Abstract
Free full text

Death Receptor–Ligand Systems in Cancer, Cell Death, and Inflammation
Abstract
The discovery of tumor necrosis factor (TNF) marked the beginning of one of the most fascinating journeys in modern biomedical research. For the moment, this journey has culminated in the development of drugs that inhibit TNF. TNF blockers have revolutionized the treatment of many chronic inflammatory diseases. Yet, the journey seems far from over. TNF is the founding member of a family of cytokines with crucial functions in cell death, inflammation, and cancer. Some of these factors, most prominently TNF, CD95L, and TRAIL, can induce cell death. The receptors that mediate this signal are therefore referred to as death receptors, even though they also activate other signals. Here I will take you on a journey into the discovery and study of death receptor-ligand systems and how this inspired new concepts in cancer therapy and our current understanding of the interplay between cell death and inflammation.
Tumour necrosis facrtor (TNF) is the founding member of the TNF superfamily (TNFSF) of cytokines. The receptors for TNFSF ligands are known as the TNF receptor superfamily (TNFRSF). Some of them are capable of inducing cell death. They can do so because they contain a cytoplasmic death domain (DD). Presence of the DD defines a subgroup of the TNFRSF known as death receptors. Despite their name, death receptors can also induce other signals. They do so by activation of transcription factors and subsequent gene activation. Indeed, soon after its discovery it became clear that the most prominent function of TNF is the induction of cytokines and chemokines, and that TNF plays a crucial role in the inflammatory cascade.
There are six human death receptors (DRs): TNF-R1 (Loetscher et al. 1990; Schall et al. 1990), CD95 (Fas/APO-1) (Itoh et al. 1991; Oehm et al. 1992), TRAIL-R1 (DR4) (Pan et al. 1997b), TRAIL-R2 (APO-2/TRICK/DR5/KILLER) (Pan et al. 1997a; Screaton et al. 1997a; Sheridan et al. 1997; Walczak et al. 1997; Wu et al. 1997), DR3 (TRAMP/APO-3) (Chinnaiyan et al. 1996; Kitson et al. 1996; Bodmer et al. 1997; Screaton et al. 1997b), and DR6 (Pan et al. 1998). They are stimulated by their respective ligands. These are TNF, CD95L (FasL), TRAIL, and TL1A, respectively (Carswell et al. 1975; Suda et al. 1993; Wiley et al. 1995; Migone et al. 2002), with the ligand for DR6 not known, or at least the one suggested (Nikolaev et al. 2009) not yet confirmed (Fig. 1). Therefore, and because the biochemistry of the TL1A/DR3 system is highly similar to that of the TNF/TNF-R1 system, this work focuses on the TNF, CD95, and TRAIL systems.

Death receptors and their ligands. The death domain-containing receptors are transmembrane proteins that contain 2–4 cysteine-rich repeats in their extracellular domain, required for ligand binding, and an intracellular death domain capable of recruiting specific adaptors that define their downstream interactors and signals (light gray for CD95/TRAIL and dark gray for TNF-R1/DR3 systems). Death receptors trigger two main signals. TNF-R1 and DR3 induce gene activation as their primary signaling output, whereas CD95, TRAIL-R1, and TRAIL-R2 induce apoptosis as their primary signal. DR6 has been proposed to be ligated by N-APP but this is unconfirmed, and DR6 signaling is altogether less well understood.
A TUMOR-NECROTIZING ACTIVITY, TNF, AND AN UNEXPECTED TURN OF FORTUNE
At the end of the nineteenth century, William Coley, at the time a young surgeon in New York, observed that in some cancer patients, especially sarcoma patients, who then contracted serious bacterial infections, the tumor necrotized and disappeared (reviewed in Hall 1997). Coley was so intrigued by this observation that over the next decades he developed and used a series of mixtures of bacterial isolates, which became known as “Coley’s toxins” for the treatment of cancer. In some of his patients he could reproduce the tumor-necrotizing activity that led him to embark on this line of research (reviewed in Nauts et al. 1946, 1953). Obviously, back then, there were no means to identify the molecules and mechanisms behind this activity and the extracts could also not be produced with the same quality and activity each time. It took until 1944 when lipopolysaccharide (LPS) was isolated from bacterial extracts and identified as the active ingredient therein responsible for triggering tumor necrosis in mice (Shear and Perrault 1944). The availability of new molecular techniques in biochemistry and genetics in the 1960s and 1970s enabled a more rigorous investigation of the tumor-necrotizing activity of LPS and led to the identification of TNF by Lloyd Old and colleagues in 1975 (Carswell et al. 1975).
In the decade following the discovery of TNF it emerged, however, that only few cancer cells died when stimulated by TNF. Instead, systemic treatment with TNF exerted a lethal inflammatory shock syndrome (reviewed in Tracey et al. 1988). Even though this observation was initially discouraging, it inspired the most important discovery of the TNF field to date: the inhibition of TNF to treat inflammatory diseases. Given that TNF can induce the above-mentioned shock syndrome, TNF blockers were first tried in the treatment of sepsis but these clinical trials failed. Around this time, Feldmann and colleagues in London showed that inhibition of TNF prevented collagen-induced arthritis, a model for rheumatoid arthritis (RA) (Williams et al. 1992). When TNF inhibition was tested clinically in RA, it turned out that a large percentage of patients benefited hugely from the treatment (reviewed in Taylor and Feldmann 2009). Today, millions of patients suffering from RA, Crohn’s disease, and psoriasis, but also other chronic inflammatory diseases, are treated with TNF blockers with success rates often higher than 50%. TNF blockers have revolutionized the therapy of chronic inflammatory diseases. Their development can be regarded as one of the most significant advances in recent biomedical research.
With respect to cancer treatment, however, TNF could not hold the early promise because of the before-mentioned systemic toxicity. Yet, in 1989, the concept of direct induction of cell death in cancer cells by an extracellular stimulus received a new lifeline: two monoclonal antibodies, anti-APO-1 and anti-Fas, were found to induce apoptosis in many different types of cancer cells by binding to a receptor on the surface of the cells they killed (Trauth et al. 1989; Yonehara et al. 1989).
