Abstract
Free full text

A Novel, Multi-Parallel, Real-Time Polymerase Chain Reaction Approach for Eight Gastrointestinal Parasites Provides Improved Diagnostic Capabilities to Resource-Limited At-Risk Populations
Abstract
Diagnosis of gastrointestinal parasites has traditionally relied on stool microscopy, which has low diagnostic sensitivity and specificity. We have developed a novel, rapid, high-throughput quantitative multi-parallel real-time polymerase chain reaction (qPCR) platform. Species-specific primers/probes were used for eight common gastrointestinal parasite pathogens: Ascaris lumbricoides, Necator americanus, Ancylostoma duodenale, Giardia lamblia, Cryptosporidium spp., Entamoeba histolytica, Trichuris trichiura, and Strongyloides stercoralis. Stool samples from 400 13-month-old children in rural Ecuador were analyzed and the qPCR was compared with a standard direct wet mount slide for stool microscopy, as were 125 8–14-year-old children before and after anthelmintic treatment. The qPCR showed higher detection rates for all parasites compared with direct microscopy, Ascaris (7.0% versus 5.5%) and for Giardia (31.5% versus 5.8%). Using an enhanced DNA extraction method, we were able to detect T. trichiura DNA. These assays will be useful to refine treatment options for affected populations, ultimately leading to better health outcomes.
Introduction
The prevalence of gastrointestinal parasites within affected populations has been underreported largely because standard stool examinations are relatively insensitive. With the development of newer multiplex polymerase chain reaction (PCR)–based assays in stool for pathogenic parasite (hookworm, Strongyloides stercoralis, Ascaris lumbricoides, Entamoeba histolytica, Cryptosporidium parvum, and Giardia lamblia) identification, it is likely that gastrointestinal parasite identification in the stool will become more sensitive and objective.1–3
Classically, the diagnosis of intestinal parasites depends on the life cycle of the parasite and its egg/larvae shedding patterns. Although microscopy and quantitative real-time PCR (qPCR) are dependent on the presence of parasite material in the sample,4 qPCR is less subjective and for several parasites (e.g., E. histolytica) more specific.2 The ability of microscopy to detect parasites is directly related to the number of organisms in stool. Although the same is true for qPCR, its benefits are in its improved sensitivity and specificity. With proper stool disruption and DNA extraction, qPCR can detect small amounts of parasite-specific DNA.5 This difference is important when calculating prevalence in a population and for elimination programs, especially where the reservoir of parasite infection has become difficult to identify. The qPCR would have an advantage in detecting low levels of infection, improving the detection of infected persons, and assessing treatment effects.
For some (but not all) gastrointestinal parasites, antibody and antigen assays are available and have varying performance characteristics.6,7 Moreover, the known limitations of these assays, their availability for only a limited number of parasites, and their cost often make them unavailable for those in resource-limited settings.8
The qPCR has been useful in the diagnosis of pathogenic microbes causing tissue infections, including amebic liver abscess, and has also been shown to have a high degree of sensitivity and specificity in the detection of enteric pathogens.2,3,9 As with other real-time PCR systems, to broaden the spectrum of enteric pathogen detection, a simple to execute, multi-parallel qPCR approach was developed that enables detection of an unlimited number of a parasite species provided that sequence information is available.
In the present study, we have used assays to identify and quantify Ascaris lumbricoides, Cryptosporidium spp. (C.parvum/C. hominis), Ancylostoma duodenale, Necator americanus, Strongyloides stercoralis, Giardia lamblia, Entamoeba histolytica, and Trichuris trichiura DNA in stool in a rapid, high-throughput fashion by using a method that we term multi-parallel qPCR. We have validated this approach for parasite identification in stool from patients in North America, India, and Mali, and then used these assays in a relatively resource-limited area of rural Ecuador.
Materials and Methods
Sample collection.
Fecal samples were collected from 400 asymptomatic children 13 months of age living in the District of Quinindé, Esmeraldas Province, Ecuador, as part of a larger ongoing birth cohort study in which direct microscopy and the Kato-Katz method were performed at the time of stool collection.10 Stool was then frozen without fixatives at −80°C until used for DNA extraction.
