Abstract
Free full text

Lamin A/C and emerin regulate MKL1/SRF activity by modulating actin dynamics
Abstract
Laminopathies, caused by mutations in the LMNA gene encoding the nuclear envelope proteins lamins A and C, represent a diverse group of diseases that include Emery-Dreifuss Muscular Dystrophy (EDMD), dilated cardiomyopathy (DCM), limb-girdle muscular dystrophy, and Hutchison-Gilford progeria syndrome (HGPS).1 The majority of LMNA mutations affect skeletal and cardiac muscle by mechanisms that remain incompletely understood. Loss of structural function and disturbed interaction of mutant lamins with (tissue-specific) transcription factors have been proposed to explain the tissue-specific phenotypes.1 We report here that lamin A/C-deficient (Lmna−/−) and Lmna N195K mutant cells have impaired nuclear translocation and downstream signaling of the mechanosensitive transcription factor megakaryoblastic leukaemia 1 (MKL1), a myocardin family member that is pivotal in cardiac development and function.2 Disturbed nucleo-cytoplasmic shuttling of MKL1 was caused by altered actin dynamics in Lmna−/− and N195K mutant cells. Ectopic expression of the nuclear envelope protein emerin, which is mislocalized in Lmna mutant cells and also linked to EDMD and DCM, restored MKL1 nuclear translocation and rescued actin dynamics in mutant cells. These findings present a novel mechanism that could provide insight into the disease etiology for the cardiac phenotype in many laminopathies, whereby lamins A/C and emerin regulate gene expression through modulation of nuclear and cytoskeletal actin polymerization.
MKL1, also known as MAL or MRTF-A, is a mechanosensitive transcription factor with important roles in the cardiovascular system.2,3 Intracellular localization of MKL1 is regulated via changes in actin polymerization.4,5 Normally, MKL1 is localized in the cytoplasm by binding to cytoplasmic G-actin and constitutive nuclear export. Mitogenic or mechanical stimulation triggers RhoA-mediated actin polymerization, liberating MKL1 from G-actin and exposing a nuclear localization sequence (NLS) within the actin-binding domain of MKL1.6,7 Increased nuclear import, coupled with decreased export, causes accumulation of MKL1 in the nucleus, where it co-activates serum response factor (SRF) to turn on genes regulating cellular motility and contractility, including vinculin, actin, and SRF itself.8 Since cells from lamin A/C-deficient mice have impaired activation of mechanosensitive genes in vitro9 and in vivo,10 we investigated whether loss of lamins A/C could affect MKL1/SRF signaling. Nuclear translocation of endogenous MKL1 in response to serum stimulation was severely abrogated in Lmna−/− mouse embryonic fibroblasts (MEFs) compared to wild-type controls (Fig. 1a, c, Suppl. Fig. 1a). We confirmed these findings by time-lapse microscopy of cells expressing MKL1-GFP (Figs. 1b, ,2a)2a) and in lamin A/C-downregulated HeLa cells (Suppl. Fig. 2a, b), indicating that impaired MKL1 translocation is a general effect of loss of lamins A/C. To test whether similar defects could also result from lamin mutations associated with DCM, we investigated cells from the LmnaN195K/N195K mouse model (subsequently referred to as Lmna N195K), which develops severe DCM but lacks skeletal muscle involvement.11 Lmna N195K MEFs (Fig. 1a–c, Suppl. Fig. 1a) and bone-marrow derived mesenchymal stem cells (Suppl. Fig. 1b) had impaired nuclear translocation of MKL1. Importantly, cardiac sections from Lmna−/− and LmnaN195K/N195K mice had significantly reduced fractions of cardiomyocytes with nuclear MKL1 (Fig. 1d, e), confirming MKL1 translocation defects in vivo and implicating altered MKL1 signaling in the development of cardiomyopathies in these animals.