PHYSIOLOGICAL AND PATHOLOGICAL FUNCTIONS OF THE CD95 SYSTEM
When the anti-APO-1 and anti-Fas antibodies and the antigens they bound to were first described, they appeared to recognize two different proteins. Anti-APO-1 was described to bind to a receptor of ~48 kD (Trauth et al. 1989), whereas anti-Fas was reported to detect a protein of ~200 kD (Yonehara et al. 1989). Yet the way the cells died when stimulated by these antibodies appeared very similar (Trauth et al. 1989; Yonehara et al. 1989), questioning whether the antibodies indeed engaged different receptors. Naturally, the next task was to identify the respective antigens. Nagata’s team in Osaka performed expression cloning, whereas Krammer’s team in Heidelberg took the classical approach of protein purification. Despite the above-described differences in the initial biochemical characterization, the same receptor of ~48 kD, now commonly referred to as Fas (APO-1) or CD95, was engaged by both antibodies (Itoh et al. 1991; Oehm et al. 1992). CD95 contained a characteristic sequence in its extracellular domain, three cysteine-rich repeats that were also present in the two then recently cloned TNF receptors (Loetscher et al. 1990; Schall et al. 1990; Smith et al. 1990). Hence, the TNF and CD95 systems were related.
When Nagata’s team mapped the gene encoding murine CD95 they realized that it localized to the same chromosomal region as the lpr mutation, and indeed, a mutation in the CD95 gene was responsible for the accumulation of lymphocytes that characterizes lpr mice (Watanabe-Fukunaga et al. 1992). T-cell accumulation in these mice had been mistaken for lymphoproliferation (lpr), hence the name of the mutation, which illustrates the focus on proliferation rather than cell death in biomedical research at the time. A similar phenotype was observed in gld (generalized lymphoproliferative disease) mice (Roths et al. 1984). Elegant bone marrow transplantations from gld to lpr mice and vice versa showed that the genes mutated in lpr and gld mice encoded proteins expressed by cells that interact with each other in vivo (Reap et al. 1993). Thus, given the CD95 gene was mutated in lpr, the gld mutation most likely affected the gene encoding the elusive CD95 ligand (CD95L/FasL). When the Nagata team identified CD95L (Suda et al. 1993), they indeed found CD95L to be mutated in gld mice (Suda and Nagata 1994; Takahashi et al. 1994).
But what caused the aberrant phenotype in lpr and gld mice? This question was solved when the Krammer team, as well as teams led by Green in San Diego, Marshak-Rothstein in Boston, and Lynch in Seattle discovered that CD95 and its ligand mediated activation-induced cell death (AICD) in T cells (Alderson et al. 1995; Brunner et al. 1995; Dhein et al. 1995; Ju et al. 1995). Thus, failure to undergo AICD results in accumulation of aberrant T cells in gld and lpr mice. Teams led by Zinkernagel and Tschopp discovered that CD95L was also responsible for the long-sought-after perforin-independent cytotoxic activity of killer T cells (Lowin et al. 1994a,b; Berke 1995; Ehl et al. 1996). Hence, the CD95 system plays crucial roles in immune homeostasis and killer T-cell activity. The realization of the latter was promising because research on CD95 was of course initially fueled by the hope that CD95 agonists could be used in cancer therapy. However, when animals were treated systemically with antibodies to CD95 or recombinant CD95L that could interact with the receptors in the treated species, rather than only with receptors on the injected xenogeneic tumor cells, this induced massive hepatocyte apoptosis, resulting in fulminant hepatitis and acute death of the animals (Ogasawara et al. 1993).
Obviously, this was as disappointing a finding regarding the possibility of using CD95 agonists in cancer therapy as the previous realization that TNF could not be used for systemic cancer therapy. Yet, like with TNF, it turned out to mark the beginning of an entirely different but equally exciting field of research. If CD95 agonists could exert such drastic toxic effects in vivo, was it possible that CD95L-induced apoptosis could be involved in acute tissue damage that occurs in various pathological conditions? Since then, ample evidence has been gathered for the involvement of the CD95 system in various situations of acute tissue damage including graft-versus-host disease (Baker et al. 1996; Braun et al. 1996; Via et al. 1996a,b; Miwa et al. 1999) and some (Galle et al. 1995; Kondo et al. 1997; Strand et al. 1998, 2004), but not all, forms of acute liver damage (Oyaizu et al. 1997). CD95L has also been reported to be implicated in tissue damage following acute myocardial infarction (Jeremias et al. 2000; Lee et al. 2003), stroke (Vogt et al. 1998; Martin-Villalba et al. 1999), and spinal cord injury (Demjen et al. 2004; Ackery et al. 2006). These findings suggest that CD95L inhibitors may be useful therapeutics in situations in which normal tissue is acutely damaged.
An additional application of CD95L inhibitors emerged from a completely different line of research. Shortly after the discovery of the CD95 system, first reports appeared that CD95 stimulation does not always induce apoptosis but can also stimulate proliferation (e.g., in CD3-activated T cells) (Alderson et al. 1993, 1994, 1995; Desbarats et al. 1999; Kennedy et al. 1999), fibroblasts (Freiberg et al. 1997), and hepatocytes following partial hepatectomy (Desbarats and Newell 2000). Intriguingly, certain cancer cells also responded to CD95 stimulation by increasing their proliferation (Owen-Schaub et al. 1993) or their motility and invasiveness (Barnhart et al. 2004). Recently, Peter and colleagues found that the protumorigenic role of CD95 seems to be a more general principle across many cancers (Chen et al. 2010). Intriguingly, on the basis of the biochemical tool we had generated to understand the role of the CD95-CD95L interaction in AICD of T cells (Dhein et al. 1995) and results obtained in a preclinical glioblastoma model (Kleber et al. 2008), a first biotherapeutic CD95L blocker was recently successfully tested in a phase II study in glioblastoma patients.