A second group of 125 asymptomatic children 8–14 years of age also had fecal samples collected as part of a separate study of anthelmintic treatment with three days of albendazole, 800 mg/day, and a single dose (200 μg/kg) of ivermectin.11 Stool samples were collected at pre-treatment and at 21 days post-treatment. An aliquot of the stool sample was fixed in ethanol and stored at −80°C until DNA was extracted.
Informed written consent was obtained from a parent or guardian of each child, and minor assent was obtained from older children in the second study. The results of stool microscopy were given to the parent/guardian and anti-parasite treatment was provided where appropriate. The study protocols were approved by the Bioethics Committee of the Universidad San Francisco de Quito, Quito, Ecuador.
Microscopy.
Stool samples were examined by using single wet mount smears in saline and by duplicate Kato-Katz slides containing each approximately 50 mg of stool. All slides were processed and examined within 24 hours of preparation. All slides were examined by two experienced parasitologists. In the case of the Kato-Katz method, slides were examined after 15 minutes for hookworm ova, then again after one hour for other helminth ova. Results were used to estimate geometric mean eggs count per gram of stool. A slide from each stool was prepared by using modified acid-fast staining for detection of Cryptosporidium spp. using standard methods.
The qPCR was compared with direct (wet mount) microscopy to evaluate the efficacy of direct microscopy in detecting any parasite. Direct microscopy is the standard of diagnosis in resource-limited areas. The Kato-Katz method was used for estimating infection intensities with A. lumbricoides and T. trichiura. Although this method is more sensitive than direct microscopy, it is more time-consuming and labor-intensive in detecting geohelminth eggs.
DNA extraction.
DNA was extracted from 50 mg stool by using the MP FastDNA for Soil Kit (MP Biochenicals, Solon, OH) according to manufacturer's instructions for all parasites except T. trichiura.12 Extractions were processed in batches of 24 samples, which was the capacity of the homogenizer instrument used (FastPrep-24; MP Biochemicals). An additional step was needed for extraction of T. trichiura DNA; the insoluble pellet from the FastDNA Soil Kit was resuspended in 200 μL of water, heated at 90°C for 10 minutes, and centrifuged at 14,000 × g for 10 minutes. The soluble fraction was then processed further by using the MP FastDNA procedure, as described by the manufacturer, in a final volume of 50 μL.
Multi-parallel qPCR.
Species-specific primers and FAM-labeled minor groove binder probes were selected (Applied Biosystems, Foster City, CA) for each of the eight parasites (Table 1) . Primer/probes were designed by using Primer Express Software version 3.0.1 (Applied Biosystems). To the best of our knowledge, all primers and probes were novel except for A. lumbricoides (forward primer), A. duodenale, N. americanus, and S. stercoralis.3,4 Each set of primers and probes was synthesized by Applied Biosystems.