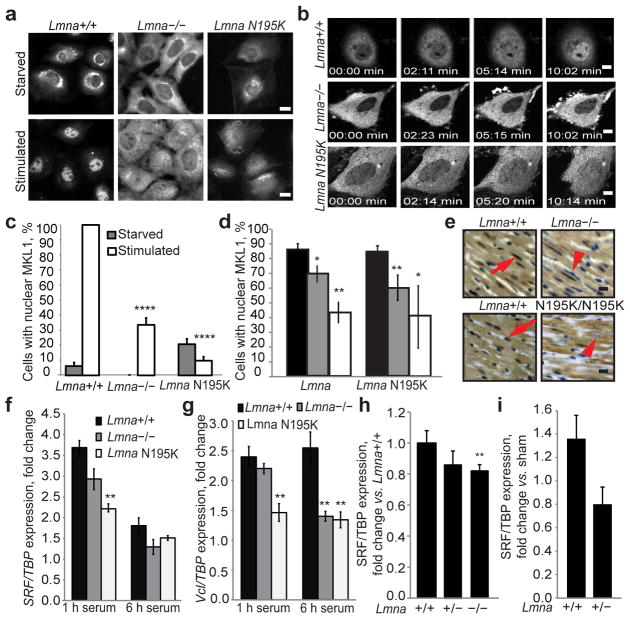
(a) Lmna−/− and Lmna N195K MEFs had a lower fraction of nuclear MKL1 after serum stimulation than Lmna+/+ cells, based on MKL1 immunofluorescence. Scale bar, 10 μm. (b) Time-lapse sequences of cells expressing MKL1-GFP stimulated with serum (see Suppl. Information for videos). Scale bar, 10 μm. (c) Quantitative analysis of MEFs with positive nuclear MKL1 staining in response to serum stimulation (N 50 per cell line). (d) Quantitative analysis of myocytes with nuclear MKL1 in cardiac sections from Lmna−/− and LmnaN195K/N195K mice as well as littermate controls (N = 3 for each). (e) Representative histological cardiac tissue sections from Lmna−/− and LmnaN195K/N195K mice and age-matched wild-type littermates stained for MKL1 (brown). Red arrows denote example of MKL1-positive nucleus; arrow head denotes an MKL1-negative nucleus. Scale bar, 20 μm. (f–g) Gene expression of serum response factor (SRF) and vinculin (Vcl) in Lmna+/+, Lmna −/− and Lmna N195K MEFs after 1 h and 6 h of serum stimulation. Values were based on 3 independent experiments and were normalized to TATA binding protein (TBP). (h) Gene expression of SRF in Lmna+/+ (N = 9), Lmna+/− (N = 11) and Lmna −/− (N = 10) cardiac tissue. Values were normalized to TBP. (i) Gene expression of SRF in Lmna+/+ (N = 5) and Lmna+/− (N = 7) cardiac tissue collected 1 week after transverse aortic constriction (TAC) surgery. Values were normalized to TBP and compared to those from sham animals. Statistical significance determined by Student’s t-test, compared to Lmna+/+ MEFs; *, indicates P ≤ 0.05; **, indicates P ≤ 0.01; ****, indicates P ≤ 0.0001. Error bars, s.e.m.
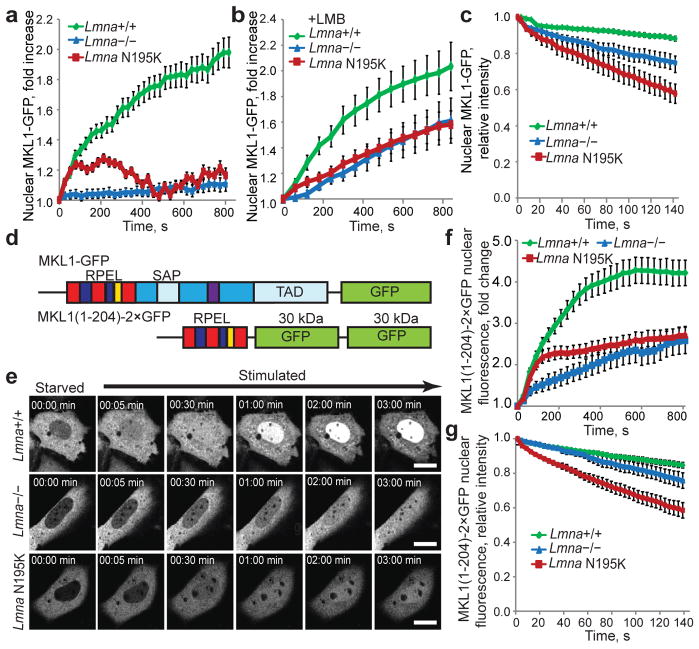
(a, b) Change in nuclear fluorescence intensity over time upon serum stimulation in Lmna+/+, Lmna−/− and Lmna N195K MEFs expressing MKL1-GFP in the absence (a) or presence of leptomycin B (b). Values were normalized to the initial nuclear fluorescence intensity before serum addition. N = 20 for each cell line. (c) Fluorescence loss in photobleaching (FLIP) experiments of MKL1-GFP to measure nuclear export. Increased loss of nuclear fluorescence indicates a higher rate of nuclear export of MKL1-GFP in lamin mutant cells. N = 10 for each cell line. (d) Schematic representation (not drawn to scale) of full length MKL1-GFP (top) and MKL1(1–204)–2×GFP (bottom), consisting of the N-terminal actin binding domain of MKL1 fused to two GFP moieties. RPEL motifs depicted in blue and red, NLS in yellow, DNA binding domain (SAP) and transcriptional activation domain (TAD) in light blue, coiled-coil domain in purple, other parts of the C-terminus in dark blue. (e) Representative frames from time-lapse series of Lmna+/+, Lmna−/− and Lmna N195K MEFs expressing MKL1(1–204)–2×GFP following serum stimulation. Scale bar, 10 μm. (f) Lmna+/+ MEFs showed rapid accumulation of MKL1(1–204)–2×GFP in the nucleus upon serum stimulation, whereas nuclear accumulation was slower in Lmna−/− and Lmna N195K cells. Nuclear fluorescence intensity was normalized to the initial nuclear fluorescence before serum stimulation. N = 60 for each cell line. (g) FLIP experiments in cell expressing MKL1(1–204)–2×GFP. Fluorescence intensity values were normalized to the initial nuclear fluorescence intensity before bleaching of a cytoplasmic region. N = 10 for each cell line. Error bars, s.e.m.
