Abstract
Free full text

Association of CD247 Polymorphisms with Rheumatoid Arthritis: A Replication Study and a Meta-Analysis
Associated Data
Abstract
Given the role of CD247 in the response of the T cells, its entailment in autoimmune diseases and in order to better clarify the role of this gene in RA susceptibility, we aimed to analyze CD247 gene variants previously associated with other autoimmune diseases (rs1052237, rs2056626 and rs864537) in a large independent European Caucasian population. However, no evidence of association was found for the analyzed CD247 single-nucleotide polymorphisms (SNPs) with RA and with the presence/absence of anti-cyclic citrullinated polypeptide. We performed a meta-analysis including previously published GWAS data from the rs864537 variant, revealing an overall genome-wide significant association between this CD247 SNP and RA with anti-CCP (OR =
= 0.90, CI 95%
0.90, CI 95% =
= 0.87–0.93, Poverall
0.87–0.93, Poverall =
= 2.1×10−10). Our results show for first time a GWAS-level association between this CD247 polymorphism and RA risk.
2.1×10−10). Our results show for first time a GWAS-level association between this CD247 polymorphism and RA risk.
Introduction
The T-cell receptor T3 zeta chain (CD3ζ), also called CD247, is essential for assembly, surface expression and signaling cascade of the T-cell receptor-CD3 (TCR/CD3) complex. Abnormalities in this pathway can result in T-cell dysfunction and development of autoimmune disorders [1]. Regarding this, previous reports have demonstrated an association of CD247 variants with systemic lupus erythematosus [2], [3] and systemic sclerosis [4], [5]. In the case of rheumatoid arthritis (RA), one meta-GWAS found no association between the CD247 gene and RA at a genome-wide significance level [6]; however, a more recent meta-analysis of GWAS data from RA and celiac disease (CeD) identified CD247 as a novel susceptibility locus for both diseases [7]. Taking into account the role of CD247 in the response of the T-cells, its entailment in autoimmune diseases and in order to better clarify the role of this gene in RA susceptibility, we aimed to analyze CD247 gene variants in a large independent European Caucasian cohort.
Materials and Methods
A large independent European Caucasian cohort containing cases from four different countries has been included in the present study. The individuals were recruited from hospitals of Spain (1,797 cases and 2,026 controls), Germany (326 cases and 379 controls), Norway (949 cases and 1,121 controls) and Australasian (488 cases and 508 controls). All cases were ascertained by the American College of Rheumatology criteria [8]. Briefly, the 75% of the patients were females, the 76% were rheumatoid factor (RF) positive and the 63% were anti-cyclic citrullinated polypeptide (anti-CCP) positive. Controls self- reported as not having RA.
All subjects provide written informed consent according to the declaration of Helsinki. The study was approved by the ethical committees from all the Spanish participating centers to Galicia, Madrid (Hospital Clínico San Carlos, Hospital La Paz and Hospital de La Princesa), Barcelona (Hospital Universitario Bellvitge) and Santander (Hospital Universitario Marqués de Valdecilla). In addition, the ethical committees of Medizinische Hochschule Hannover from Germany, the Regional Committees for Research Ethics in Eastern and Southern Norway and the Multi-region Ethics Committee and the Lower South Ethics Committee of New Zealand also approved the study.
Three CD247 single-nucleotide polymorphisms (SNPs) previously associated with different autoimmune diseases including RA were selected: rs1052237 [2], [3], rs2056626 [5] and rs864537 [7], and genotyped using TaqMan assays. A total of 350 patients were genotyped twice to verify the genotyping consistency, showing 99% identical genotypes.
In order to increase the robustness of our analyses, we conducted a meta-analysis with previously published data. The searching of the articles was performed on MEDLINE and PUBMED electronic databases. Studies for the meta-analysis were selected if they met the following conditions: 1) RA patients fulfilled the ACR criteria of 1988; 2) data were collected in European Caucasian populations; 3) they included any of the three polymorphisms selected in the present study. Given these considerations, only two articles were included in our study [6], [7].
