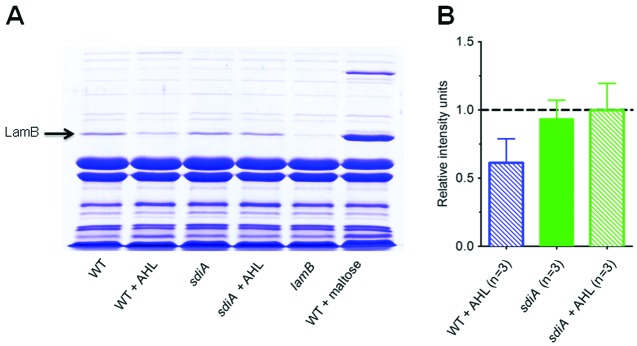
AHL signaling induces downregulation of the λ receptor LamB. (A) Outer membrane protein preparations were separated by SDS-PAGE and stained with Coomassie blue. Outer membrane proteins from wild-type E. coli, AHL receptor mutant (sdiA mutant), and λ receptor mutant (lamB mutant) grown to an OD600 of 1.0 with 5 µM AHLs or 0.4% maltose are shown as indicated below the lanes. (B) Quantification of LamB protein. Band intensities of the protein band were quantified using ImageJ software and normalized to the intensity of a LamB band that is not affected by AHLs. To enable pooling of the data from different gels, the intensity of the LamB band in untreated wild-type cells was set at 1 in each gel, and the intensities of the remaining bands relative to that of the untreated wild-type cells are shown. The number of independent outer membrane protein preparations tested (n) is indicated. Each error bar indicates 1 standard deviation from the mean.
AHL signaling dramatically improves E. coli’s chances of surviving a virulent phage attack.
What is the physiological significance of the modest reduction in adsorption rate? To evaluate the effect of quorum sensing on E. coli survival, we exposed E. coli cells grown in the presence or absence of AHL signals to a virulent variant of λ, λvir. If the λvir phages were allowed to complete multiple life cycles, they would eventually kill all wild-type cells, and the only surviving cells would be λ-resistant mutants that have lost the ability to express lamB (23). To address the survival of nonmutant cells, we exposed the cell cultures to a 20-fold excess of λvir for only 50 min, corresponding to just under one phage generation time under our laboratory conditions. After 50 min, the cations in the medium were chelated to inactivate any free phage and prevent additional rounds of infection. Uninfected cells were counted as those that grew to form a colony on petri plates the next day.
Figure 4 shows that the number of E. coli cells surviving λvir infection is 3 to 4 times higher in cultures that are grown in the presence of AHLs than in those grown in the absence of AHLs (blue bars). In sdiA mutant cultures, the presence of AHLs do not increase the chance of surviving the virulent infection, thus demonstrating that the AHL effect is entirely dependent on SdiA (green bars). The surviving cells can be divided into two groups, those that are λ sensitive but were not infected by a phage for the duration of the experiment and those that carry a mutation that makes them genetically resistant to infection by λ. To assess whether the surviving cells in the experiment shown in Fig. 4 are mostly λ-resistant mutants or nonmutant cells that have avoided λvir infection, we took advantage of the genetic linkage between the λ resistance phenotype and that of the ability to utilize maltose as a carbon source (24, 25). lamB encodes the maltoporin, and its expression is coregulated with the genes required for the uptake and utilization of maltose. Hence, about 80% of λ-resistant mutants have concurrently lost the ability to utilize maltose as a carbon source (23). These mal mutants can be easily distinguished from mal+ cells, as the former form white colonies on maltose-MacConkey indicator agar, while the latter form red colonies. When the surviving cells were plated on maltose-MacConkey agar, less than 1% of the colonies were white, indicating that the vast majority of surviving cells at the end of the first lytic cycle are nonmutant cells which have simply avoided infection by λvir, presumably due to the display of a reduced number of LamB receptors on their cell surface at the time of phage invasion. The decreased susceptibility to phage infection is transient, as the vast majority of daughter cells in colonies arising from the survivor cells are again sensitive to infection by λvir, as determined by cross-streaking those colonies with a high-titer λvir lysate. Thus, AHL quorum sensing through SdiA leads to a severalfold increase in the fraction of an E. coli population that is not resistant, but transiently less susceptible, to λ phage attack.