To characterize the consequences of altered MKL1 translocation, we assessed expression of select MKL1/SRF target genes. Lmna−/− and Lmna N195K MEFs had impaired serum-induced expression of SRF and vinculin (Fig. 1f, g) and had fewer focal adhesions than wild-type controls (Suppl. Fig. 3c, d); expression of an SRF-dependent luciferase reporter was also significantly reduced (Suppl. Fig. 3e). Cardiac tissues from Lmna−/− mice had lower SRF and actin transcript levels than those of wild-type littermates, and activation of SRF expression in response to left ventricular pressure-overload was impaired in Lmna+/− mice (Fig. 1h, i; Suppl. Fig. 3a, b), demonstrating disturbed MKL1-SRF mechanosignaling in vivo.
Experiments with an NLS-GFP-NES reporter construct consisting of GFP fused to an NLS and a nuclear export sequence (NES) revealed that general nuclear import and export were preserved in Lmna−/− and Lmna N195K cells (Suppl. Fig. 4), as were levels and localization of the nuclear transport factor Ran and its regulator, RCC1 (Suppl. Fig. 2c–f). We then devised experiments to independently assess nuclear import and export of MKL1. Nuclear import was measured by monitoring nuclear accumulation of MKL1-GFP while blocking nuclear export with leptomycin B.8 Lmna−/− and Lmna N195K mutant cells had significantly reduced nuclear import of MKL1 in response to serum stimulation than wild-type controls (Fig. 2b), which we confirmed with photoactivatable MKL1-PAGFP (Suppl. Fig. 5). Fluorescence loss in photobleaching (FLIP) experiments revealed that lamin mutant MEFs had a significantly faster decrease in nuclear MKL1-GFP (Fig. 2c) than wild-type cells, suggesting increased nuclear export of MKL1 in Lmna−/− and mutant MEFs.
Nuclear import and export of MKL1 are regulated by actin polymerization,8 requiring interaction between MKL1 and G-actin via three N-terminal RPEL motifs.12 We expressed a truncated MKL1 construct, MKL1(1-204)-2×GFP, which contains the RPEL motifs but lacks the transcriptional domains (Fig. 2d) and recapitulates the actin-binding characteristics and serum-inducible translocation of full-length MKL1.12 Nuclear accumulation of MKL1(1-204)-2×GFP was substantially lower in Lmna−/− and Lmna N195K cells than in wild-type cells (Fig. 2e, f), suggesting that impaired nuclear translocation of MKL1 was caused by disturbed actin dynamics in the lamin mutant cells. As seen with full-length MKL1-GFP, FLIP studies showed that nuclear export of MKL1(1-204)-2×GFP was significantly increased in the mutant cells (Fig. 2g). In contrast, abrogation of G-actin binding by mutating all three RPEL motifs (MKL1(1-204)XXX-2×GFP) or disrupting the interaction between G-actin and MKL1 with cytochalasin D resulted in nuclear accumulation of MKL1(1-204)XXX-2×GFP (Suppl. Fig. 6a) and endogenous MKL1 (Suppl. Fig. 6b), respectively, in all cell types, indicating that MKL1 can enter the nucleus of Lmna−/− and Lmna N195K cells when decoupled from actin dynamics.
We subsequently compared actin organization between mutant and wild-type cells. Fluorescence recovery after photobleaching (FRAP) revealed that nuclear actin, which modulates nuclear export of MKL1,8 was more mobile in Lmna−/− cells than in wild-type controls (Suppl. Fig. 7). Lmna−/− and Lmna N195K MEFs also had a larger fraction of highly mobile cytoplasmic actin (Fig. 3a). Furthermore, Lmna−/− and Lmna N195K cells were slower to reassemble stress fibers after disruption of actin filaments with cytochalasin D (Fig. 3b, c). In addition, whereas wild-type cells increased their ratio of F-actin to G-actin upon serum stimulation, Lmna−/− and Lmna N195K MEFs had a consistently weaker response (Fig. 3d). These findings indicate that actin polymerization is disturbed in Lmna−/− and Lmna N195K cells and that altered actin organization may be responsible for the impaired nuclear translocation of MKL1.