The Hardy-Weinberg equilibrium (HWE) was tested by the means of χ2 or Fisher’s exact test when necessary. The case-control association study was performed by 2×2 contingency table with χ2 to obtain P-values, odds ratios (OR) and 95% confidence intervals (CI). Combined ORs were calculated according to a fixed-effects model (Mantel–Haenszel meta-analysis) and the heterogeneity of ORs among all populations was calculated by Breslow-Day test. P values lower than 0.0007 were regarded as significant after applying the Bonferroni correction. Statistical analyses were carried out with Plink v1.7 [9] (see URL: http://pngu.mgh.harvard.edu/~purcell/plink/) and StatsDirect software v2.4.6.
The estimation of the power of the study to detect an effect of a polymorphism in disease susceptibility was performed using the the CaTS Power Calculator software [10] (see URL: http://www.sph.umich.edu/csg/abecasis/CaTS).
Results and Discussion
The estimated power of the study was 61% for rs1052237 and 82% for rs2056626 and rs864537 to detect ORs =
= 1.10 at the 5% significant level. The genotyping call-rate success was 98.3% in cases and 99.0% in controls for rs1052231, 97.9% in cases and 96.9% in controls for rs864537 and 97.6% in cases and 96.4% in controls for rs2056626. Genotype frequencies were in HWE in cases and controls. The distribution of genotypic and allelic frequencies for each population is shown in Table S1. We did not observe any significant association after the multiple testing correction was applied. However, the meta-analysis of the four cohorts using the Mantel-Hanzel method under fixed effects model showed no evidence of association of the three CD247 SNPs and RA (rs1052231: OR
1.10 at the 5% significant level. The genotyping call-rate success was 98.3% in cases and 99.0% in controls for rs1052231, 97.9% in cases and 96.9% in controls for rs864537 and 97.6% in cases and 96.4% in controls for rs2056626. Genotype frequencies were in HWE in cases and controls. The distribution of genotypic and allelic frequencies for each population is shown in Table S1. We did not observe any significant association after the multiple testing correction was applied. However, the meta-analysis of the four cohorts using the Mantel-Hanzel method under fixed effects model showed no evidence of association of the three CD247 SNPs and RA (rs1052231: OR =
= 1.02, CI 95%
1.02, CI 95% =
= 0.94–1.20, p
0.94–1.20, p =
= 0.595; rs864537: OR
0.595; rs864537: OR =
= 0.97, CI 95%
0.97, CI 95% =
= 0.91–1.03, p
0.91–1.03, p =
= 0.330; rs2056626: OR
0.330; rs2056626: OR =
= 0.97, CI 95%
0.97, CI 95% =
= 0.91–1.04, p
0.91–1.04, p =
= 0.348) (see
Table 1
). The combinability test according to the Breslow-Day method showed no significant heterogeneity among the ORs of the different cohorts. Moreover, no evidence of association was observed after stratification for RF and anti-CCP status (
Table 1
and Supplementary
Table 1
).
0.348) (see
Table 1
). The combinability test according to the Breslow-Day method showed no significant heterogeneity among the ORs of the different cohorts. Moreover, no evidence of association was observed after stratification for RF and anti-CCP status (
Table 1
and Supplementary
Table 1
).
