Abstract
Summary
EcoGene.org is a genome database and website dedicated to Escherichia coli K-12 substrain MG1655 that is revised daily using information derived from the biomedical literature and in-house analysis. EcoGene is a major source of annotation updates for the MG1655 Genbank record, one of only a few Genbank genome records that are updated by a community effort. The Reference Sequence (RefSeq) database, built by The National Center for Biotechnology Information, comprises a set of duplicate Genbank genome records that can be modified by the NCBI staff annotators. EcoGene-RefSeq is being developed as a stand-alone internet resource to facilitate the usage of EcoGene-based tools on any of the >2400 completed prokaryotic genome records that are currently available at the RefSeq database.Availability
The web interface of EcoGene-RefSeq is available at http://www.ecogene.org/refseq.Contact
[email protected] or [email protected].Free full text

EcoGene-RefSeq: EcoGene tools applied to the RefSeq prokaryotic genomes
Abstract
Summary: EcoGene.org is a genome database and website dedicated to Escherichia coli K-12 substrain MG1655 that is revised daily using information derived from the biomedical literature and in-house analysis. EcoGene is a major source of annotation updates for the MG1655 Genbank record, one of only a few Genbank genome records that are updated by a community effort. The Reference Sequence (RefSeq) database, built by The National Center for Biotechnology Information, comprises a set of duplicate Genbank genome records that can be modified by the NCBI staff annotators. EcoGene-RefSeq is being developed as a stand-alone internet resource to facilitate the usage of EcoGene-based tools on any of the >2400 completed prokaryotic genome records that are currently available at the RefSeq database.
Availability: The web interface of EcoGene-RefSeq is available at http://www.ecogene.org/refseq.
Contact: ude.imaim.dem@ddurk or [email protected]
1 INTRODUCTION
New sequencing technologies have significantly increased the volume of genomic sequence data that are being generated. The NCBI RefSeq collection is a curated sequence database providing a comprehensive, non-redundant and annotated set of sequences representing naturally occurring DNA, RNA and proteins (Pruitt et al., 2012). Included are taxonomically diverse sequences from plasmids, organelles, viruses, archaea, bacteria and eukaryotes. The Escherichia coli K-12 MG1655 genome was the third sequenced genome and is represented by the Genbank U00096 complete genome record (Blattner et al., 1997), which has been extensively revised since its original submission (Riley et al., 2006; Rudd, 2000; Zhou and Rudd, 2013). EcoGene was developed to maintain, display, query and document the revised genome and proteome sequences and annotations. EcoGene is the modern version of the historical E.coli K-12 genetic maps (Rudd, 1998). A suite of customized tools has been developed for EcoGene, providing functionality that is unavailable elsewhere. We are now making these tools available for viewing and retrieving genomic maps and sequences for any prokaryotic genome sequence through EcoGene-RefSeq.
The applications ported from EcoGene to EcoGene-Refseq include (i) PrimerPairs, a tool for automatically designing genome-wide sets of primers to engineer either a clone library or a deletion strain library (Zhou and Rudd, 2011), (ii) Search and Download, a search interface for querying and downloading gene information, (iii) GenePages, web pages displaying individual genes as well as dynamic gene maps and restriction sites maps for genome navigation and (iv) Cross Reference Mapping and Download, a tool for accessing many additional gene identifiers. EcoGene-RefSeq is powered by the open source content management platform Drupal and supported by a MySQL database. All data stored in the EcoGene-RefSeq MySQL database are faithfully parsed from RefSeq for efficient retrieval.