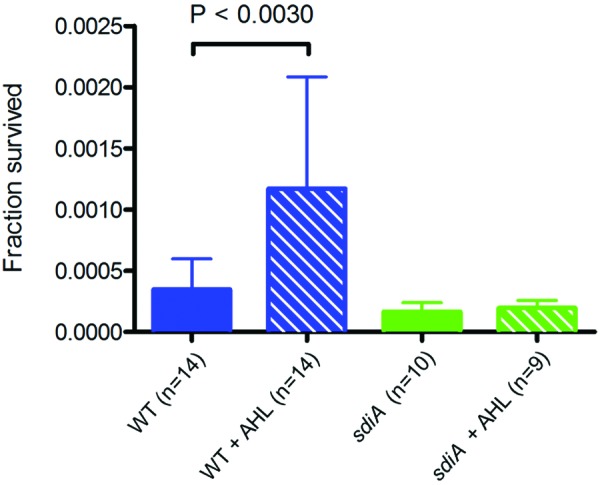
AHLs dramatically enhance E. coli’s chances of surviving attack by the lytic phage λvir. Wild-type and sdiA mutant E. coli cells were grown to an OD600 of 1.0 in the presence or absence of 5 µM AHLs. λvir was added at an average phage input of 20 phages per cell. Fifty minutes after the addition of phage, an aliquot of the culture was diluted in M63-salts containing 50 mM sodium citrate to inactivate the free phage. The figure shows the number of colonies formed by cells that survived 50 min in the presence of λvir relative to the number of CFU immediately prior to phage addition. The number of independent cultures tested (n) is indicated. Each error bar indicates 1 standard deviation from the mean. Reported differences were evaluated using a Student’s t test.
AHL quorum-sensing signals also protect E. coli from phage χ.
AHL signals could also protect E. coli from phages that recognize other receptors than LamB. One such example is phage chi (χ), which is known to infect E. coli and other enteric bacteria through the flagellum (26–28). χ phages were added to wild-type E. coli cells grown in the presence or absence of AHLs, and the proportion of free, nonadsorbed phages was monitored over time. Figure 5 shows that growth in the presence of AHLs significantly reduces the rate of adsorption of phage χ to wild-type cells. Analogous to the λ case, the AHL effect depends on the AHL receptor SdiA, as the effect is not observed in sdiA mutant cells.
AHL signaling protects E. coli from χ phage adsorption. Wild-type E. coli was grown to an OD600 of 0.75 in the presence or absence of 5 µM AHLs. To arrest growth and prevent phage multiplication, chloramphenicol was added prior to the addition of phage χ. The numbers of free (nonadsorbed) χ phages were measured as PFU on a lawn of sensitive, motile bacteria. The number of independent cultures tested (n) is indicated. Each error bar represents 1 standard deviation from the mean. Reported differences were evaluated using a two-way analysis of variance (ANOVA).
As we find that AHLs mediate an antiphage defense against both phage λ and phage χ, which have different mechanisms of infection, we suggest that quorum sensing may generically activate antiphage strategies in E. coli and potentially other bacterial species. The generality of this quorum-sensing response is under investigation in our laboratory.
DISCUSSION
Quorum sensing is crucial for the survival and fitness of numerous microorganisms, and many variations on the canonical LuxI-LuxR quorum-sensing system have evolved that allow bacteria to engage in communication with their own and other species, enable quorum quenching and the spread of misinformation, and even allow crosskingdom signaling with eukaryotic hosts (reviewed in reference 29). Here, we suggest that bacteria have additionally evolved to use quorum sensing as a means of regulating their interactions with the most abundant biological entities on Earth, namely, the viruses that prey on them. We propose that quorum-sensing signals, as a measure of population density, may be utilized to estimate the local risk of phage infection and to regulate antiviral defense strategies accordingly.
As a first approach to investigate our hypothesis, we evaluated the effect of AHL quorum sensing on phage-host interactions in the classical model system of phage λ and E. coli K-12. We found that AHL quorum sensing leads to downregulation of the number of LamB λ receptors on the cell surface, which in turn increases the number of survivor E. coli cells in a population following a potent phage attack. In addition, we find that AHLs protect E. coli against infection by the broad-host-range phage χ. Importantly, the observed AHL effects depend entirely on the AHL receptor SdiA. The requirement for SdiA demonstrates that the reduced phage adsorption rates in AHL-treated cultures occur as a consequence of SdiA-AHL-mediated intracellular regulation, not as a side effect of unexpected physical or chemical changes in the medium caused by the addition of synthetic AHLs.