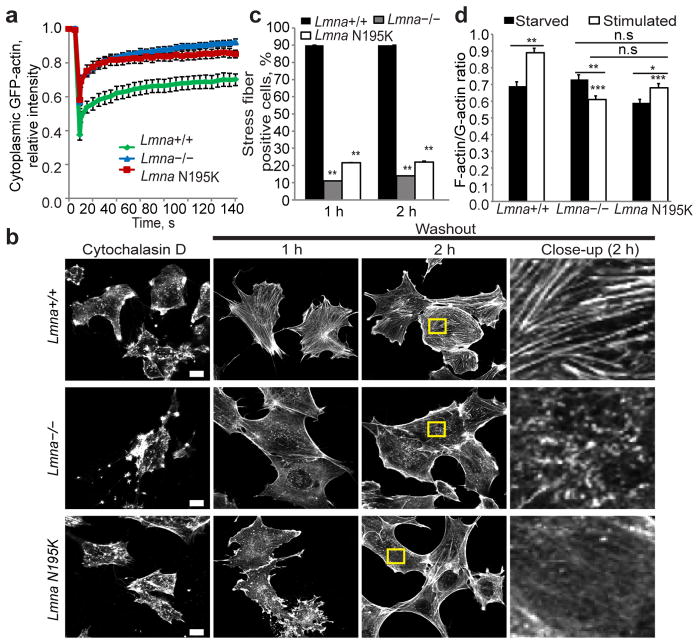
(a) Fluorescence recovery after photobleaching (FRAP) studies with GFP-actin revealed increased cytoplasmic actin mobility in Lmna−/− and Lmna N195K cells relative to Lmna+/+ controls. N = 20 for each cell line. (b) Representative images of Lmna+/+, Lmna−/− and Lmna N195K MEFs stained for actin-stress fibers with phalloidin after cytochalasin D washout. Right column contains close-up images of the regions marked by the yellow rectangle. Scale bar, 10 μm. (c) Lmna+/+ MEFs had a larger fraction of cells containing stress fibers at 1 h and 2 h after cytochalasin D washout than Lmna−/− and Lmna N195K cells. N = 50 for each cell line. (d) Comparison of F-actin/G-actin ratio in starved and serum-stimulated Lmna+/+, Lmna−/− and Lmna N195K MEFs based on phalloidin (F-actin) and DNase1 (G-actin) staining. Difference in the F-actin/G-actin ratio in serum-starved cells were not statistically significant (n.s.); N = 35 for each cell line. *, indicates P ≤ 0.05; **, indicates P ≤ 0.01; ***, indicates P≤ 0.001; all comparisons relative to corresponding Lmna+/+ cells unless indicated otherwise by horizontal bars. Error bars, s.e.m.
What causes disturbed actin organization in lamin mutant cells? Since lamins contribute to nucleo-cytoskeletal coupling and impaired nucleo-cytoskeletal coupling can disturb perinuclear actin organization, we tested whether disrupting nucleo-cytoskeletal coupling with dominant-negative nesprin mutants (DN KASH) could reproduce defects in MKL1 translocation. However, expression of DN KASH had no effect on MKL1 localization (Suppl. Fig. 8). Emerin, an inner nuclear membrane protein associated with X-linked EDMD,13 is an actin pointed-end capping protein that promotes actin polymerization in vitro14 and requires lamin A/C for proper localization. In Lmna−/− and Lmna N195K MEFs, emerin was more mobile and mislocalized from the nuclear envelope (Fig. 4a; Suppl. Fig. 9a). Emerin-deficient (Emd−/Y) MEFs displayed the same impaired nuclear translocation of MKL1 as lamin mutant cells, which could be rescued by re-introduction of exogenous emerin (Fig. 4b–d). FRAP studies in Emd−/Y MEFs demonstrated that exogenous emerin completely restored actin mobility to levels of wild-type cells (Fig. 4e, f). Ectopic expression of emerin also dramatically improved nuclear translocation of MKL1 in Lmna−/− and Lmna N195K cells (Fig. 4d, Suppl. Fig. 9b) by increasing the amount of emerin available at the nuclear envelope. In contrast, expression of emerin mutants unable to bind actin and to promote actin polymerization14 failed to restore nuclear translocation of MKL1 and caused dominant-negative defects in wild-type cells (Fig. 4d). These data indicate that emerin is a crucial modulator of actin polymerization and that loss of emerin from the nuclear envelope causes disturbed actin dynamics and impaired MKL1 signaling.
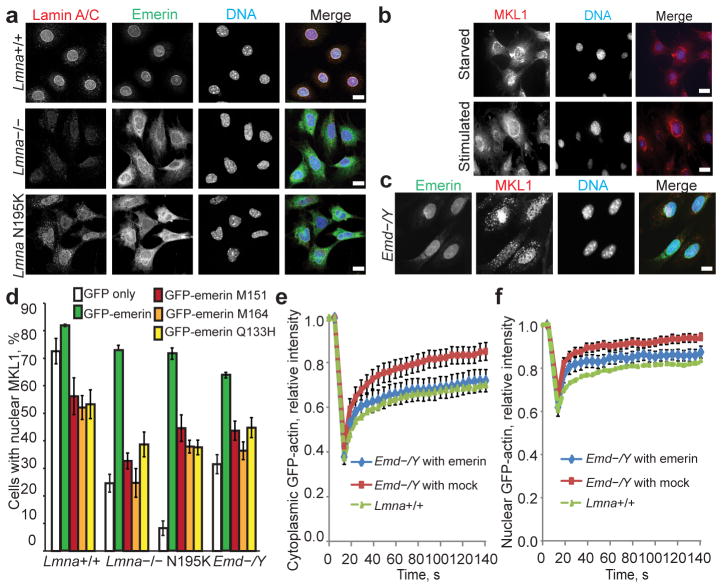
(a) Representative immunofluorescence images showing mislocalization of emerin from the nuclear envelope in Lmna−/− and Lmna N195K MEFs. Scale bar, 10 μm. (b) Emd−/Y MEFs had the same defects in MKL1 translocation as lamin mutant cells (compare with Fig. 1a). Scale bar, 10 μm. (c) Stable expression of HA-emerin in Emd−/Y MEFs restored normal nuclear MKL1 localization (71.1±6.02%) in response to serum stimulation. Scale bar, 10 μm. (d) Quantification of nuclear MKL1 localization upon serum stimulation in Lmna+/+, Lmna−/−, Lmna N195K, and Emd−/Y MEFs transiently expressing GFP-emerin, emerin mutants that do not bind to actin (GFP-M151, GFP-M164, GFP-Q133H), or GFP vector alone. Cells were categorized as either having ‘nuclear’ or ‘diffuse/cytoplasmic’ localization of MKL1. Expression of GFP-emerin restored serum-induced nuclear localization of MKL1 in Lmna−/−, Lmna N195K and Emd−/Y cells. N = 50 for each cell line. Statistical significance determined by One-way ANOVA (P ≤ 0.001) with Dunnett Multiple Comparison Post Test. Each group was compared to Lmna+/+ expressing GFP-emerin. (e–f) FRAP analysis of GFP-actin mobility in the cytoplasm (e) and in the nucleus (f) of Emd−/Y MEFs stably expressing either HA-emerin or a mock control. N = 10 for each cell line. Lmna+/+ data reproduced from Fig. 3a for comparison. Error bars, s.e.m.