Table 1
Sequence information for eight parasites tested by using a high-throughput quantitative multi-parallel real-time polymerase chain reaction*
| Parasite | Forward primer sequence, 5′→3′ | Target region | GenBank accession no. |
|---|---|---|---|
| Reverse primer sequence, 5′→3′ | |||
| Probe sequence (FAM), 5′→3′ | |||
| Ascaris lumbricoides4 | TGCACATAAGTACTATTTGCGCGTAT | ITS-1 | AB571301.1 |
| CCGCCGACTGCTATTACATCA | |||
| GAGCCACATAGTAAATT | |||
| Cryptosporidium parvum/hominis | AACTTCACGTGTGTTTGCCAAT | DNA J-like protein | XM_625506.1 |
| CCAATCACAGAATCATCAGAATCG | |||
| CATATGAAGTTATAGGGATACCAG | |||
| Ancylostoma duodenale3 | GAATGACAGCAAACTCGTTGTTG | ITS-2 | EU344797.1 |
| ATACTAGCCACTGCCGAAACGT | |||
| ATCGTTTACCGACTTTAG | |||
| Necator americanus3 | CTGTTTGTCGAACGGTACTTGC | ITS-2 | AJ001599.1 |
| ATAACAGCGTGCACATGTTGC | |||
| CTGTACTACGCATTGTATAC | |||
| Strongyloides stercoralis3 | GAATTCCAAGTAAACGTAAGTCATTAGC | 18S rRNA | AF279916.2 |
| TGCCTCTGGATATTGCTCAGTTC | |||
| ACACACCGGCCGTCGCTGC | |||
| Giardia lamblia | CATGCATGCCCGCTCA | 16S rRNA | AJ293299.1 |
| AGCGGTGTCCGGCTAGC | |||
| AGGACAACGGTTGCAC | |||
| Entamoeba histolytica | GTTTGTATTAGTACAAAATGGCCAATTC | 18S rRNA | X75434.1 |
| TCGTGGCATCCTAACTCACTTAGA | |||
| CAATGAATTGAGAAATGACA | |||
| Trichuris trichiura | TCCGAACGGCGGATCA | ITS-1 | FM991956.1 |
| CTCGAGTGTCACGTCGTCCTT | |||
| TTGGCTCGTAGGTCGTT |
All qPCRs were conducted in 96-well MicroAmp optical plates (Applied Biosystems). All reactions were performed in a total volume of 7 μL containing 3.5 μL of Taqman™ fast mix (Applied Biosystems), 2 μL of template DNA, and 1.5 μL of species-specific primers (final concentration = 900 μM) and FAM-labeled minor grove binder probes (final concentration = 100 μM). Samples were run on an ABI 7900HT Fast Real-Time PCR System (Applied Biosystems) in North America or on an ABI 7500 apparatus in Ecuador by using default parameters and 40 cycles. The qPCR results were considered negative if cycle threshold (Ct) values were > 38. This threshold was determined by measuring the detectable limits of each assay in which serial dilutions of appropriate parasite plasmid were included. The Ct value was set at 38, and this value was the limit of our standard curves (additional plasmid dilutions were undetectable). All unknown samples were tested in duplicate.
For quantification, plasmids containing the target sequences were synthesized (Genscript, Piscataway, NJ). TOP10 competent E. coli cells (Life Technologies, Grand Island, NY) were transformed with the individual plasmids and the plasmids were purified by using the QIAprep Spin Miniprep Kit (QIAGEN, Valencia, CA). The purified plasmid DNA was quantified by using a ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE). Plasmids were then diluted to a fixed concentration, and serial 10-fold dilutions were made for use in a standard curve format to which unknown samples were compared. The standards served as positive controls to ensure amplification and wells with no template were used as the negative controls. All controls were tested in triplicate.
These assays were validated by using genomic DNA spiked into known negative stool and/or stools from patients known to be parasite positive by standard stool examination (performed by Clinical Center Microbiology Laboratory, National Institutes of Health, Bethesda, MD) by using standard techniques (e.g., direct microscopy, modified acid-fast staining for Cryptosporidium, and agar plate migration for S. stercoralis).6 Validation using similar techniques had been performed for all parasites except T. trichiura.2,3 In addition, stools cryopreserved in the United States, Mali, or India (as part of other studies) that had multiple negative results for stool examinations for helminth eggs and larvae were tested by using qPCR. All samples showed negative results.
Plates were prepared before DNA extraction with all reaction mixtures (standards and controls) minus 2 μL to enable patient samples to be added. Plates were prepared in Ecuador and stored at −40°C with airtight aluminum foil covers. Each plate held eight standards, controls and space for 36 unknown samples to be processed in duplicate. After DNA extraction, samples could be loaded and processed by using fast chemistry parameters (30 minutes), which provided a rapid and efficient system from extraction to results.
Statistical analysis.
All statistical analysis was performed by using Prism version 5.0d (GraphPad, La Jolla, CA). Comparisons of microscopy with qPCR were analyzed by using Fisher's exact test, and P values < 0.05 was considered significant. Correlations were estimated by calculation of Spearman's rank correlation coefficients. The non-parametric Wilcoxon-signed rank test was used for paired analyses. Specificity was calculated by using a binary classification test comparing a single qPCR result with the geometric mean of two Kato-Katz results.
