Abstract
Free full text

The suil suppressor locus in Saccharomyces cerevisiae encodes a translation factor that functions during tRNA(iMet) recognition of the start codon.
Abstract
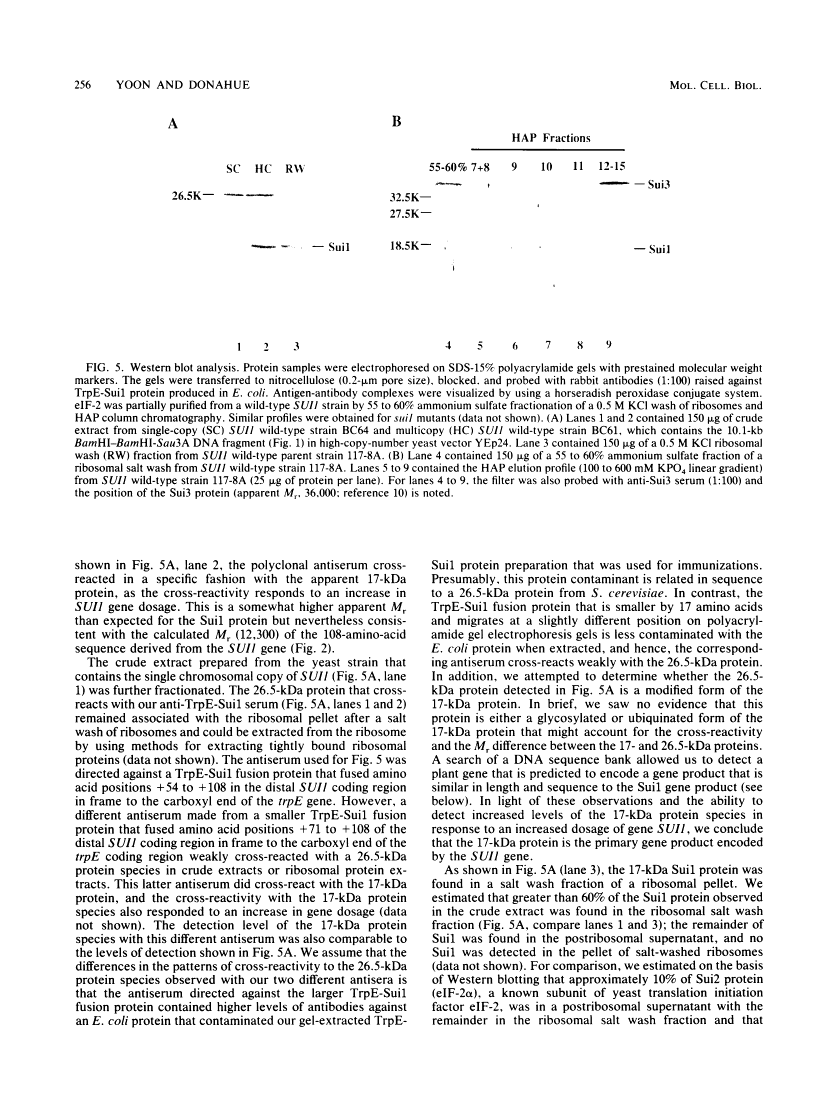
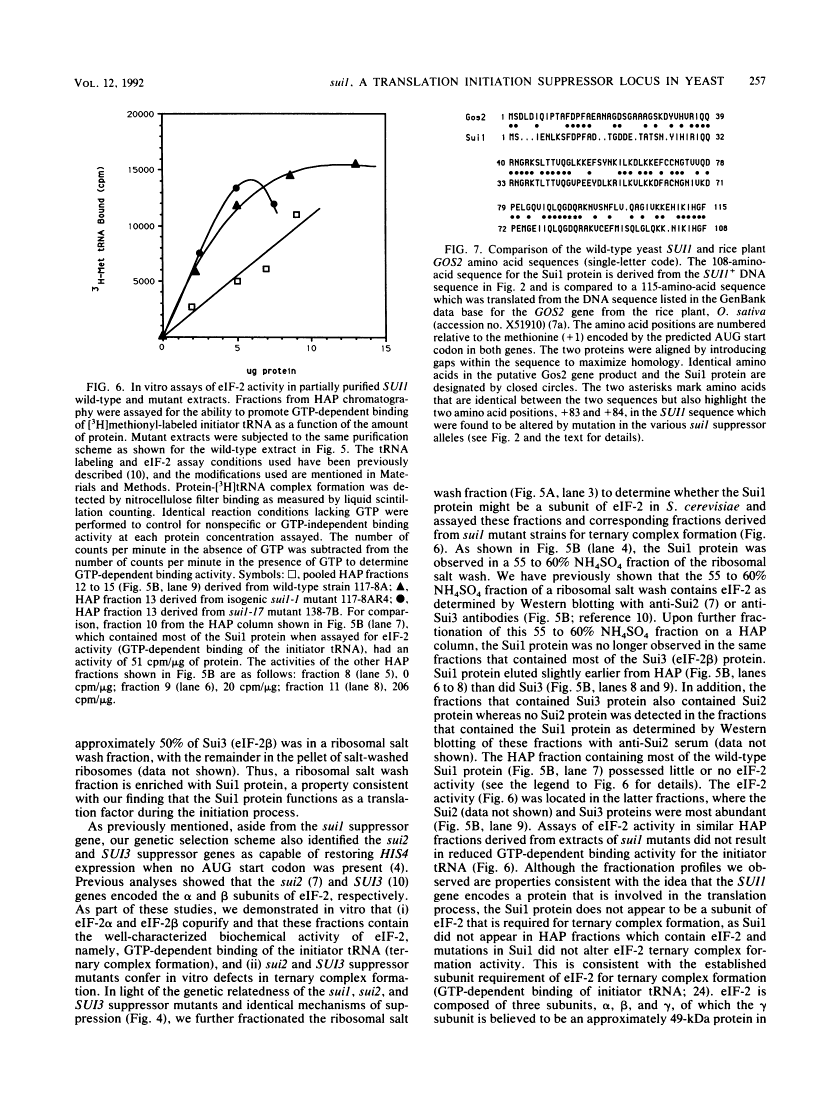
We initiated a genetic reversion analysis at the HIS4 locus to identify components of the translation initiation complex that are important for ribosomal recognition of an initiator codon. Three unlinked suppressor loci, suil, sui2, and SUI3, that restore expression of both HIS4 and HIS4-lacZ in the absence of an AUG initiator codon were identified. In previous studies, it was demonstrated that the sui2 and SUI3 genes encode mutated forms of the alpha and beta subunits, respectively, of eukaryotic translation initiation factor 2 (eIF-2). In this report, we describe the molecular and biochemical characterizations of the sui1 suppressor locus. The DNA sequence of the SUI1+ gene shows that it encodes a protein of 108 amino acids with a calculated Mr of 12,300. The sui1 suppressor genes all contain single base pair changes that alter a single amino acid within this 108-amino-acid sequence. sui1 suppressor strains that are temperature sensitive for growth on enriched medium have altered polysome profiles at the restrictive temperature typical of those caused by alteration of a protein that functions during the translation initiation process. Gene disruption experiments showed that the SUI1+ gene encodes an essential protein, and antibodies directed against the SUI1+ coding region identified a protein with the predicted Mr in a ribosomal salt wash fraction. As observed for sui2 and SUI3 suppression events, protein sequence analysis of His4-beta-galactosidase fusion proteins produced by sui1 suppression events indicated that a UUG codon is used as the site of translation initiation in the absence of an AUG start codon in HIS4. Changing the penultimate proline codon 3' to UUG at his4 to a Phe codon (UUC) blocks aminopeptidase cleavage of the amino-terminal amino acid of the His4-beta-galactosidase protein, as noted by the appearance of Met in the first cycle of the Edman degradation reaction. The appearance of Met in the first cycle, as noted, in either a sui1 or a SUI3 suppressor strain showed that the mechanism of suppression is the same for both suppressor genes and allows the initiator tRNA to mismatch base pair with the UUG codon. This suggests that the Sui1 gene product performs a function similar to that of the beta subunit of eIF-2 as encoded by the SUI3 gene. However, the Sui1 gene product does not appear to be a required subunit of eIF-2 on the basis of purification schemes designed to identify the GTP-dependent binding activity of eIF-2 for the initiator tRNA. In addition, suppressor mutations in the sui1 gene, in contrast to suppressor mutations in the sui2 or SUI3 gene, do not alter the GTP-dependent binding activity of the eIF-2. The simplest interpretation of these studies is that the sui1 suppressor gene defines an additional factor that functions in concert with eIF-2 to enable tRNAiMet to establish ribosomal recognition of an AUG initiator codon.