Taken together, our data suggest a novel mechanism for nuclear envelope proteins to regulate MKL1-SRF signaling by modulating actin polymerization. We hypothesize that emerin primarily affects nuclear actin polymerization, which controls nuclear export and transcriptional activity of MKL1.2,8 Disturbed MKL1-SRF signaling could then further affect cytoskeletal actin, as MKL1/SRF are master regulators for numerous cytoskeletal proteins, including actin and actin-binding proteins, consistent with the reduced cytoskeletal stiffness reported in Lmna−/− MEFs.9,15,16 Given the low levels of emerin at the outer nuclear membrane17 and the fact that the fraction of emerin at the endoplasmic reticulum increases in Lmna−/− and Lmna N195K cells (Suppl. Fig. 10), it is likely that emerin has only a limited direct effect on cytoplasmic actin polymerization. Nonetheless, we cannot exclude that emerin (and lamins) may have additional effects on MKL1. For example, direct interaction of lamins A/C with nuclear G-actin18 could further contribute to the altered actin dynamics in Lmna−/− and Lmna N195K cells, as lamins, together with emerin and spectrin IIa, may form a nuclear cortical actin network.14 Furthermore, emerin can inhibit or reduce the nuclear accumulation of other transcription factors, including β-catenin, Lmo7 and phospho-ERK1/2.19
Lmna−/− and LmnaN195K/N195K mice develop DCM and have defects in cytoskeletal organization and focal adhesions,9,20,21 consistent with impaired MKL1-SRF signaling.22 Underscoring the crucial role of MKL1-SRF in cardiac function, cardiac-specific deletion of SRF in adult mice results in DCM.23 Considering the striking similarity of the cardiac phenotype observed in these animals with those in EDMD and DCM patients, we propose that impaired MKL1-SRF signaling and the resulting alterations in cytoskeletal organization may play a pivotal role in the development of cardiac defects and muscle phenotypes in various laminopathies. Surprisingly, although Emd−/Y cells have obvious defects in nuclear stability and mechanotransduction signaling,24,25 emerin-deficient mice—unlike human patients with emerin mutations—lack an overt muscular phenotype,26 indicating additional layers of complexity, species-specific differences, and possible compensation in the Emd−/Y mice. Nonetheless, Emd−/Y animals show delays in muscle repair,26 consistent with a role of MKL1 in satellite cells and skeletal muscle regeneration27 and providing additional support for the involvement of impaired MKL1 signaling in nuclear envelopathies.
While our findings further illustrate the wide-ranging impacts of mutations in nuclear envelope proteins, they also provide a glimmer of hope to affected patients. Treatment of lamin and emerin mutant mice with MAPK inhibitors can reduce cardiac and skeletal phenotypes,28 which may be attributed at least in part to the effect on MKL1 signaling, as inhibiting ERK1/2 activity is expected to increase nuclear localization of MKL1 by reducing its nuclear export.20 These findings encourage further approaches to correct impaired MKL1/SRF signaling to ameliorate the devastating cardiac disease associated with many laminopathies.
Full Methods (Online only)
Plasmids
MKL1-GFP was a kind gift from Dr. Andres Kapus (U of Toronto, Canada); MKL1(1–204)–2×GFP, MKL1(1–204)XXX–2×GFP, NLS-GFP-NES and MKL1-PAGFP were obtained from Dr. Maria Vartiainen (University of Helsinki, Finland). The NLS-GFP-NES reporter construct consists of GFP fused to an NLS and an importin α/β-dependent nuclear export sequence (NES), utilizing the same import/export mechanism as MKL1.30 The MKL1(1-204)XXX-2×GFP construct contains alanine substitutions in all three of the MKL1 RPEL motifs, which abrogates binding to G-actin and leads to constitutively nuclear localization independent of actin polymerization.12 GFP-RCC1 was obtained from Dr. Brian Paschal. GFP-actin was from Dr. Frank Gertler (Massachusetts Institute of Technology) and emerin-GFP were obtained from Dr. Howard Worman (Columbia University).