Results
Validation and specificity of qPCR for eight parasites.
Each primer/probe combination had 100% specificity for its respective parasite. Using genomic DNA for A. lumbricoides, A. duodenale, N. americanus, S. stercoralis, G. lamblia, and E. histolytica, we showed that our assays specifically amplified the appropriate target genomic DNA but there was no amplification of DNA from the other parasites. For example, qPCR using the primers and probes for A. lumbricoides detected A. lumbricoides DNA but failed to amplify DNA from A. duodenale, N. americanus, S. stercoralis, G. lamblia, and E. histolytica. In a limited laboratory setting, all eight parasite DNA plasmids (used to generate standard curves for each assay) were analyzed by using every prime/probe combination individually and as combined mixtures and no cross-reactivity seen. Furthermore, using cryopreserved stool samples from > 300 persons with negative stool examinations results performed as part of other studies in the United States, Mali and India, we found that there were no false positive results in any of the assays.
In addition, 10 patients with positive stool examination results at the Clinical Center Microbiology Laboratory of the National Institutes of Health were tested by qPCR concurrently, showed 100% congruency (A. lumbricoides, C. parvum/C. hominis, A. duodenale, S. stercoralis, G. lamblia, E. histolytica, and T. trichiura). One patient had an Entamoeba sp. detected in stool, but was negative for E. histolytica by the qPCR described in this report. Subsequent testing for E. histolytica stool antigen using an enzyme immunoassay (Focus Diagnostics, Cypress, CA) suggested that this parasite was the non-pathogenic species E. dispar. The limit of our qPCR was 0.001 fg/μL of extracted DNA for each of the eight parasite DNAs.
Comparison of qPCR with microscopy.
For all qPCRs, negative controls showed no amplification. For each parasite sample in which there was specific amplification, the Ct values were compared with a standard curve for which absolute DNA concentrations were known and concentrations of DNA were derived by interpolation from a given standard. As shown in Table 2, based on data from the 400 children 13 months of age, qPCR was more sensitive than direct microscopy for A. lumbricoides, A. duodenale (the only hookworm species detected in this study), S. stercoralis, G. lamblia (P = 0.0001), and T. trichiura (P = 0.0335). For A. lumbricoides, PCR identified 28 (7.0%) of 400 positive samples whereas direct smears identified 22 (5.5%) of 400 positive samples. The qPCR identified an additional six A. lumbricoides-positive samples with a specificity of 97.8% and a negative predictive value of 96.7%. For T. trichiura (3% versus 0.75%) nine additional positive samples were detected (12 of 400 versus 3 of 400; P = 0.0335), which indicated a specificity of 99.2% and a negative predictive value of 99.2%.
Table 2
Comparison of gastrointestinal parasite detection in 400 13-month-old children, Ecuador*
| Parasite | No. positive by multi-parallel qPCR (%) | No. positive by direct microscopy (%) | DNA concentrations (fg/μL) in stool positive by microscopy, median (range) | DNA concentrations (fg/μL) in stool negative by microscopy, median (range) |
|---|---|---|---|---|
| Ascaris lumbricoides | 28/400 (7.0) | 22/400 (5.5) | 7.14 (0.043–252.5) | 0.031 (0.0016–4.7) |
| Cryptosporidium parvum/hominis† | 21/400 (5.3) | 0/400 | 0‡ | 0.17 (0.049–15.6) |
| Ancylostoma duodenale | 2/400 (0.5) | 0/400 | 0‡ | 37.5 (23.9–51.2) |
| Necator americanus | 0/400 | 0/400 | 0‡ | 0‡ |
| Strongyloides stercoralis | 3/400 (0.75) | 0/400 | 0‡ | 0.089 (0.0144–0.163) |
| Giardia lamblia† | 126/400 (31.5) | 23/400 (5.8) | 24.3 (0.9–359) | 5.0 (0.0457–47.9) |
| Entamoeba histolytica§ | 4/400 (1.0) | 6/400 (1.5) | 0‡ | 0.46 (0.036–0.966) |
| Trichuris trichiura† | 12/400 (3.0) | 3/400 (0.75) | 0.014 (0.0035–0.032) | 0.013 (0.0025–0.175) |
Although specificity in the validation test was 100%, we also report field data specificity and negative predictive values for A. lumbricoides and T. trichiura. The difference between qPCR and microscopy was seen even more dramatically for G. lamblia (31.5% versus 5.8%) with 103 additional positive samples for G. lamblia (126 of 400 versus 23 of 400; P = 0.001). For C. parvum/C. hominis, specific DNA was detected in 5.3% of the children, and none of the samples was positive by modified acid-fast staining (P = 0.0001). For the hookworm species, two children had A. duodenale DNA (no N. americanus DNA was detected) although no hookworm was seen by microscopy. Three children had S. stercoralis DNA in their stool, but no S. stercoralis parasites were found by microscopy. For Entamoeba, our assay is specific for E. histolytica, and we detected E. histolytica DNA in 4 of 400 children. Six children had E. histolytica/dispar cysts by microscopy, but none were positive by qPCR.