Table 1
| Samples | Minor allele (%) | Allele test* | ||||||
| Sets | N | Genotype (%) | P-value | OR [95% CI] | PBD | |||
| rs1052231 | T/T | T/A | A/A | A | ||||
| Controls | 3994 | 69.1 | 27.6 | 2.9 | 16.7 | |||
| RA | 3499 | 69.1 | 27.7 | 3.2 | 17.0 | 0.595 | 1.02 [0.94–1.20] | 0.117 |
| RA RF + | 2094 | 68.5 | 28.2 | 3.3 | 17.4 | 0.463 | 1.04 [0.94–1.15] | 0.080 |
| RA RF − | 991 | 69.1 | 28.0 | 2.9 | 16.9 | 0.792 | 1.02 [0.89–1.16] | 0.506 |
| RA anti-CCP + | 1702 | 69.2 | 27.8 | 3.0 | 16.9 | 0.852 | 0.99 [0.88–1.10] | 0.142 |
| RA anti-CCP − | 1013 | 68.2 | 29.2 | 2.6 | 17.2 | 0.451 | 0.95 [0.83–1.08] | 0.192 |
| rs864537 | A/A | A/G | G/G | G | ||||
| Controls | 3910 | 38.3 | 46.8 | 14.9 | 38.3 | |||
| RA | 3485 | 39.0 | 46.8 | 14.2 | 37.6 | 0.33 | 0.97 [0.91–1.03] | 0.648 |
| RA RF + | 2087 | 39.5 | 47.1 | 13.4 | 37.8 | 0.060 | 0.93 [0.86–1.00] | 0.509 |
| RA RF − | 982 | 37.8 | 46.6 | 15.6 | 38.4 | 0.406 | 1.04 [0.94–1.56] | 0.931 |
| RA anti-CCP + | 1699 | 38.8 | 47.5 | 13.7 | 37.5 | 0.457 | 0.97 [0.89–1.05] | 0.482 |
| RA anti-CCP − | 1011 | 40.5 | 45.5 | 14.0 | 36.8 | 0.309 | 0.95 [0.86–1.05] | 0.844 |
| rs2056626 | T/T | T/G | G/G | G | ||||
| Controls | 3888 | 38.6 | 46.6 | 14.8 | 38.1 | |||
| RA | 3473 | 39.2 | 46.8 | 14.0 | 37.4 | 0.348 | 0.97 [0.91–1.04] | 0.051 |
| RA RF + | 2082 | 39.9 | 45.9 | 14.3 | 37.9 | 0.294 | 0.96 [0.87–1.04] | 0.031 |
| RA RF − | 979 | 38.4 | 47.3 | 14.3 | 38.1 | 0.803 | 1.01 [0.91–1.12] | 0.545 |
| RA anti-CCP + | 1683 | 40.6 | 45.1 | 14.3 | 36.9 | 0.323 | 0.96 [0.88–1.04] | 0.031 |
| RA anti-CCP − | 1005 | 39.4 | 47.2 | 13.4 | 37.0 | 0.712 | 0.98 [0.89–1.09] | 0.956 |
In addition to these analyses, we performed a meta-analysis including published GWAS data for rs864537 [6], [7]. Considering that these studies included auto-antibody-positive RA individuals, we performed the combined analysis including only RA patients with anti-CCP. The data of these GWASs together with our combined data sets revealed an overall genome-wide significant association between rs864537 and RA with anti-CCP (OR =
= 0.90, Ci 95%
0.90, Ci 95% =
= 0.87–0.93, Poverall
0.87–0.93, Poverall =
= 2.1×10−10) (Figure 1).
2.1×10−10) (Figure 1).
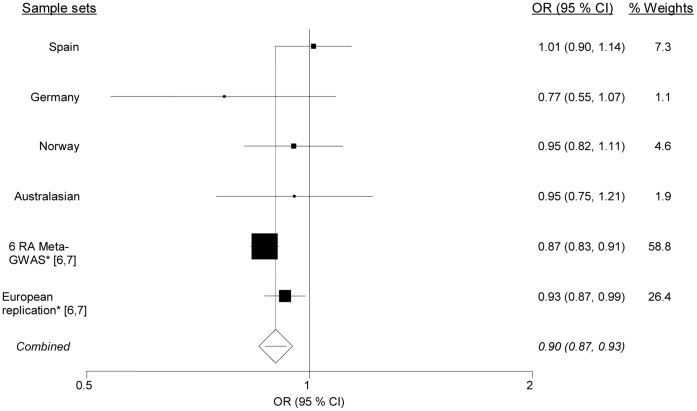
The combined analysis included a total of 10,763 RA patients with anti-CPP and 28,879 controls. There was no evidence of heterogeneity between sample sets (Breslow-Day test for heterogeneity p =
= 0.12).
0.12).
The present study represents a good example of how increasing the study cohort may be necessary to detect genetic associations of weak effect size at GWAS level. In this regard, the study performed by Stahl et al. detected no association of CD247 with RA at GWAS level [6], and the CD247 association only reached genome-wide significance level when data of two autoimmune diseases, RA and CeD, were combined [7]. Therefore, the current study described for first time at GWAS-level association of CD247 and RA risk using a meta-analysis of a large European Caucasian case-control sample set.