It will be fascinating to see how clinical testing of CD95L inhibition in cancer, but also the various other diseases outlined above, will unfold. It appears though that, as with TNF, the medical applications in the CD95 system are significantly more likely to derive from antagonizing the ligand, rather than stimulating the receptor.
THE TRAIL APOPTOSIS SYSTEM AND ITS POTENTIAL FOR CANCER THERAPY
Shortly after it became clear that also CD95L could not hold TNF’s promise, a new TNF family member with an interesting activity profile was discovered. In 1995, Wiley and colleagues at Immunex in Seattle and Ashkenazi and colleagues at Genentech in San Francisco, independently found an expressed sequence tag (EST) in the public database with homology to CD95L, which was even annotated as such. The TNF-related apoptosis-inducing ligand (TRAIL), or Apo2L, as these groups named it, respectively, killed many cancer, but not normal, cells in vitro (Wiley et al. 1995; Pitti et al. 1996). So, was TRAIL going to finally fulfill TNF’s promise?
In 1999, we at Immunex and Ashkenazi’s team at Genentech found that systemic treatment of tumor-bearing mice with recombinant TRAIL killed tumor cells in vivo (Ashkenazi et al. 1999; Walczak et al. 1999). Importantly, a highly active form of TRAIL, capable of inducing apoptosis in mouse cells, neither induced a systemic shock syndrome, as seen with TNF, nor fulminant toxic effects owing to apoptosis induction, as observed with CD95L (Walczak et al. 1999). This meant that with TRAIL a TNF-like cytokine could be used systemically to kill tumor cells without being toxic.
Given TRAIL’s potential to induce apoptosis in cancer but not normal cells, the receptor responsible for mediating this activity appeared to be an attractive target for the development of agonists capable of cross-linking it. The result of the race for the cloning of this receptor was quite surprising as TRAIL has five receptors of which two can mediate apoptosis.
At the time, in 1996 and 1997, many new human genes with homology to already known members of interesting protein families, like TRAIL itself, were found in EST databases. The EST “data mining” approach of a privately owned database led to the discovery of the first apoptosis-inducing receptor for TRAIL, now referred to as TRAIL-R1 or DR4, by the Dixit team (Pan et al. 1997a). Using a biochemical purification approach with TRAIL as bait we identified a different apoptosis-inducing receptor for TRAIL, TRAIL-R2 (Walczak et al. 1997). Shortly thereafter, TRAIL-R2 was also discovered by a number of other groups when its sequence appeared in public and private EST databases, and it received several other names including DR5, TRICK2, and KILLER (Pan et al. 1997a; Screaton et al. 1997a; Sheridan et al. 1997; Wu et al. 1997). In the following weeks and months, work by a number of groups led to the identification of two additional receptors for TRAIL, TRAIL-R3 (DcR1) and TRAIL-R4 (DcR2), which cannot induce apoptosis because they lack an intracellular DD (reviewed in Walczak and Krammer 2000). At first, these receptors were thought to exert a decoy function for TRAIL (hence the name “decoy receptor” [DcR]) by being expressed on normal but not cancer cells. When the expression patterns of the TRAIL-Rs were analyzed, it became clear, however, that this concept did not hold. To this day we know little about the physiological function of these nondeath receptors for TRAIL. Finally, osteoprotegerin (OPG), a soluble, high-affinity receptor for RANKL, another member of the TNFSF, was found to interact with TRAIL, yet with lower affinity. It is rather unlikely that this interaction is relevant in vivo because mice overexpressing TRAIL do not show any bone-related phenotype, which would have been expected if TRAIL were capable of interacting with OPG in vivo, because in mice deficient for OPG, osteoclastogenesis is not inhibited and these mice therefore develop a strong bone-related phenotype (Simonet et al. 1997). In summary, TRAIL turned out to be the most promiscuous TNFSF member.
TRAIL-R1 and TRAIL-R2 share 58% sequence homology and so far it has not been possible to identify significant distinct functions of one receptor versus the other. They both trigger apoptosis via the same pathway and, at present, even no differences to the CD95-engaged pathway are known. The big quest, therefore, is to identify the biochemical basis for the diametrically different outcome of the triggering of the TRAIL-Rs versus CD95 in normal cells, as the unraveling of this may lead the way to more targeted and patient-tailored applications of TRAIL in cancer treatment.
Based on the encouraging preclinical results with recombinant TRAIL in vivo (Ashkenazi et al. 1999; Walczak et al. 1999), Immunex and Genentech joined forces to explore the clinical potential of a first recombinant form of TRAIL in cancer treatment. Even though some clinical benefit has been detected in the various clinical trials in which this form of TRAIL has now been tested, the results of these trials were not nearly as striking as initially hoped. Apart from this form of TRAIL, “agonistic” antibodies to TRAIL-R1 and TRAIL-R2 have now been tested clinically (reviewed in Newsom-Davis et al. 2009).
Overall, the results of these studies are, however, disappointing. One reason for this is the absence of biomarkers to select patients who are likely to respond to TRAIL receptor agonist therapy, either alone or in combination with a particular TRAIL-sensitizing drug. To effectively use TRAIL-comprising cancer therapies, it will be crucial, however, to identify such biomarkers.
A perhaps more important reason for the lack of clear clinical activity of the currently developed TRAIL receptor agonists likely lies in their very nature, which simply renders them rather weak inducers of apoptosis. The recombinant form of TRAIL that has been developed clinically displays rather low apoptosis-inducing activity when compared with higher activity forms, perhaps also because the formulation of this ligand may not exactly match the biological moiety. Even though TRAIL had been suggested to be hepatotoxic (Lawrence et al. 2001), which may explain the hesitation to develop a more active form of TRAIL, we found that highly active forms of TRAIL are neither toxic to mice (Walczak et al. 1999) nor to primary human hepatocytes, even in combination with various TRAIL-sensitizing drugs (Ganten et al. 2005, 2006), suggesting that development and clinical application of high-activity forms of TRAIL and other TRAIL receptor agonists should be possible.