To assess whether molecular diagnostic technique could be considered quantitative, egg counts determined by the Kato-Katz method were compared with the DNA quantitation used for the qPCR for A. lumbricoides and T. trichiura (Figure 1). There was a statistically significant correlation between egg counts measured by the Kato-Katz method and the DNA quantified by qPCR for A. lumbricoides (r = 0.713, P < 0.05) and T. trichiura (r = 0.743, P < 0.05) (Figure 1).
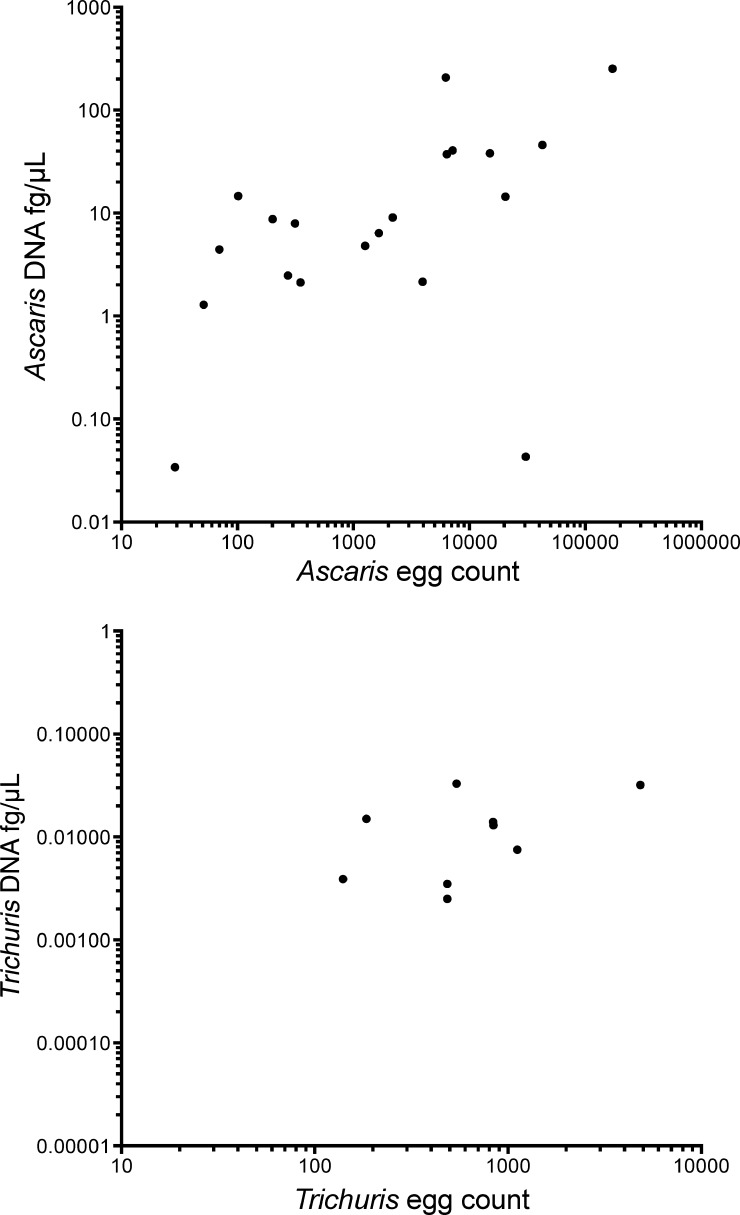
Correlation of parasite DNA concentration (fg/μL) with egg count by Kato-Katz method for Ascaris lumbricoides (top) (r = 0.713, P < 0.0001) and Trichuris trichiura (bottom) (r = 0.743, P < 0.0001) for 400 13-month-old-children, Ecuador. Only samples positive by high-throughput quantitative multi-parallel real-time polymerase chain reaction are shown.