Lamin and emerin mouse models
Lmna−/− animals along with heterozygous (Lmna+/−) and wild-type (Lmna+/+) littermates were obtained from crossing Lmna+− animals. Similarly, LmnaN195K/N195K and heterozygous (LmnaN195K/+) and wild-type (Lmna+/+) littermates resulted from breeding LmnaN195K/+ animals. Lmna−/− mice develop severe muscular dystrophy and cardiomyopathy and die at 4 to 8 weeks of age.29 LmnaN195K/N195K mice develop severe dilated cardiomyopathy and die prematurely around 12 to 16 weeks of age.11 Genotype was determined by polymerase chain reaction of genomic tail DNA, as described previously.29 The mouse colonies were derived from breeders provided by Dr. Colin Stewart (Institute of Medical Biology, Singapore).11,26,29 All mice were maintained in the animal facility at Cornell University (Ithaca, NY) following protocols approved by the Cornell University Institutional Animal Care and Use Committee (IACUC).
Cell lines, transfection and drug treatment
Immortalized mouse embryonic fibroblasts (MEFs) from Lmna+/+, Lmna−/−, LmnaN195K/N195K, and Emd−/Y mice were a kind gift from Dr. Colin Stewart (Institute of Medical Biology, Singapore). Cells were maintained in Dulbecco’s Eagle’s Modified Media (DMEM) with GlutaMax (Gibco, Invitrogen) containing 10% fetal bovine serum (FBS) (PAA), and 1% Penicillin/Streptomycin at 37°C in a humidified atmosphere with 5% carbon dioxide. Serum starvation was done by withdrawing serum and incubating the cells in 0.3% FBS for 24 h. Starved cells were stimulated with DMEM with 15% FBS. Transient transfection was carried out using Lipofectamine® Reagent (Invitrogen) according to manufacturer’s protocol. 1 μg of plasmid DNA was used per transfection reaction. For cytochalasin D (Sigma Aldrich, MO) treatment, a stock of 1 mM was prepared by reconstituting the drug in DMSO and a working concentration of 1 μM was used for all experiments. For leptomycin B (Sigma Aldrich, MO) treatment, a working concentration of 5 nM was used for all experiments.
Antibodies and immunofluorescence staining
Goat anti-MKL1 (C-19 and H-180), goat anti-actin (C-11) conjugated with HRP and goat anti-lamin A/C (N-18) antibodies were purchased from Santa Cruz Biotechnology. Mouse anti-Ran antibody (610341) was purchased from BD Biosciences. Mouse anti-emerin antibody (NCL-emerin) was a product from Novocastra. Mouse anti-paxillin (05-417) was purchased from Millipore. Rabbit anti-tubulin (ab6046) was purchased from Abcam. Secondary antibodies conjugated to Alexa-488 or Alexa-568 fluorophores were purchased from Molecular Probes (Invitrogen). For immunofluorescence staining, cells grown at subconfluency were collected and fixed with 4% paraformaldehyde/phosphate-buffered saline (PBS) and permeabilized with 0.2% Triton X-100 in phosphate-buffered saline for 10 min at room temperature. cells grown at similar subconfluency for all cell lines tested. Primary antibodies in 4% bovine serum albumin in Tris-buffered saline (TBS) with 0.05% Tween 20 were incubated for 1 h at room temperature or overnight at 4°C. Cells were washed three times and incubated with appropriate secondary antibodies for 1 h at room temperature. Slides were mounted in Prolong Gold Anti-Fade medium with 4′,6-diamidino-2-phenylindole (DAPI) (Molecular Probes, Invitrogen). Images were collected and analyzed on a Zeiss LSM 700 confocal microscope (Carl Zeiss Inc., Germany). Images were captured using identical exposure times for each cell line.
Immunohistochemistry
Hearts from 4-week-old Lmna−/− mice and 8-week-old LmnaN195K/N195K mice along with wild-type and heterozygous littermate controls were harvested and fixed in 4% paraformaldehyde in PBS prior to processing for paraffin embedding and cutting. The sections were deparaffinized with xylene and rehydrated. Antigen retrieval was performed by incubation with sodium citrate buffer (10mM Sodium Citrate, 0.05% Tween 20, pH 6.0) (Invitrogen Inc) at 95°C for 20 min. The sections were then incubated with 3% hydrogen peroxide (H2O2) for 20 min to quench endogenous peroxidase activity. Blocking of non-specific sites was done by incubating the slices with 10% horse serum in PBS for 20 min. Anti-MKL1 antibody (Santa Cruz Biotechnologies) was used at 1:50 dilution at 4°C for 12–16 h to detect endogenous MKL1. A biotinylated anti-goat IgG secondary antibody (Vector Laboratories Inc, CA) was used at 1:400 dilution at room temperature for 1 h. The biotinylated secondary antibody was then detected using the VECTASTAIN® ABC system (Vector Laboratories Inc, CA) which utilizes a preformed macromolecular complex between avidin and biotinylated horseradish peroxidase. A working solution of 3, 3′-diaminobenzidine (DAB) was used as a substrate for the peroxidase. The sections were then counterstained with hematoxylin before air-drying and mounting. Images were acquired using a Zeiss Aviovert 200 inverted microscope (Carl Zeiss, Germany) equipped with a 20x objective with an AxioCam ICc1. The number of cells positive for nuclear MKL1 was counted manually on at least 5 random microscopic fields per section and normalized to the total number of cardiac myocytes.