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (2.9M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Click on the image to see a larger version.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahmad MF, Nasrin N, Bagchi MK, Chakravarty I, Gupta NK. A comparative study of the characteristics of eIF-2 and eIF-2-ancillary factor activities from yeast Saccharomyces cerevisiae and rabbit reticulocytes. J Biol Chem. 1985 Jun 10;260(11):6960–6965. [Abstract] [Google Scholar]
- Baim SB, Pietras DF, Eustice DC, Sherman F. A mutation allowing an mRNA secondary structure diminishes translation of Saccharomyces cerevisiae iso-1-cytochrome c. Mol Cell Biol. 1985 Aug;5(8):1839–1846. [Europe PMC free article] [Abstract] [Google Scholar]
- Burgin AB, Parodos K, Lane DJ, Pace NR. The excision of intervening sequences from Salmonella 23S ribosomal RNA. Cell. 1990 Feb 9;60(3):405–414. [Abstract] [Google Scholar]
- Castilho-Valavicius B, Yoon H, Donahue TF. Genetic characterization of the Saccharomyces cerevisiae translational initiation suppressors sui1, sui2 and SUI3 and their effects on HIS4 expression. Genetics. 1990 Mar;124(3):483–495. [Europe PMC free article] [Abstract] [Google Scholar]
- Cigan AM, Feng L, Donahue TF. tRNAi(met) functions in directing the scanning ribosome to the start site of translation. Science. 1988 Oct 7;242(4875):93–97. [Abstract] [Google Scholar]
- Cigan AM, Pabich EK, Donahue TF. Mutational analysis of the HIS4 translational initiator region in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jul;8(7):2964–2975. [Europe PMC free article] [Abstract] [Google Scholar]
- Cigan AM, Pabich EK, Feng L, Donahue TF. Yeast translation initiation suppressor sui2 encodes the alpha subunit of eukaryotic initiation factor 2 and shares sequence identity with the human alpha subunit. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2784–2788. [Europe PMC free article] [Abstract] [Google Scholar]
- Dieckmann CL, Tzagoloff A. Assembly of the mitochondrial membrane system. CBP6, a yeast nuclear gene necessary for synthesis of cytochrome b. J Biol Chem. 1985 Feb 10;260(3):1513–1520. [Abstract] [Google Scholar]
- Donahue TF, Cigan AM. Genetic selection for mutations that reduce or abolish ribosomal recognition of the HIS4 translational initiator region. Mol Cell Biol. 1988 Jul;8(7):2955–2963. [Europe PMC free article] [Abstract] [Google Scholar]
- Donahue TF, Cigan AM, Pabich EK, Valavicius BC. Mutations at a Zn(II) finger motif in the yeast eIF-2 beta gene alter ribosomal start-site selection during the scanning process. Cell. 1988 Aug 26;54(5):621–632. [Abstract] [Google Scholar]