The comparison between DNA concentrations measured by qPCR in positive and microscopy-negative samples resulted in lower DNA concentrations for the negative group for all parasites (except A. duodenale), but this difference was not statistically significant (Table 2).
Detecting polyparasitism by qPCR.
Not only was qPCR able to detect single parasite species more readily (an overall 18.5% increase in detection; P < 0.05), but it also was much more likely to detect two or more concurrent infections in a given sample than was microscopy (P < 0.05) (Figure 2A). The qPCR was able to distinguish between patients without parasites and those with polyparasitism more accurately and with greater detection rates than direct smear microscopy (61.0% to 87.3%). The use of direct smears led to underreporting of the presence of any parasite (Figure 2A). When looking at specific co-infections, qPCR was able to detect G. lamblia/A. lumbricoides co-infections more often than direct microscopy (P < 0.05). It also detected A. lumbricoides/T. trichiura and G. lamblia/T. trichiura co-infections more frequently than by direct microscopy. The qPCR was also able to identify the one quadruple infection seen among the 400 patients, whereas direct microscopy failed to do so (Figure 2B).
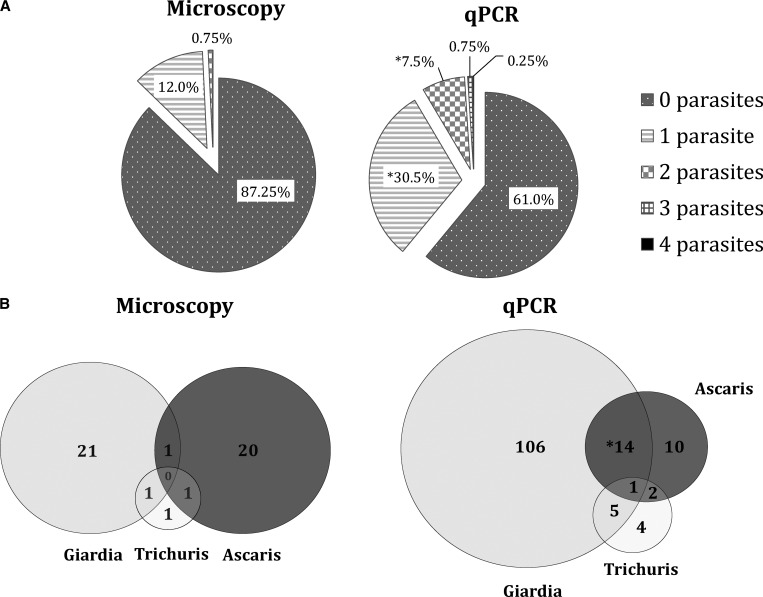
Detection of polyparasitism by quantitative multi-parallel real-time polymerase chain reaction (qPCR) and microscopy in 400 13-month-old children, Ecuador. A, Proportion of stools positive for 0–4 parasites by either microscopy or qPCR. Asterisk indicates statistical significance (P < 0.05) difference in detection between microscopy and qPCR. B, Improved detection of the three most prevalent infections showing the distribution (in a Venn diagram) of these parasites based on microscopy or qPCR. Asterisk indicates a statistically significant (P < 0.05) difference in detection between microscopy and PCR.