With respect to the antibodies, the failure likely has a different cause. Antibodies of the immunoglobulin G (IgG) type contain two antigen-binding sites. Triggering of TNFRSF members, however, requires cross-linking of at least three receptors for agonistic activity. Hence, IgG antibodies cannot do this without a cross-linking platform. This was first shown by the Krammer team for antibodies to CD95 (Dhein et al. 1992) and recently by the Genentech team for their clinically developed TRAIL-R2-specific antibody (Wilson et al. 2011).
As disappointing as it is that more than a decade after the demonstration of the tumor-specific apoptosis-inducing potential of a highly active form of TRAIL in vivo (Walczak et al. 1999) we still do not have a high-activity TRAIL receptor agonist in clinical use, it seems that this issue could soon be overcome with second-generation, high-activity TRAIL receptor agonists currently being bound for the clinic. It will be exciting to see how they fare, and whether TRAIL will finally be able to hold the promise that TNF and CD95L could not keep.
CD95- AND TRAIL-INDUCED APOPTOSIS
A prime goal for scientists working on CD95 was to elucidate how receptor cross-linking led to apoptosis induction in target cells. Nagata and colleagues made the first step by showing that apoptosis induction by CD95 required a part of the cytoplasmic domain, the DD (Itoh and Nagata 1993). They, as well as Goeddel and colleagues (Tartaglia et al. 1993), identified a similar DD in TNF-R1.
Discovery of FADD, Caspase-8, and the Death-Inducing Signaling Complex
Using yeast two-hybrid (Y2H), Dixit’s team in Ann Arbor and Wallach’s team in Rehovot discovered a protein they called FADD and MORT1, respectively, as a CD95-DD-interacting factor (Boldin et al. 1995a,b; Chinnaiyan et al. 1995). Apart from a DD that interacted with the CD95-DD, FADD also contained a second, DD-like domain, termed death-effector domain (DED). In the meantime, Kischkel and colleagues from the team led by Krammer and Peter biochemically studied the signaling complex that forms when CD95 is cross-linked to induce apoptosis. They dubbed this complex death-inducing signaling complex (DISC) and found that FADD and one additional, unidentified factor formed part of the CD95 DISC (Kischkel et al. 1995). Collaborating with Mann’s team in Heidelberg, they determined the identity of the elusive second factor and, together with Dixit’s team, were able to show how it works (Muzio et al. 1996). The identified factor was homologous to IL-1-converting enzyme (ICE) (Cerretti et al. 1992; Thornberry et al. 1992). CED-3, one of the key factors identified by Horvitz and colleagues in what later became a Nobel Prize-winning effort to define the genes required for cell death in Caenorhabditis elegans, was also homologous to ICE (Yuan and Horvitz 1990; Yuan et al. 1993). Consequently, the protein was named FLICE for FADD-like ICE. FLICE is FADD-like because it contains two DEDs that are homologous to the DED of FADD. The same protein was discovered by Wallach’s team in a Y2H screen with the DED of MORT1 (FADD). They named the protein MACH (Boldin et al. 1996). The protein formerly known as FLICE or MACH is now referred to as caspase-8. Caspases are cysteine-dependent aspartate-directed proteases (i.e., proteases that cleave target proteins at aspartic acid residues and use cysteine in their active center). There are 14 mammalian caspases (11 in humans and 10 in rodents) which, interestingly, have been classified as “apoptotic” or “inflammatory” (Creagh and Martin 2001; Salvesen and Riedl 2008; Pop and Salvesen 2009).
The mechanism of DISC formation and CD95-induced apoptosis identified is as follows: cross-linking of CD95 results in recruitment of FADD, which in turn recruits caspase-8 to form the DISC. Caspase-8 is then activated at the DISC, resulting in cleavage of downstream substrates. One of them is caspase-3, the mammalian homolog of CED-3 and crucial effector caspase in the apoptosis pathway (Creagh and Martin 2001). Once caspase-3 is activated it cleaves many vital cellular proteins responsible for the characteristic biochemical and morphological hallmarks of apoptosis. Thus, the discovery of caspase-8 and its function at the DISC provided the missing link between CD95 cross-linking and the known part of the apoptosis pathway.
In the following years the process of DISC formation and downstream as well as regulatory events of CD95-induced apoptosis were further refined. Besides FADD and caspase-8, two related proteins, the cellular FLICE-like inhibitory protein (cFLIP) and caspase-10 (Irmler et al. 1997; Thome et al. 1997; Kischkel et al. 2001; Sprick et al. 2002), which both also contain DEDs, are recruited to the CD95 DISC. Although cFLIP lacks a catalytic cysteine in its active center and therefore blocks CD95-induced apoptosis, caspase-10 is highly homologous to caspase-8. To this day, however, the function of caspase-10 remains unclear (Kischkel et al. 2001; Sprick et al. 2002). Recently, the stoichiometry of the CD95 DISC was addressed. Surprisingly, these studies proposed that three receptors recruit one FADD protein and this FADD molecule recruits six to 10 DED-containing proteins (Fig. 2) (Dickens et al. 2012; Schleich et al. 2012).