Although many aspects of E. coli biology are very well understood, the role of AHL quorum sensing in E. coli has remained elusive (30). First, E. coli does not produce AHL molecules and therefore presumably uses the AHL receptor SdiA exclusively to pick up signals released from other AHL-producing bacteria (10). Second, very few genes have reproducibly been shown to be regulated by the AHL-SdiA complex in E. coli K-12 (12–14). One of the regulons that are consistently found to be downregulated by SdiA is the flagellar genes (12, 14, 31, 32), which supports our finding that AHLs reduce the risk of χ infection, as phage χ infects its host through the flagellum. Interestingly, SdiA-mediated transcriptional regulation of LamB is supported by results from Van Houdt et al. who found that the AHL variant N-hexanoyl-l-homoserine lactone (C6-HSL) downregulates malT at 30°C in E. coli MG1655, in a plasmid-based promoter trap screen (19). As MalT is a positive regulator of lamB (33), downregulation of malT would lead to decreased lamB transcription. The expression of another gene in the MalT regulon, malE, was additionally found to be repressed by plasmid-based expression of sdiA in E. coli K-12 grown at 37°C (32). Hence, there are several indications that SdiA may downregulate λ receptor levels via transcriptional repression of malT, but whether the SdiA-AHL complex directly binds and represses the malT promoter has yet to be elucidated.
The question remains why AHLs that are not produced by E. coli affect E. coli’s phage defense. Many phages appear to have extreme host specificity, infecting only the bacterial strain with which they were isolated and thus only species-specific, or even strain-specific, quorum-sensing signals would be relevant for regulating phage-host interactions. However, the standard methods for phage isolation may have been biased to favor phages of limited host range (35), and broad-host-range phages are readily isolated from environmental samples (34, 35). Moreover, phages may adapt to new host strains at relatively high rates (36). The issue of host specificity in mixed-species microbial communities is still poorly understood, but it is clear that the subject is more complex than suggested by the traditional one-phage−one-host view. Thus, in its natural habitats, E. coli may encounter a variety of broad-host-range phages. By exploiting quorum-sensing signals released from other bacterial species to reduce its susceptibility to phage infection, E. coli may be able to protect itself against these phages.
The evolutionary dynamics between phage and host has been described as an arms race, where antagonistic interactions between host and parasite result in ever increasing phage infectivity and bacterial resistance. However, increased bacterial resistance comes at a price. Hall et al. reported that arms race evolution results in decreased relative fitness, and that over time, this dynamics is replaced by fluctuating selection, potentially due to costs of generalism and mutational limitations (5). Gómez and Buckling (4) found that in a natural microbial environment, bacterium-phage coevolution is characterized by fluctuating selection and thus generates bacteria that are more resistant to present phages than to past and future viruses. They suggest that the evolution of resistance toward contemporary phages is favored, as this is less costly in terms of growth rate than general resistance mechanisms (4). In keeping with this theory, we argue that quorum sensing allows bacteria to spare the cost of maintaining a constantly elevated phage defense. Thus, a quorum-sensing-regulated phage defense mechanism could potentially play a key role for bacterium-phage coevolution in natural environments.
Phage-host systems show unusual prey-predator dynamics because the consumption of one prey leads to the generation of tens to hundreds of new predators. Given this, it is remarkable that virulent phages do not always drive their host population to extinction. To explain phage-host coexistence, various mechanisms have been suggested that would generate a minority group of host cells that are protected from infection, either due to spatial separation (37), decreased lysis in stationary-phase cells (38), or stochastic fluctuations in phage receptor expression (39). We propose that by subjecting genes encoding phage receptors (lamB and flagellar genes) to quorum-sensing control, E. coli has evolved the ability to increase the subpopulation of phage-tolerant receptor-free cells at times where the risk of phage infection is elevated, despite the cost associated with temporary loss of the primary function of these phage receptors (uptake of maltodextrins and motility). Quorum-sensing-mediated downregulation of phage susceptibility may well turn out to be an important factor in understanding phage-host coexistence. The phenomenon bears a striking similarity to the formation of population heterogeneity in other contexts, such as the generation of competent subpopulations, sporulating subpopulations, or subpopulations that persist despite antibiotic treatment. Notably, quorum sensing has been found to upregulate the generation of all of these minority groups (40–43).