- Donahue TF, Farabaugh PJ, Fink GR. The nucleotide sequence of the HIS4 region of yeast. Gene. 1982 Apr;18(1):47–59. [Abstract] [Google Scholar]
- Ernst H, Duncan RF, Hershey JW. Cloning and sequencing of complementary DNAs encoding the alpha-subunit of translational initiation factor eIF-2. Characterization of the protein and its messenger RNA. J Biol Chem. 1987 Jan 25;262(3):1206–1212. [Abstract] [Google Scholar]
- Gupta NK, Woodley CL, Chen YC, Bose KK. Protein synthesis in rabbit reticulocytes. Assays, purification, and properties of different ribosomal factors and their roles in peptide chain initiation. J Biol Chem. 1973 Jun 25;248(12):4500–4511. [Abstract] [Google Scholar]
- Hartwell LH, McLaughlin CS. A mutant of yeast apparently defective in the initiation of protein synthesis. Proc Natl Acad Sci U S A. 1969 Feb;62(2):468–474. [Europe PMC free article] [Abstract] [Google Scholar]
- Hershey JW. Translational control in mammalian cells. Annu Rev Biochem. 1991;60:717–755. [Abstract] [Google Scholar]
- Hewick RM, Hunkapiller MW, Hood LE, Dreyer WJ. A gas-liquid solid phase peptide and protein sequenator. J Biol Chem. 1981 Aug 10;256(15):7990–7997. [Abstract] [Google Scholar]
- Kozak M. How do eucaryotic ribosomes select initiation regions in messenger RNA? Cell. 1978 Dec;15(4):1109–1123. [Abstract] [Google Scholar]
- Kozak M. Evaluation of the "scanning model" for initiation of protein synthesis in eucaryotes. Cell. 1980 Nov;22(1 Pt 1):7–8. [Abstract] [Google Scholar]
- Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. [Abstract] [Google Scholar]
- Orr-Weaver TL, Szostak JW, Rothstein RJ. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 1983;101:228–245. [Abstract] [Google Scholar]
- Pathak VK, Nielsen PJ, Trachsel H, Hershey JW. Structure of the beta subunit of translational initiation factor eIF-2. Cell. 1988 Aug 26;54(5):633–639. [Abstract] [Google Scholar]
- Petes TD, Hereford LM, Skryabin KG. Characterization of two types of yeast ribosomal DNA genes. J Bacteriol. 1978 Apr;134(1):295–305. [Europe PMC free article] [Abstract] [Google Scholar]
- Rothstein RJ. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. [Abstract] [Google Scholar]
- Nomenclature of initiation, elongation and termination factors for translation in eukaryotes. Recommendations 1988. Nomenclature Committee of the International Union of Biochemistry (NC-IUB). Eur J Biochem. 1989 Dec 8;186(1-2):1–3. [Abstract] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. [Europe PMC free article] [Abstract] [Google Scholar]
- Schnier J, Schwelberger HG, Smit-McBride Z, Kang HA, Hershey JW. Translation initiation factor 5A and its hypusine modification are essential for cell viability in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1991 Jun;11(6):3105–3114. [Europe PMC free article] [Abstract] [Google Scholar]