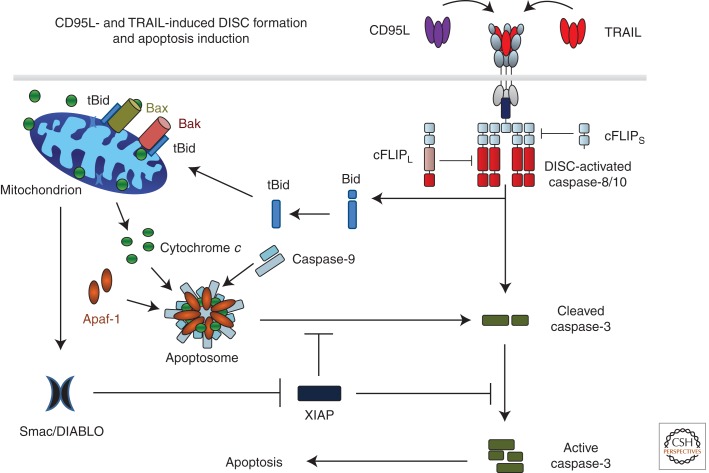
CD95L- and TRAIL-induced DISC formation and apoptosis induction. Binding of CD95L or TRAIL to their cognate receptors leads to receptor trimerization and formation of the death-inducing signaling complex (DISC). The DD of one FADD molecule interacts with the DDs of the three cross-linked receptors. Subsequently, procaspases 8 and 10 are recruited by interaction of their DED with that of FADD. Heterodimers between cFLIPL and caspase-8 or -10 are not inactive as proteolytic enzymes but their proteolytic activity is edited as compared with caspase-8 or -10 homodimers. cFLIPS in turn inhibits caspase activity at the DISC by preventing dimerization of caspase-8/10/cFLIPL. Interestingly, one FADD molecule recruits six to 10 DED-containing caspase-8, -10, and cFLIP proteins. DISC-activated caspase-8/-10 cleaves caspase-3, enabling autoactivation of caspase-3, which renders the enzyme fully active. This latter step can, however, be blocked by X-linked inhibitor of apoptosis protein (XIAP). DISC-activated caspase-8/-10 also cleaves Bid (tBID). Interaction of tBid with Bak and Bax in the mitochondrial outer membrane induces Bax/Bak oligomerization, resulting in mitochondrial outer-membrane permeabilization (MOMP) so that cytochrome c and Smac/DIABLO are released from the mitochondrial intermembrane space to the cytosol. Cytochrome c, together with Apaf-1 and caspase-9, forms the apoptosome, the activation platform for caspase-9, which can, however, also be inhibited by XIAP. Smac/DIABLO binds to XIAP, which releases caspase-3 and -9 from XIAP-imposed inhibition. Activation of these caspases enables execution of apoptotic cell death.
Connecting the Death Receptor and the Mitochondrial Apoptosis Pathways
The proapoptotic BH3-only family member Bid was identified as a second critical substrate of caspase-8 for engagement of the cellular apoptosis machinery (Gross et al. 1999; Yin et al. 1999). Caspase-8-cleaved, truncated Bid (tBid) translocates from the cytosol to mitochondria where it induces mitochondrial outer membrane permeabilization (MOMP), thereby activating the mitochondrial apoptosis pathway. These processes are discussed in detail in Green (2013). In the context of this work, however, there are two crucial points about these processes: (1) cleavage of Bid by caspase-8 provides the missing link between the death receptor and mitochondrial apoptosis pathways, and (2) MOMP does not only induce release of cytochrome c but also of the second mitochondrial activator of caspases (SMAC, also known as DIABLO). Whereas cytochrome c release triggers apoptosome formation and activation of caspase-9, cytosolic SMAC binds to, and thereby neutralizes, the X-linked inhibitor of apoptosis protein (XIAP). XIAP, in turn, normally binds to, and thereby inhibits, caspase-8-cleaved caspases 3 and 9. Hence, SMAC releases these caspases from their XIAP-imposed inhibition and cell death can finally ensue (Fig. 2).
Requirement for tBid-induced MOMP distinguishes so-called type I from type II cells. Type I cells do not require MOMP to undergo CD95-induced apoptosis—even though it may nevertheless occur—whereas type II cells do (reviewed in Peter and Krammer 1998). In cells expressing a high XIAP/caspase-3 ratio, full activation of caspase-3 following caspase-8-mediated cleavage is blocked. Therefore, XIAP is a crucial factor for categorizing cells as type I or type II regarding CD95-mediated apoptosis (Jost et al. 2009). Differences in CD95 DISC formation, first thought to be solely responsible for the type I/type II distinction (reviewed in Peter and Krammer 1998), likely contribute to this categorization through different caspase activity outputs of the DISC in type I versus type II cells. Hence, the balance between DISC-generated caspase activity and XIAP/caspase-3 ratio is decisive for categorization of a given cell as type I or type II.
TRAIL-Induced Apoptosis
Once the TRAIL receptors were cloned, the pathway of TRAIL-induced apoptosis was investigated. Despite the initial claim that TRAIL would not use FADD as adaptor (Marsters et al. 1996; Pan et al. 1997b), we and others showed that both TRAIL-R1 and TRAIL-R2 use FADD and caspase-8 (Walczak et al. 1997; Kischkel et al. 2000, 2001; Sprick et al. 2000, 2002), just like CD95 (Fig. 2). It has been a conundrum that agonists of CD95 are so toxic, whereas even highly active TRAIL receptor agonists are not (Walczak et al. 1999), even when combined with TRAIL-apoptosis-sensitizing drugs (Ganten et al. 2005, 2006; Koschny et al. 2007). Solving this conundrum will be an important step forward in understanding the biology of death receptors in general but particularly for the use of TRAIL receptor agonists in cancer therapy.
TNF-/TNF-R1-INDUCED GENE ACTIVATION AND CELL DEATH
TNF has two cellular receptors. Whereas TNF-R1 is expressed in many tissues, TNF-R2 is almost exclusively present on lymphoid and endothelial cells. TNF-R1 contains a DD, whereas TNF-R2 does not. TNF-R1 initiates the majority of TNF-induced biological activities, including induction of cell death (Wajant et al. 2003).
The TNF-R1 Signaling Complex (TNF–RSC)
Binding of TNF to TNF-R1 triggers a series of intracellular events, but the primary output is activation of NF-κB and the mitogen-activated protein kinases (MAPKs) c-Jun amino-terminal kinase (JNK) and p38. The resulting gene activation is crucial for the inflammatory response to infection. Induction of cell death by TNF is a secondary signal that is only triggered when the gene-activatory signals are too weak or otherwise perturbed.