Phage-host interactions are understood better in the classical model system of bacteriophage λ and E. coli K-12 than in any other system. However, despite its paradigmatic status, new knowledge continues to be revealed as exemplified here by a novel and important role for quorum sensing in regulating phage susceptibility. Given the incompleteness of our understanding of this relatively simple system, one can only speculate how far we must be from an in-depth understanding of phage-host interactions in complex natural niches, such as the human gut. The launch of the human microbiome project marks a renaissance in studies of the contributions of the human bacterial flora to well-being and promises to deliver new treatments for lifestyle diseases through engineering alterations in the gut flora (reviewed in references 44, 45, and 46). However, for these efforts to succeed, a comprehensive understanding of the factors that naturally shape the size, composition, structure, and development of microbial communities is required. On the basis of our finding in the E. coli-λ model system, we hypothesize that an important, but so far overlooked, determinant of phage-host dynamics could be bacterial cell-cell communication.
MATERIALS AND METHODS
Bacterial strains and bacteriophages.
The bacterial strains used in this study are all derivatives of Escherichia coli K-12. The bacterial strains, phages, and plasmids used in this study are listed in Table S1 in the supplemental material.
Strain construction.
A single λi434 lysogen of E. coli BW25113, identified by the PCR assay described by Powell et al. (47), gave rise to strain JMØ3. Gene replacement of the open reading frames (ORFs) of lamB, sdiA, and rcsA with an antibiotic resistance cassette was performed by the method of Datsenko and Wanner (48). Antibiotic-resistant transformants were screened by PCR with primers flanking the ORF to identify transformants with gene replacements of the expected size. The PCR primers used to amplify the antibiotic resistance cassette are listed in Table S2 in the supplemental material.
AHL preparation.
A mixture of 6 AHLs (N-(butyl, heptanoyl, hexanoyl, ketocaproyl, octanoyl, and tetradecanoyl)-dl-homoserine lactones) were dissolved in ethyl acetate acidified by 0.1% acetic acid (EA) and stored at −20°C. Prior to each experiment, the AHL mix or the equivalent volume of EA alone was added to glass culture tubes to a final AHL concentration of 5 µM and incubated with shaking at room temperature until completely dry as in reference 20.
Bacterial growth conditions.
For all experiments, E. coli BW25113 and its derivatives were grown from single colonies in TB medium (15 g tryptone and 5 g NaCl per liter) at 30°C, shaking at 220 rpm. Exponentially growing cultures were diluted a minimum of 1,000-fold into TB medium in glass tubes coated with the AHL mixture or control tubes treated with EA as described above and allowed to reach the desired cell density. Cell densities were measured by determining the optical density at 600 nm (OD600) on an Ultraspec 2100 Pro (Amersham Biosciences). One OD600 unit corresponds to a cell density of 1.1 × 109 CFU/ml.
Measurement of free phage in lysogenic cultures.
E. coli BW25113 single lysogens of λi434 and otherwise isogenic lamB, sdiA, and rcsA mutant strains were grown to an OD600 of 1.2, and cells were pelleted by centrifugation for 5 min at 7,500 × g. A drop of chloroform was added to the supernatants. The concentration of free phages in the supernatant was quantified as the number of PFU on a lawn of λ-sensitive, maltose-induced E. coli MG1655 as described previously (49). Three independently constructed rcsA mutants were included in the study to verify the AHL-dependent phenotype observed in this mutant.
35S labeling of bacteriophage λ.
Single colonies of E. coli S2775 were grown for 5 to 6 h at 30°C in sulfate-free M63 (30.00 g KH2PO4, 91.71 g K2HPO4·3H2O, 16.19 g NH4Cl, 6.54 mg FeCl2·4H2O, 9.68 mg Na3citrate per liter) supplemented with Na2SO4 to a final concentration of 276 µM, followed by a 15-min incubation at 42°C to induce the temperature-sensitive prophage. To remove sulfate, cells were washed twice in sulfate-free M63, and the growth medium was supplemented with 500 µCi 35S-labeled methionine and 2 µg unlabeled methionine. The cultures were incubated with shaking for 2 h at 42°C to allow phage proliferation. Then, the lysate was cleared by the addition of a few drops of chloroform followed by centrifugation for 10 min at 8,000 × g to remove cell debris. The supernatant was transferred to a fresh tube and subjected to cesium chloride banding to purify the phages as previously described (50).
Adsorption assay of 35S-labeled phages.