- Schreier MH, Erni B, Staehelin T. Initiation of mammalian protein synthesis. I. Purification and characterization of seven initiation factors. J Mol Biol. 1977 Nov;116(4):727–753. [Abstract] [Google Scholar]
- Thomas A, Goumans H, Voorma HO, Benne R. The mechanism of action of eukaryotic initiation factor 4C in protein synthesis. Eur J Biochem. 1980;107(1):39–45. [Abstract] [Google Scholar]
Associated Data
Articles from Molecular and Cellular Biology are provided here courtesy of Taylor & Francis
Full text links
Read article at publisher's site: https://doi.org/10.1128/mcb.12.1.248-260.1992
Free after 4 months at mcb.asm.org
http://mcb.asm.org/cgi/reprint/12/1/248
Free to read at mcb.asm.org
http://mcb.asm.org/cgi/content/abstract/12/1/248
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Translational fidelity screens in mammalian cells reveal eIF3 and eIF4G2 as regulators of start codon selectivity.
Nucleic Acids Res, 51(12):6355-6369, 01 Jul 2023
Cited by: 5 articles | PMID: 37144468 | PMCID: PMC10325891
A C. elegans model of C9orf72-associated ALS/FTD uncovers a conserved role for eIF2D in RAN translation.
Nat Commun, 12(1):6025, 15 Oct 2021
Cited by: 20 articles | PMID: 34654821 | PMCID: PMC8519953
The Role of the MCTS1 and DENR Proteins in Regulating the Mechanisms Associated with Malignant Cell Transformation.
Acta Naturae, 13(2):98-105, 01 Apr 2021
Cited by: 1 article | PMID: 34377560 | PMCID: PMC8327141
Dynamic competition between SARS-CoV-2 NSP1 and mRNA on the human ribosome inhibits translation initiation.
Proc Natl Acad Sci U S A, 118(6):e2017715118, 01 Feb 2021
Cited by: 112 articles | PMID: 33479166 | PMCID: PMC8017934
Toward a Kinetic Understanding of Eukaryotic Translation.
Cold Spring Harb Perspect Biol, 11(2):a032706, 01 Feb 2019
Cited by: 33 articles | PMID: 29959192 | PMCID: PMC6360857
Review Free full text in Europe PMC
Go to all (122) article citations
Other citations
Wikipedia
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Yeast translation initiation suppressor sui2 encodes the alpha subunit of eukaryotic initiation factor 2 and shares sequence identity with the human alpha subunit.
Proc Natl Acad Sci U S A, 86(8):2784-2788, 01 Apr 1989
Cited by: 107 articles | PMID: 2649894 | PMCID: PMC287003
Genetic characterization of the Saccharomyces cerevisiae translational initiation suppressors sui1, sui2 and SUI3 and their effects on HIS4 expression.
Genetics, 124(3):483-495, 01 Mar 1990
Cited by: 47 articles | PMID: 2179049 | PMCID: PMC1203942
Mutations at a Zn(II) finger motif in the yeast eIF-2 beta gene alter ribosomal start-site selection during the scanning process.
Cell, 54(5):621-632, 01 Aug 1988
Cited by: 162 articles | PMID: 3136928
Gene-specific translational control of the yeast GCN4 gene by phosphorylation of eukaryotic initiation factor 2.
Mol Microbiol, 10(2):215-223, 01 Oct 1993
Cited by: 96 articles | PMID: 7934812
Review