Binding of TNF to TNF-R1 induces receptor oligomerization and formation of the TNF-R1 signaling complex (TNF–RSC), which is initiated by recruitment of the DD-containing proteins TRADD and RIP1 to the DD of the receptor (Fig. 3). TRADD then recruits TRAF2, which in turn serves as a recruitment platform for cIAP1 and cIAP2. cIAPs are E3 ubiquitin ligases and, following their recruitment to the TNF–RSC, they polyubiquitinate RIP1, which enables activation of NF-κB and MAPKs. Ubiquitin chains can be formed via linkages of the ubiquitin subunits on different ε-amino groups of the seven lysines present in ubiquitin or via the α-amino group at the amino terminus of ubiquitin, the latter creating linear ubiquitin chains (Rieser et al. 2012). Until recently, it was thought that polyubiquitin chains involved in TNF signaling are either linked via the ε-amino groups of lysine 63 (K63) or K48 of ubiquitin. Recently, however, we showed that linear ubiquitin chains are also present on components of the TNF–RSC and are crucial in providing the physiological TNF signaling output (Gerlach et al. 2011). A protein complex termed LUBAC, for linear ubiquitin chain assembly complex, is recruited to the TNF–RSC in a cIAP-activity-dependent manner and required for activation of NF-κB and MAPKs to the full physiologically required extent. This is most likely owing to the fact that NEMO binds more strongly to linear than K63-linked ubiquitin chains (Lo et al. 2009; Rahighi et al. 2009). Besides linear and K63 ubiquitin linkages, we also found K11 and K48 linkages on RIP1 in the native TNF–RSC (Gerlach et al. 2011). Hence, different forms of ubiquitin linkages cooperate to achieve the physiological signaling output of the TNF–RSC. This suggested the following model: Rather than one long K63-linked chain on RIP1, differently linked—possibly rather short—chains, which are placed in exact positions on various TNF–RSC components, enable precise construction of this protein complex, regarding both positioning of recruited functional units and timing of complex construction and deconstruction (Walczak 2011). Thereby, the functional units of the NEMO/IKK and TAB/TAK complexes are recruited at exact times and into predefined positions within the complex so that the TNF–RSC works as designed and gene expression by NF-κB and AP-1 is activated exactly as intended and physiologically required (Fig. 3).
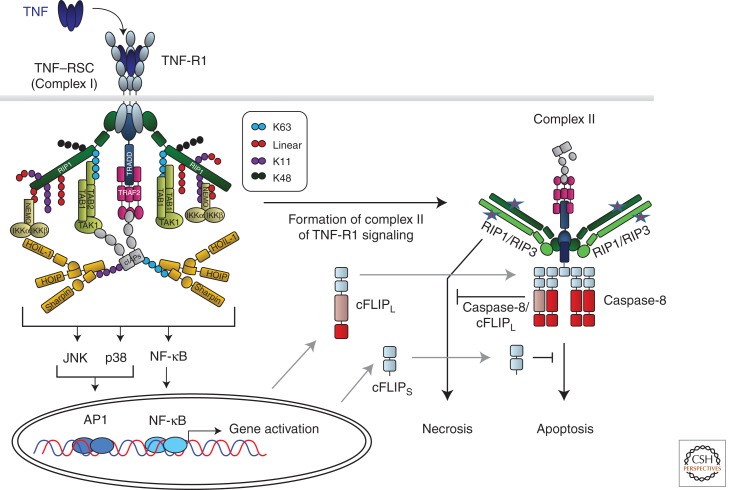
TNF-R1-induced gene activation and cell death signaling. Cross-linking of TNF-R1 by TNF results in formation of the TNF-R1 signaling complex (TNF–RSC). Cross-linked TNF-R1 recruits TRADD and RIP1 to the DD of the receptor. Subsequently, TRADD recruits TRAF2, which in turn provides the platform for cIAP1/2. cIAPs then place ubiquitin chains, linked via different interubiquitin linkages, on various TNF–RSC components. cIAP-Mediated ubiquitination is required to recruit LUBAC. Once recruited, LUBAC places linearly linked ubiquitin linkages on RIP1 and NEMO. Together, the different cIAP- and LUBAC-generated ubiquitin chains, placed in defined positions and sequences on specific components of the TNF–RSC, enable the physiologically required gene-activatory capacity of this complex by mediating the exact positioning of both the IKK and TAB/TAK complexes in the TNF–RSC. The different ubiquitin linkages are indicated in different colors. The depicted chain lengths, the sequence of the different linkages in them, and their exact positioning on different TNF–RSC components are only shown as examples in this model of the TNF–RSC as they are currently mostly unknown. Most likely involving the action of deubiquitinases (DUBs; not depicted here), the TNF–RSC releases TRADD, together with RIP1 and other cytoplasmic constituents of the complex, into the cytosol. This secondary complex, complex II, recruits FADD, caspase-8/10, and, when expressed, the different isoforms of cFLIP and RIP3. RIP1/3-induced necrosis from complex II is counteracted by the activity of the caspase-8/cFLIPL heteromer, and FADD/caspase-8-mediated apoptosis by cFLIPS and possibly also by cFLIPL. Depending on the relative presence of the components in complex II, it can therefore either initiate FADD/caspase-8-dependent apoptosis or RIP1/RIP3-kinase-activity-dependent necrosis, or, when cFLIP and perhaps other, currently unknown inhibitory factors are present in the complex at sufficiently high levels, its cell-death-inducing capacity may be entirely inhibited.
Disassembly of the complex is mainly mediated by so-called deubiquitinases (DUBs), enzymes that cleave ubiquitin from target proteins. It appears that certain DUBs, including the TNF–RSC-recruited A20, Cezanne, and CYLD, act in a ubiquitin-linkage-specific manner (i.e., they cleave certain types of interubiquitin linkages but not others) (Komander et al. 2009b). Their recruitment occurs later than that of the E3s and the functional units responsible for activating gene induction in response to TNF. At least with respect to A20, TNF-induced gene activation is decisive for the appearance of this protein in the complex (Gerlach et al. 2011). We are, however, only beginning to understand which DUBs cleave which interubiquitin linkages (Komander et al. 2009a), let alone in the context of particular RSCs or on specific components thereof. Likely, exact positioning of DUBs within RSCs and the timing thereof will be crucial.