Western blotting
Cells were lysed using radioimmunoprecipitation assay (RIPA) buffer with freshly added protease inhibitor cocktail. The cells were scraped off using a cell scraper and incubated on ice for 30 min. The lysate was then cleared by centrifuging at 13,200 rpm at 4°C. Protein concentration was estimated by Bradford assay. 30 μg of protein resuspended in Laemmli sample buffer was loaded per sample. Cytoplasmic and nuclear fractions were prepared using the Pierce NE-PER® Nuclear and Cytoplasmic Extraction Kit according to manufacturer’s instructions. Denatured proteins were resolved on 4–15% Nu-PAGE bis-tris polyacrylamide gels and blotted to a polyvinylidene fluoride (PVDF) membrane. Blocking was done with incubation in 10% non-fat dry milk in tris-buffered saline (TBS) with 0.1% Tween-20. The membrane was then probed with primary antibodies in 5% milk in TBST at 4°C overnight and sequentially detected with horseradish peroxidase conjugated secondary antibodies. The signal was revealed by autoradiography using enhanced chemiluminescence (ECL) (Pierce, Thermo Fisher Scientific Inc. Hampshire, UK).
F- and G-actin assays
For the cytochalasin D washout experiment, MEFs were treated with 1 μM Cytochalasin D (Sigma Aldrich, MO) for 30 min; subsequent drug washout was performed by rinsing the cells with three changes of medium. Cells were fixed with 4% paraformaldehyde/phosphate-buffered saline at 1 h or 2 h after washout and then permeabilized with 0.2% Triton-X100 in phosphate-buffered saline. Stress fibers were visualized using Phalloidin-Alexa 568. For fluorescence labeling of F- and G-actin, starved and stimulated Lmna+/+, Lmna−/− and Lmna N195K MEFs were fixed and permeabilized as described above. Cells were then stained with Phalloidin-Alexa 568 to label F-actin and DNase1-Alexa 488 (Invitrogen, Carlsbad, CA) to label G-actin.31 Slides were mounted in Prolong Gold Anti-Fade medium with DAPI (Invitrogen). Images were collected and analyzed on a Zeiss LSM 700 confocal microscope (Carl Zeiss Inc., Germany). Images were captured using identical exposure times for each cell line.
Real-time PCR
Total RNA from cell lines was extracted using the Qiagen RNeasy Kit (Qiagen) according to manufacturer’s instructions. Total RNA from cryo-preserved tissues was extracted using TRIzol® Reagent (Life Technologies, Invitrogen Inc.) according to manufacturer’s instructions. RNA was reverse-transcribed to cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Real-time PCR was carried out using SYBR-Green technology (Applied Biosystems) in a total volume of 25 μL. Gene expression for SRF (Serum Response Factor) and Vcl (Vinculin) was quantified. Values were normalized to an endogenous control, TATA binding protein (TBP), and compared to unstimulated samples with the ΔΔCt method. Data are based on results from 3 independent experiments.
SRE-luciferase assay
SRF forms a complex over the SRE/CArG element upon receiving upstream signals from the MAPK pathway and/or the RhoA pathway. SRE activity was measured using the Dual-Glo SRE-Luciferase Assay (Promega) according to manufacturer’s instructions. Briefly, Lmna+/+, Lmna−/− and Lmna N195K MEFs were transfected with either the SRE reporter construct or positive and negative controls. Dual-luciferase data from starved and serum-stimulated transfected cells were then collected on a luminometer. The fold change of SRE activity for each cell line was determined by comparing normalized luciferease activities of the reporter in stimulated versus starved samples.
Time-lapse microscopy and photobleaching experiments
For live cell imaging, a Zeiss LSM 700 confocal microscope (Carl Zeiss Inc. Germany) equipped with a 63× oil immersion objective (Carl Zeiss Inc. Germany) was used. Cells were maintained at 37°C in HEPES-buffered DMEM for the duration of the time-lapse acquisition. Images were recorded at 30 sec or 1 min intervals and analyzed using the Zen software (Carl Zeiss Inc. Germany). For photobleaching experiments, cells were plated on a coverslip and mounted onto a glass slide with a depression containing culture media. Fluorescence loss in photobleaching (FLIP) experiments were performed on a Zeiss LSM 700 confocal microscope (Carl Zeiss Inc. Germany) using the 488 nm laser line. Cells had been serum-stimulated for 30 min prior to the experiments. Relative loss in nuclear fluorescence during continuous photobleaching of cytoplasmic MKL1-GFP was computed by normalizing nuclear fluorescence intensity to pre-bleach values (t = 0). Increased loss of nuclear fluorescence indicates a higher rate of nuclear export of MKL1-GFP. For FLIP experiments, two single scans were acquired, followed by repeated photobleaching using a single bleach pulse at intervals of 1 s for 200 iterations in defined regions of approximately 30 μm2 in the cytoplasm. Single section images were then collected at 1 s intervals. For imaging, the laser power was attenuated to 2% of the bleach intensity. The relative fluorescence intensity in a region of interest was determined by normalizing fluorescence intensity in the region to the total fluorescence in the same region during prebleach. This method provides a means of quantifying nuclear export as the cytoplasmic pool of fluorescent protein is rapidly bleached and subsequent loss of fluorescence signal from the nucleus reflects nuclear export. For FRAP experiments, the cells were scanned two times before photobleaching by scanning the region of interest 80 times at 100% laser intensity of a 488 nm laser line. Single section images were then collected at 5 s intervals with laser power attenuated to 2% of the bleach intensity. The fluorescence intensity at the region of interest at each time point was normalized to the change in total fluorescence due to bleaching and imaging, as described previously.32 For actin FRAP experiments, cells were photobleached in defined ~2 μm diameter nuclear and cytoplasmic regions with 2 μm thickness.33 Values were normalized to the whole cell fluorescence or nuclear fluorescence at each time point, for cytoplasmic and nuclear actin FRAP, respectively. For photoactivation experiments, cells expressing MKL1-PAGFP were plated on glass-bottom dishes and starved for 24 h prior to stimulation with 15% FBS in phenol-red free DMEM for 30 minutes. Imaging was performed at 37°C using an LSM 700 confocal microscope (Carl Zeiss Inc., Germany). Photoactivation of cytoplasmic MKL1-PAGFP was carried out using the 405 nm laser at 50% laser power for 30 iterations. Sequential imaging after photoactivation was performed using a 63× oil immersion objective using excitation from a 488 nm laser line with 2% laser intensity. The increase of fluorescence was normalized to the initial fluorescence of the cytoplasmic activation area.