Wild-type and lamB and sdiA mutant cells were grown in the presence or absence of AHLs to an OD600 of 1.0, and chloramphenicol was added to a final concentration of 0.05 mg/ml to prevent further growth. To begin the assay, 1.6 × 108 PFU/ml of 35S-labeled phages were added to the shaking cell culture, and aliquots were filtered 2, 4, 6, 8, and 10 min after the addition of phage. Filtration through a 0.45-µm membrane filter (Durapore membrane filter [catalog no. HVLP02500; Millipore]) served to retain bacterial cells and any adsorbed phages on the filter, while allowing free phages to pass through. The radioactivity retained on the filters was quantified using a PerkinElmer Wallac 1414 liquid scintillation counter, counting each filter for 20 min. For each cell culture, an unfiltered sample aliquot was counted to quantify the total radioactivity of the phage-cell mixture. Adsorption values in Fig. 2 are calculated as follows: (counts on filter minus the average counts retained on filters with lamB mutant cells)/(total counts of sample aliquot).
Survival assay.
Cultures of wild-type and sdiA mutant cells were grown in the presence or absence of AHLs to an OD600 of 1.0. λvir was added at an average phage input of 20 phages per cell. Samples were collected immediately before and 50 min after phage addition and diluted into ice-cold M63-salts containing 50 mM sodium citrate to inactivate any free phages. The dilutions were plated on 0.4% maltose-MacConkey plates containing 5 mM sodium citrate and incubated at 30°C overnight. Surviving cells were counted as red and white colonies the following day. Two hundred colonies were cross-streaked against λvir to determine whether survival was due to transient phage tolerance (λvir-sensitive colony) or inherited phage resistance (λvir-resistant colony).
Outer membrane protein preparations and SDS-PAGE.
Cultures were grown in the presence or absence of AHLs to an OD600 of 1.0, harvested, and chilled on ice. All steps were carried out cold, except when noted otherwise. Cell pellets were collected by centrifugation for 5 min at 6,000 × g at 4°C. Pellets were washed in 1 ml of 50 mM Tris HCl (pH 8) and resuspended in 20% sucrose and 100 mM Tris HCl (pH 8). Lysozyme and EDTA were added to final concentrations of 0.4 mg/ml and 10 mM, respectively. Samples were incubated at 4°C overnight. The next day, the samples were overlaid by an equal volume of 2% Triton X-100, 10 mM MgCl2, and 100 mM Tris HCl (pH 8) and sonicated until clear. Outer membrane proteins were collected by centrifugation for 22.5 min at 16,000 × g at 4°C. Membrane particles were washed once in 50 mM Tris HCl (pH 8), 5 mM MgCl2, and 1% Triton X-100 and spun down at 16,000 × g and 4°C for 15 min. Membrane particles were washed twice with 50 mM Tris HCl (pH 8) and 5 mM MgCl2 to remove Triton X-100. Pellets were resuspended at room temperature in 100 mM Tris HCl (pH 8) and 2% SDS. Protein concentrations were determined using a NanoDrop 1000 (Thermo Scientific). This protocol was adapted from references 51 and 52. Proteins were separated on a 10% SDS-polyacrylamide gel and stained with Coomassie blue. Protein band intensities were quantified using ImageJ and normalized to a band that was constantly expressed regardless of the presence of AHLs.
χ phage assay.
Wild-type cells were grown in the presence or absence of AHLs to an OD600 of 0.75, and chloramphenicol was added to a final concentration of 0.05 mg/ml to prevent further growth. χ phage lysate was added to a final concentration of 1.6 × 108 PFU/ml to begin the assay. At each time point, a culture aliquot was diluted into ice cold TMG (0.121% Tris base, 0.12% MgSO4, 0.01% gelatin [pH 7.4]) to prevent further adsorption, and cells were removed by centrifugation for 5 min at 7,500 × g. Chloroform was added to the supernatant, and free, nonadsorbed χ phages were quantified by plating on motile, χ-sensitive cells, and counting PFUs. Sensitive chloramphenicol-resistant cells were prepared by growing E. coli KX1440 cells to an OD600 of 0.75. χ phage dilutions were mixed with sensitive cells, incubated 10 min at 37°C, plated with TB motility agar (0.4% agar) on a solid TB plate, and incubated at 30°C overnight.
Statistical analyses.
Reported differences were evaluated using a Student’s t test for individual measurements (Fig. 1 and 4) or a two-way analysis of variance (ANOVA) for data containing repeated measurements of the same cultures (Fig. 2 and 5). The analyses were carried out with GraphPad Prism 5 software.