A reduced density of synovial T-cell surface CD3ζ has been observed in RA patients [11], [12], [13], suggesting a decrease in TCR signaling, thus favoring positive selection of autoreactive T effector cells in the thymus [14]. It is unclear what is the cause of the low expression of CD3ζ in synovial T cells and several possible mechanisms should be considered [1], [11], [12], [13]. Regarding this, splicing variants of CD247 have been demonstrated to lead to downregulated protein expression in SLE patients [1], [15]. It is interesting to note that the risk variant of CD247 rs864537 has also been correlated with cis gene expression of CD247 [7]. Further functional studies should shed more light on the role of this genetic variant in the susceptibility to RA.
Acknowledgments
We thank Sofía Vargas, Sonia Garcia and Siri Flåm for their excellent technical assistance. We thank Banco Nacional de ADN (University of Salamanca, Spain) and the Norwegian Bone Marrow Donor Registry who supplied part of control DNA samples, and all patients and donors for their collaboration. We also thank Nicola Dalbeth, Peter Gow, Peter Jones, Andrew Harrison, John Highton and Lisa Stamp for their role in recruiting New Zealand RA patients.
Funding Statement
This work was supported by two Spanish grants from RETICS Program, RD12/0009/0004 (RIER) from Instituto de Salud Carlos III (ISCIII), within the VI PN de I+D+i 2008–2011 (FEDER) and also by grants from the European IMI BTCure Program. MT was supported by Ministerio Economia y Competividad through the program Juan de la Cierva (JCI-2010-08227). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
Articles from PLOS ONE are provided here courtesy of PLOS
Full text links
Read article at publisher's site: https://doi.org/10.1371/journal.pone.0068295
Read article for free, from open access legal sources, via Unpaywall:
https://journals.plos.org/plosone/article/file?id=10.1371/journal.pone.0068295&type=printable
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Effects of immunorelated gene polymorphisms on trastuzumab targeting breast cancer cell <i>in vitro</i>.
Pharmacogenomics, 25(10-11):461-468, 11 Oct 2024
Cited by: 0 articles | PMID: 39392082
Immunogenetics of Systemic Sclerosis.
Genes (Basel), 15(5):586, 05 May 2024
Cited by: 0 articles | PMID: 38790215 | PMCID: PMC11121022
Review Free full text in Europe PMC
Identification of biomarkers for abdominal aortic aneurysm in Behçet's disease via mendelian randomization and integrated bioinformatics analyses.
J Cell Mol Med, 28(10):e18398, 01 May 2024
Cited by: 1 article | PMID: 38785203 | PMCID: PMC11117452
A simple genotyping method for CD247 3'-untranslated region polymorphism rs1052231 and characterization of a reference cell panel.
HLA, 98(3):218-222, 16 Jul 2021
Cited by: 2 articles | PMID: 34233083 | PMCID: PMC9291556
Bioinformatics and system biology approaches to identify the diseasome and comorbidities complexities of SARS-CoV-2 infection with the digestive tract disorders.
Brief Bioinform, 22(6):bbab126, 01 Nov 2021
Cited by: 8 articles | PMID: 33993223 | PMCID: PMC8194728
Go to all (12) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
SNPs (4)
- (5 citations) dbSNP - rs864537
- (4 citations) dbSNP - rs2056626
- (2 citations) dbSNP - rs1052237
- (2 citations) dbSNP - rs1052231
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
TCR-CD3ζ gene polymorphisms and expression profile in rheumatoid arthritis.
Autoimmunity, 49(7):466-471, 26 Apr 2016
Cited by: 6 articles | PMID: 27118209
Association Study of CD226 and CD247 Genes Single Nucleotide Polymorphisms in Iranian Patients with Systemic Sclerosis.
Iran J Allergy Asthma Immunol, 16(6):471-479, 01 Dec 2017
Cited by: 2 articles | PMID: 29338153
Association of PTPN22 rs2476601 and STAT4 rs7574865 polymorphisms with rheumatoid arthritis: A meta-analysis update.
Immunobiology, 220(8):1012-1024, 28 Apr 2015
Cited by: 17 articles | PMID: 25963842