2 RESULTS
2.1 Data parsing
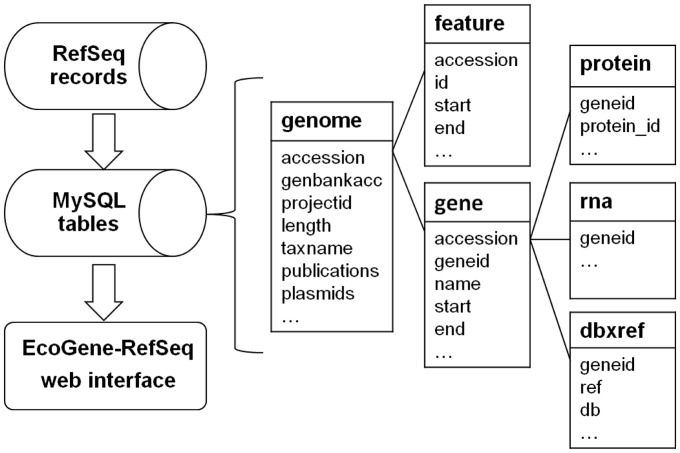
The RefSeq database is made freely available and can be accessed through several methods, including FTP downloading, internet query and script. In our implementation, only completed prokaryotic genomes, including bacterial and archaeal species, are considered. Project information about the frequently updated completed prokaryotic genomes is obtained from the genome report at NCBI’s ftp site (ftp://ftp.ncbi.nlm.nih.gov/genomes/GENOME_REPORTS/). The report contains detailed genome project information grouped by major taxonomic division. These taxonomic division reports include information on genome data submitted to the primary archival sequence data that are exchanged among members of the International Nucleotide Sequence Database Collaboration (INSDC) and genome data represented in NCBI’s RefSeq dataset. Detailed genome records that are used to parse data elements into EcoGene-RefSeq database are obtained from ftp://ftp.ncbi.nih.gov/genomes/Bacteria/. RefSeq offers several different types of files to store genomic records, one of which we use is the Genetic Feature Format Version 3(GFF3). Hypertext preprocessor (PHP, http://www.php.net) scripts were written to parse, extract, reformat, construct data elements and interact with a MySQL database (http://www.mysql.com/) for storage and querying. The EcoGene-RefSeq MySQL database is designed to efficiently store and query the parsed data as shown in Figure 1.
All data in EcoGene-RefSeq are faithfully parsed from RefSeq, and no attempt is made to curate or re-annotate. The EcoGene-RefSeq MySQL database is updated daily from RefSeq records to include newly added records, and it is also refreshed monthly to reflect annotation changes of the genomes. Currently there are 2268 bacterial genomes and 148 archaeal genomes in the EcoGene-RefSeq database.
2.2 Usage
A web interface is provided for searching and viewing all the completed prokaryotic genomes stored in the EcoGene-RefSeq database. Genome records can be searched by domain, type, name, RefSeq or Genbank accession numbers. Each genome sequence has a summary page reporting the stable accession numbers, plasmid information and the related publications from the BioProject (Barrett et al., 2012). The summary page also provides the numbers of proteins genes, RNA genes and pseudogenes with internal linkers to access each of these categories directly at the Search and Download page, allowing the user to download details of each gene in the category. A set of web-based applications are ported from EcoGene to all prokaryotic genomes stored in the EcoGene-RefSeq database, including:
2.2.1 Search and Download
This interface allows for retrieval of a list of genes by querying gene names, IDs, products and other fields, which can subsequently be downloaded and applied to other applications using only these user-specified genes. For example, the user can upload the specified genes to PrimerPairs and get a desired primer pair subset.
2.2.2 Gene Index and GenePage
The Gene Index page, provided for each genome, is an alphabetical index to the individual GenePages. The GenePage contains text information about DNA sequence and gene product with external linkers to their sources at NCBI and UniProtKB. In addition, three regional maps (a dynamic Gene Map, a Feature Map and an interactive restriction Sites Map) are created in the Portable Network Graphic (PNG) format that can be saved for use in publications and presentations. The Gene Map is an interactive display that depicts a default 10 kb region of genomic DNA with user ability to zoom for a shorter more detailed region and click internal linkers to other nearby GenePages. The Feature Map depicts IS elements and intergenic repeats. The Site Map depicts all the restriction sites for up to seven user-specified restriction enzymes, with three restriction enzymes (BamHI, EcoRI and HindIII) used as the default selection. Every gene in EcoGene-RefSeq has a GenePage accessible through the internal linkers at the Search and Download page, Gene Index page or through a URL using the genome’s RefSeq accession number and unique GeneID assigned by NCBI.
2.2.3 PrimerPairs
PrimerPairs is a web application that allows for automatic genome-wide polymerase chain reaction primer design enabling the deletion or cloning of all genes in a genome (Zhou and Rudd, 2011). The DNA fragments these primers amplify can be used to implement a genome re-engineering strategy using complementary in vitro cloning and in vivo re-combineering. The integration of a primer design tool with a completed genome database increases the level of quality control. PrimerPairs can automatically detect and correct overlapping deletion primers because of integration with the genome annotation in the EcoGene-Refseq database.