SUPPLEMENTAL MATERIAL
Figure S1
Decrease in free phage over time in wild-type and sdiA mutant cultures grown in the presence of 5 µM AHLs or absence of AHLs. The lines indicate the regression exponential curves of the form y = e−ax for each data set. The value of the slope a relative to the slope a of the wild-type regression curve is reported as the relative adsorption rate in Fig. 2 of the article. The adsorption rates reported in the text of the article are also calculated from the slope a, corrected for the number of cells and the volume of the sample aliquot. The fraction of free phages y(t) was calculated as y(t) = 1 − [(z(t) − L)/(T × M)], where z(t) is the counts per minute adsorbed on the filter at time t, L is the average counts per minute adsorbed to filters with lamB mutant cells, and T is the total counts per minute of an unfiltered aliquot of the phage-bacterium mixture. With this nomenclature, the adsorption measure plotted in Fig. 2 of the article is equal to (z(t) − L)/T. As the total counts per minute of an aliquot of the phage-bacterium mixture (T) consist of both counts from 35S incorporated in lambda phages and 35S incorporated in protein impurities present in the lysate, it was necessary to define a parameter, M, which is the fraction of T that can be contributed to phage-incorporated 35S. M was estimated as the value between 0 and 1 that gives the best collective fit of the four data sets to exponential curves, as measured by the average R2 value of the four data sets that is closest to 1. We found that M = 0.461 gave the best fit, with an average R2 value of 0.981. Download
ACKNOWLEDGMENTS
This work was supported in part by a research grant to S.L.S. from the Novo Nordisk Foundation. N.M.H.-K. was supported by the Danish National Research Foundation through the Center for Models of Life. We are grateful to Stanley Brown and Steen Pedersen for their help with generating the radioactive phage lysate. Special thanks to Linda Hove Christensen, Julia Madsen-Østerbye, and Samantha Steen for their contributions in the project’s earliest stages. We thank Stanley Brown, Saeed Tavazoie, and Karina Xavier for bacterial strains and phages.
Footnotes
Citation Høyland-Kroghsbo NM, Mærkedahl RB, Svenningsen SL. 2013. A quorum-sensing-induced bacteriophage defense mechanism. mBio 4(1):e00362-12. 10.1128/mBio.00362-12.
REFERENCES
Articles from mBio are provided here courtesy of American Society for Microbiology (ASM)
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/mbio.00362-12
Article citations
Growth stage-related capsular polysaccharide translocon Wza in Vibrio splendidus modifies phage vB_VspM_VS2 susceptibility.
Commun Biol, 7(1):1338, 16 Oct 2024
Cited by: 0 articles | PMID: 39414953 | PMCID: PMC11484964
Essential phage component induces resistance of bacterial community.
Sci Adv, 10(36):eadp5057, 04 Sep 2024
Cited by: 0 articles | PMID: 39231230 | PMCID: PMC11373596
The LuxO-OpaR quorum-sensing cascade differentially controls Vibriophage VP882 lysis-lysogeny decision making in liquid and on surfaces.
PLoS Genet, 20(7):e1011243, 30 Jul 2024
Cited by: 0 articles | PMID: 39078816 | PMCID: PMC11315295
Quorum sensing positively regulates CPS-dependent Autographiviridae phage infection in Vibrio alginolyticus.
Appl Environ Microbiol, 90(8):e0221023, 29 Jul 2024
Cited by: 0 articles | PMID: 39072624
Salmonella enteritidis acquires phage resistance through a point mutation in rfbD but loses some of its environmental adaptability.
Vet Res, 55(1):85, 05 Jul 2024
Cited by: 0 articles | PMID: 38970094 | PMCID: PMC11227202
Go to all (116) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Quorum Sensing Determines the Choice of Antiphage Defense Strategy in Vibrio anguillarum.
mBio, 6(3):e00627, 16 Jun 2015
Cited by: 67 articles | PMID: 26081633 | PMCID: PMC4471561
Acyl-homoserine lactones can induce virus production in lysogenic bacteria: an alternative paradigm for prophage induction.
Appl Environ Microbiol, 75(22):7142-7152, 25 Sep 2009
Cited by: 60 articles | PMID: 19783745 | PMCID: PMC2786502
Phage-Encoded LuxR-Type Receptors Responsive to Host-Produced Bacterial Quorum-Sensing Autoinducers.
mBio, 10(2):e00638-19, 09 Apr 2019
Cited by: 31 articles | PMID: 30967469 | PMCID: PMC6456758
Beneficial effects of bacteria-plant communication based on quorum sensing molecules of the N-acyl homoserine lactone group.
Plant Mol Biol, 90(6):605-612, 22 Feb 2016
Cited by: 59 articles | PMID: 26898296
Review