Complex II of TNF-R1 Signaling: Cell Death in Two Flavors
When the TNF–RSC is deconstructed it does not disintegrate into its individual constituents. Micheau and Tschopp discovered that, following formation of the TNF–RSC, a second complex appears intracellularly that contains TRADD and some, but not all, of the other cytoplasmic components of the TNF–RSC. Additionally the cytoplasmic DISC components FADD, caspase-8, caspase-10, and cFLIP are also recruited to complex II of TNF-R1 signaling (Micheau and Tschopp 2003). It is, hence, an “intracellular DISC” whose formation temporally follows TNF–RSC (complex I) formation, and which generates the cell-death-inducing signals of TNF. However, when complex I works properly, complex II does not get to induce cell death because its signaling output is counteracted by the gene-activatory signals generated at complex I as these increase expression of prosurvival proteins, including the two isoforms of cFLIP, cFLIPL, and cFLIPS. Thus, if complex I is not decomposed prematurely and its output reaches physiological levels, complex II-induced cell death is prevented (Fig. 3).
It has long been known that TNF cannot only induce apoptotic but also necrotic cell death (reviewed in Vandenabeele et al. 2010). It emerged only recently, however, that TNF-induced necrosis is the result of a regulatable process that requires the kinase activities of both RIP1 and RIP3 (Holler et al. 2000; Hitomi et al. 2008; Cho et al. 2009; He et al. 2009; Zhang et al. 2009). For a long time it was a mystery why deficiency in caspase-8 or FADD is embryonically lethal if all they did was induce apoptosis. Recently, however, a plausible explanation was offered: Concomitant deletion of RIP3 or RIP1 rescued mice deficient for caspase-8 or FADD from embryonic lethality (Kaiser et al. 2011; Oberst et al. 2011; Zhang et al. 2011). This means, on the one hand, that RIP1 and RIP3 mediate lethality induced by deficiency in caspase-8 or FADD, but on the other, that the presence of caspase-8 and FADD prevents this. Thus, besides their role in apoptosis induction, FADD and caspase-8 also act together to prevent RIP1/RIP3-induced lethality. Interestingly, cFLIP deficiency is also lethal (Yeh et al. 2000). This lethality could only be reversed by concomitant deficiency in RIP3 and FADD but not when either factor was absent individually (Dillon et al. 2012). Thus, cFLIP is required for caspase-8 and FADD to block RIP1/RIP3-induced lethality (van Raam and Salvesen 2012), but also to interfere with caspase-8-induced lethality. It appears that the different isoforms of cFLIP may play distinct roles in these processes as cFLIPS interferes with apoptosis and cFLIPL with necrosis and, at least at high expression levels, also with apoptosis (Kavuri et al. 2011). Hence, RIP1/RIP3- and caspase-8/FADD/cFLIP-mediated signals control each other at complex II. Interestingly, recent studies have shown that inflammation in mice deficient for caspase-8 or FADD in either skin or gut (Kovalenko et al. 2009), does not occur when RIP3 is absent (Welz et al. 2011; Gunther et al. 2012). Whether this is solely because of prevention of RIP3-induced necrosis, or whether other RIP3-mediated signals may also play a role in preventing inflammation induced by deficiency in DISC components (Vince et al. 2012; Wallach et al. 2012), remains to be resolved.
In summary, our current model of the cell-death-inducing activities at complex II is as follows: FADD and caspase-8 induce apoptosis, which is prevented in the presence of sufficient amounts of cFLIP that are also required, together with FADD as adaptor and caspase-8 as an enzymatic partner, to interfere with RIP1/3-induced necrosis. There must be a fine balance between the two types of cell death induced by this complex, and the expression levels of cFLIP isoforms likely play an important part in tipping it one way or the other (Fig. 3).
INTEGRATING CELL DEATH AND INFLAMMATION
TRAIL and CD95L cannot only induce apoptosis but also proinflammatory, proliferative, and promigratory signals (Ehrhardt et al. 2003; Budd et al. 2006; Peter et al. 2007; Wilson et al. 2009; Lemke et al. 2010; Roder et al. 2011). Some of these signals occur in a FADD/caspase-8-independent manner but the mechanisms are still debated (Green 2010). One suggestion has been that the Src-family kinase Yes and the p85 subunit of PI3 kinase associate with the membrane-proximal portion of CD95’s cytoplasmic domain on cross-linking in glioblastoma cells and that the resulting activation of the PKB/AKT pathway could be responsible for the protumorigenic signals induced by CD95L in glioblastoma (Sancho-Martinez and Martin-Villalba 2009; Parrish et al. 2013).
More is known about the DD-requiring, nonapoptotic signals induced by CD95L or TRAIL. Interestingly, Ashkenazi and colleagues found that release of FADD from the TRAIL DISC triggers formation of an intracellular complex II by recruiting TRAF2, cIAP1/2, RIP1, NEMO, and most likely several other factors required for activation of NF-κB, MAPKs, and consequently, gene induction (Varfolomeev et al. 2005). Also in this case induction of signals from complex II does not occur, or at least only to a limited extent, when the primary signal induced by complex I, in this case the TRAIL DISC, results in cell death. This suggests the following model: Although the output of the primary complex of TNF-R1 signaling is gene activation, its second complex can induce two rather different forms of cell death. In the case of CD95 and TRAIL, the signaling output of the DISC is apoptosis, whereas that of complex II is gene activation (Fig. 4).

Comparison of CD95/TRAIL-R1/R2 and TNF-R1/DR3 signaling. For both the proapoptotic CD95 and TRAIL systems as well as the proinflammatory TNF and DR3 systems, the complex defined as complex I is the protein complex that forms at the plasma membrane and exerts the primary function of the respective receptor (i.e., apoptosis for CD95 and TRAIL-R1/R2 and gene activation via NF-κB and MAPK by TNF-R1 and DR3). The two primary complexes dissociate from the DD of the respective receptor and recruit additional proteins from the cytosol to form complex II, which triggers the respective secondary signal. In the case of CD95 and TRAIL-R1/R2 the second signal is gene activation via the NF-κB and MAPK pathways; in the case of TNF-R1/DR3 it is induction of necrosis or apoptosis. The signaling outputs of the respective secondary complexes are prevented or attenuated in case the respective primary complexes reach theirs.