2.2.4 Cross Reference Mapping
The Cross Reference Mapping and Download page is created for user access to many additional accession numbers and other gene identifiers, such as gene name and the NCBI Gene ID that are collected in RefSeq records. The cross references facilitate both hyperlink construction and the integration of experimental and bioinformatics results.
3 CONCLUSION
EcoGene-RefSeq is developed to facilitate the usage of a set of tools available at EcoGene with any prokaryotic genome. In the future, we can add capabilities into EcoGene-RefSeq, including manual curation tools enabling an individual or interested group to build and re-annotate an EcoGene-like database for any prokaryotic genome.
Funding: National Institutes of Health [1-R01-GM58560].
Conflict of Interest: none declared.
REFERENCES
- Barrett T, et al. BioProject and BioSample databases at NCBI: facilitating capture and organization of metadata. Nucleic Acids Res. 2012;40:D57–D63. [Europe PMC free article] [Abstract] [Google Scholar]
- Blattner FR, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. [Abstract] [Google Scholar]
- Pruitt K, et al. NCBI Reference Sequence (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40:130–135. [Europe PMC free article] [Abstract] [Google Scholar]
- Riley M, et al. Escherichia coli K-12: a cooperatively developed annotation snapshot-2005. Nucleic Acids Res. 2006;34:1–9. [Europe PMC free article] [Abstract] [Google Scholar]
- Rudd KE. Linkage map of Escherichia coli K-12, edition 10: the physical map. Microbiol. Mol. Biol. Rev. 1998;62:985–1019. [Europe PMC free article] [Abstract] [Google Scholar]
- Rudd KE. EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res. 2000;28:60–64. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhou J, Rudd KE. Bacterial Genome Reengineering, Methods in Molecular Biology. Vol. 765. New Jersey: Springer; 2011. [Abstract] [Google Scholar]
- Zhou J, Rudd KE. EcoGene 3.0. Nucleic Acids Res. 2013;41:D613–D624. [Europe PMC free article] [Abstract] [Google Scholar]
Articles from Bioinformatics are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/bioinformatics/btt302
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/bioinformatics/article-pdf/29/15/1917/735945/btt302.pdf
Citations & impact
Impact metrics
Article citations
Mycobacterium smegmatis does not display functional redundancy in nitrate reductase enzymes.
PLoS One, 16(1):e0245745, 20 Jan 2021
Cited by: 2 articles | PMID: 33471823 | PMCID: PMC7816997
Transcriptome analysis to understand the effects of the toxoflavin and tropolone produced by phytopathogenic Burkholderia on Escherichia coli.
J Microbiol, 57(9):781-794, 27 Aug 2019
Cited by: 4 articles | PMID: 31452043
Unveiling the Hybrid Genome Structure of Escherichia coli RR1 (HB101 RecA+).
Front Microbiol, 8:585, 04 Apr 2017
Cited by: 3 articles | PMID: 28421066 | PMCID: PMC5379014
NCBI prokaryotic genome annotation pipeline.
Nucleic Acids Res, 44(14):6614-6624, 24 Jun 2016
Cited by: 3630 articles | PMID: 27342282 | PMCID: PMC5001611
Comprehensive identification of translation start sites by tetracycline-inhibited ribosome profiling.
DNA Res, 23(3):193-201, 23 Mar 2016
Cited by: 46 articles | PMID: 27013550 | PMCID: PMC4909307
Go to all (7) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences
- (1 citation) ENA - U00096
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
EcoGene 3.0.
Nucleic Acids Res, 41(database issue):D613-24, 28 Nov 2012
Cited by: 137 articles | PMID: 23197660 | PMCID: PMC3531124
EcoGene: a genome sequence database for Escherichia coli K-12.
Nucleic Acids Res, 28(1):60-64, 01 Jan 2000
Cited by: 166 articles | PMID: 10592181 | PMCID: PMC102481
RefSeq and the prokaryotic genome annotation pipeline in the age of metagenomes.
Nucleic Acids Res, 52(d1):D762-D769, 01 Jan 2024
Cited by: 15 articles | PMID: 37962425 | PMCID: PMC10767926
The EcoCyc Database.
EcoSal Plus, 8(1), 01 Nov 2018
Cited by: 59 articles | PMID: 30406744 | PMCID: PMC6504970
Review Free full text in Europe PMC