Statistical analysis
Statistical analysis was performed using GraphPad Prism (GraphPad Software Inc., La Jolla, CA, USA). Data is presented as mean ± s.e.m. unless stated otherwise. Two-tailed unpaired t-test and One-way ANOVA were used as detailed in respective figure legends. Statistical significance was defined as P < 0.05. All results are derived from three independent experiments.
Acknowledgments
We thank Dr. Colin Stewart for the mouse models and cell lines. We thank Joe Gannon for TAC surgeries and Mihaela Cupesi for collecting the cardiac samples from the pressure-overload model. This work was supported by National Institutes of Health awards [R01 NS059348 and R01 HL082792]; the Department of Defense Breast Cancer Idea Award [BC102152]; an award from the Progeria Research Foundation; and a postdoctoral fellowship from the American Heart Association to D.J. [AHA award 09POST2320042]. The work in the lab of MKV is funded by the Academy of Finland and the Sigrid Juselius Foundation.
Footnotes
Full methods and any associated references are available in the online version of the paper.
This file contains Supplementary Figures 1–10, Supplementary References, and Supplementary Videos 1–9.
Author Contributions
C.Y.H., D.E.J., and J.L. conceived and designed the overall project, with valuable help from M.V. C.Y.H. and D.E.J. performed the experiments. C.Y.H., D.E.J., and J.L. analyzed data. C.Y.H. and J.L. wrote the paper.
Competing financial interests
The authors declare no competing financial interests.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nature12105
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3666313?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Role of lamin A/C on dendritic cell function in antiviral immunity.
Cell Mol Life Sci, 81(1):400, 12 Sep 2024
Cited by: 0 articles | PMID: 39264480 | PMCID: PMC11393282
SIRT1 Ameliorates Lamin A/C Deficiency-Induced Cardiac Dysfunction by Promoting Mitochondrial Bioenergetics.
JACC Basic Transl Sci, 9(10):1211-1230, 31 Jul 2024
Cited by: 0 articles | PMID: 39534638 | PMCID: PMC11551877
Cypher/ZASP drives cardiomyocyte maturation via actin-mediated MRTFA-SRF signalling.
Theranostics, 14(11):4462-4480, 22 Jul 2024
Cited by: 0 articles | PMID: 39113806 | PMCID: PMC11303069
Sensing the squeeze: nuclear mechanotransduction in health and disease.
Nucleus, 15(1):2374854, 01 Jul 2024
Cited by: 0 articles | PMID: 38951951 | PMCID: PMC11221475
Review Free full text in Europe PMC
Altered extracellular matrix-related pathways accelerate the transition from normal to prefibroid myometrium in Black women.
Am J Obstet Gynecol, 231(3):324.e1-324.e12, 31 May 2024
Cited by: 0 articles | PMID: 38825029
Go to all (287) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Decreased mechanical stiffness in LMNA-/- cells is caused by defective nucleo-cytoskeletal integrity: implications for the development of laminopathies.
Hum Mol Genet, 13(21):2567-2580, 14 Sep 2004
Cited by: 238 articles | PMID: 15367494
Effect of pathogenic mis-sense mutations in lamin A on its interaction with emerin in vivo.
J Cell Sci, 116(pt 14):3027-3035, 03 Jun 2003
Cited by: 52 articles | PMID: 12783988
Nesprin-1 and actin contribute to nuclear and cytoskeletal defects in lamin A/C-deficient cardiomyopathy.
J Mol Cell Cardiol, 50(3):479-486, 13 Dec 2010
Cited by: 25 articles | PMID: 21156181
Emery-Dreifuss muscular dystrophy: focal point nuclear envelope.
Curr Opin Neurol, 32(5):728-734, 01 Oct 2019
Cited by: 10 articles | PMID: 31460960 | PMCID: PMC7550203
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NCI NIH HHS (1)
Grant ID: BC102152
NHLBI NIH HHS (1)
Grant ID: R01 HL082792
NINDS NIH HHS (1)
Grant ID: R01 NS059348