Why are specific tasks separated into different, sequentially acting signaling complexes? One explanation could be that, when the first signal prevails, the second one is not required or should at least be minimized. If, however, the primary signal does not reach its intended end point, signaling by the second complex is initiated. If a physiological extracellular stimulus (i.e., TRAIL or CD95L) triggers the apoptosis machinery in a cell but it does not die, this should alarm the organism because infectious agents often inhibit apoptosis. Hence, an alarm signal capable of stimulating immunity should be induced now. By activating proinflammatory gene induction, and perhaps necrosis, complex II of the CD95 and TRAIL systems can do exactly that (Fig. 4).
In the case of TNF-R1, and most likely DR3, it is the other way around. When proper gene activation cannot be achieved by complex I, complex II induces cell death. TNF-induced cell death can either be apoptotic or necrotic. It is unclear which type of TNF-induced cell death prevails when its gene-activatory pathway is perturbed, and it is likely that this differs with the type of perturbation. It is, however, likely that there will be an inflammatory component in it. Thereby, a biological outcome similar to the originally intended one could be achieved (i.e., the creation of a proinflammatory environment), yet by going down a very different path than originally intended (Fig. 4).
Interestingly, inflammation occurs in both autoimmunity and cancer. TNF has been shown to be intimately involved in both processes (Balkwill 2009; Taylor and Feldmann 2009). Yet, whereas autoimmunity-associated inflammation is immunostimulatory, cancer-related inflammation suppresses immunity. It is tempting to speculate that the two cell death modalities induced by TNF and its relatives, together with their gene-activatory capacity, are decisive in determining whether inflammation will stimulate or suppress immunity. Whatever the outcome will be, it is now clear that cell death and inflammation are more closely linked than previously thought and in fact seem to represent two sides of the very same coin.
CONCLUDING REMARKS AND OUTLOOK
Studying the TNF, CD95, and TRAIL death receptor ligands has provided tremendous insight into the biochemistry and function of cell death and inflammation. Importantly, however, this research has also yielded three classes of drugs: TNF blockers, CD95L inhibitors, and TRAIL receptor agonists. TNF blockers have transformed the treatment of chronic inflammatory diseases. CD95L inhibitors and TRAIL receptor agonists have now entered the clinic and it will be exciting to see how their clinical potential unfolds in the future.
The understanding of death receptor-mediated signaling pathways has been at the forefront of many fascinating developments in cell biology, most importantly in the fields of cancer, inflammation, and cell death. Among the promising future areas in death receptor signaling are the specific sensitization of cancer cells to TRAIL, the elucidation of the pathways of regulated necrosis and CD95-mediated protumorigenic signaling, the deciphering of the ubiquitin code in death receptor signaling, and the reciprocal regulation of apoptosis and necrosis. It is likely that studies into these processes will, at times, provide surprising results. That, however, comes as no surprise because the journey from Coley’s discovery of a tumor-necrotizing activity to today’s research on the biology of death receptor–ligand systems has been packed with surprises, making this journey an exciting one that promises to continue for some time to come.
Footnotes
Editors: Eric H. Baehrecke, Douglas R. Green, Sally Kornbluth, and Guy S. Salvesen
Additional Perspectives on Cell Survival and Cell Death available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
Articles from Cold Spring Harbor Perspectives in Biology are provided here courtesy of Cold Spring Harbor Laboratory Press
Full text links
Read article at publisher's site: https://doi.org/10.1101/cshperspect.a008698
Read article for free, from open access legal sources, via Unpaywall:
http://cshperspectives.cshlp.org/content/5/5/a008698.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1101/cshperspect.a008698
Article citations
From Crypts to Cancer: A Holistic Perspective on Colorectal Carcinogenesis and Therapeutic Strategies.
Int J Mol Sci, 25(17):9463, 30 Aug 2024
Cited by: 0 articles | PMID: 39273409 | PMCID: PMC11395697
Review Free full text in Europe PMC
MicroRNAs in Hepatocellular Carcinoma Pathogenesis: Insights into Mechanisms and Therapeutic Opportunities.
Int J Mol Sci, 25(17):9393, 29 Aug 2024
Cited by: 0 articles | PMID: 39273339 | PMCID: PMC11395074
Review Free full text in Europe PMC
In-depth analysis of the interplay between oncogenic mutations and NK cell-mediated cancer surveillance in solid tumors.
Oncoimmunology, 13(1):2379062, 18 Jul 2024
Cited by: 0 articles | PMID: 39036370 | PMCID: PMC11259085
Review Free full text in Europe PMC
The Interplay between Obesity and Inflammation.
Life (Basel), 14(7):856, 08 Jul 2024
Cited by: 3 articles | PMID: 39063610 | PMCID: PMC11277997
Review Free full text in Europe PMC
Orchestrating Stress Responses in Multiple Sclerosis: A Role for Astrocytic IFNγ Signaling.
Int J Mol Sci, 25(14):7524, 09 Jul 2024
Cited by: 1 article | PMID: 39062765 | PMCID: PMC11276796
Review Free full text in Europe PMC
Go to all (118) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Receptor-mediated endocytosis is not required for tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis.
J Biol Chem, 282(17):12831-12841, 27 Feb 2007
Cited by: 79 articles | PMID: 17327223
Death receptor pathways mediate targeted and non-targeted effects of ionizing radiations in breast cancer cells.
Carcinogenesis, 30(3):432-439, 06 Jan 2009
Cited by: 41 articles | PMID: 19126655 | PMCID: PMC2650794
Cellular FLICE-inhibitory protein (cFLIP) isoforms block CD95- and TRAIL death receptor-induced gene induction irrespective of processing of caspase-8 or cFLIP in the death-inducing signaling complex.
J Biol Chem, 286(19):16631-16646, 22 Mar 2011
Cited by: 57 articles | PMID: 21454681 | PMCID: PMC3089506
Targeting death receptors to fight cancer: from biological rational to clinical implementation.
Curr Med Chem, 17(25):2713-2728, 01 Jan 2010
Cited by: 10 articles | PMID: 20586721
Review