Post-treatment results after empiric therapy.
In a separate cohort of 125 asymptomatic children 8–14 years of age whose stool samples were collected before and after empiric treatment with albendazole and ivermectin, we were able to assess the utility of qPCR in following parasite burden after treatment (Figure 3). Comparison of microscopy and qPCR yielded similar data as obtained for the other study sample. Twenty-one days after therapy, all persons with positive qPCR results for A. lumbricoides before treatment were completely below the threshold of detection at 21 days, a change that was highly significant (P < 0.0001), and two children initially negative were positive on day 21. For G. lamblia, 35.1% of the patients remained positive after albendazole/ivermectin therapy, although for some, there was a non-significant decrease in measured DNA levels.
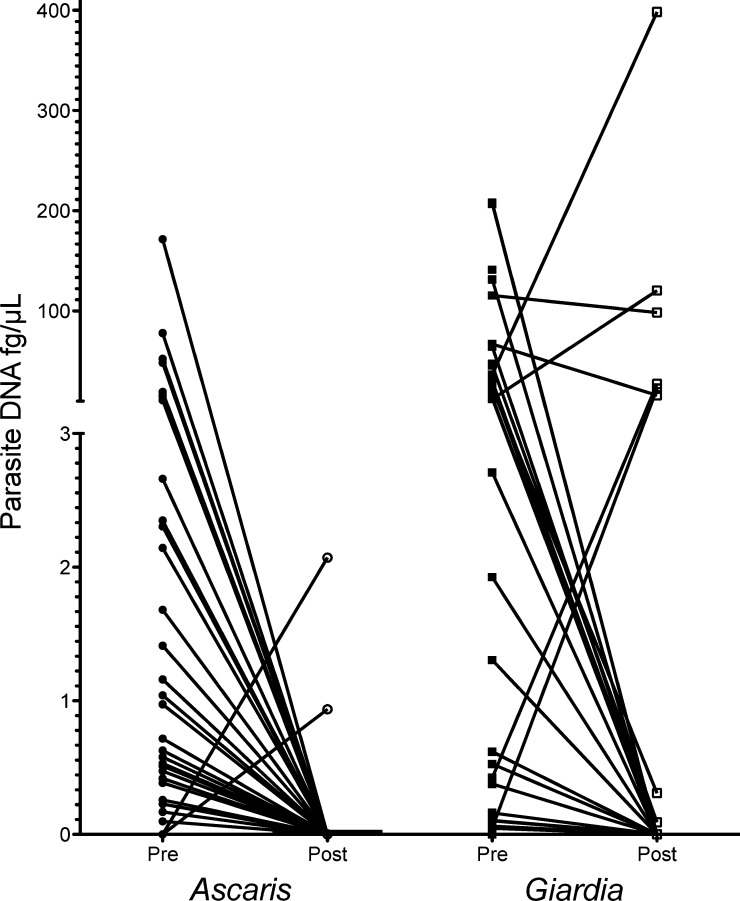
Prevalence of Ascaris lunbricoides and Giardia lamblia before (pre) and 21 days after (post) albendazole/ivermectin treatment in 125 children 8–14 years of age, Ecuador. A. lumbricoides DNA decreased to undetectable levels post-treatment with the exception of two samples. A total of 35.1% of children remained positive for G. lamblia after empiric treatment.
Discussion
A novel high throughput quantitative assay for the identification of gastrointestinal parasites has been developed and implemented in a resource-limited setting. Although molecular approaches to stool parasite pathogens have been described in other studies using multiplex assays, they are limited often by upfront technical problems and cost.2,3 Multiplex real-time PCR and this multi-parallel qPCR system were designed to compliment poorly sensitive microscopy and enable higher rates of detection, especially when used in epidemiologic studies.
Although direct observation of a stool sample in a wet smear, Kato-Katz slides, and stool concentration methods have been used for diagnosing diarrheal illness and for evaluation of eosinophilia, the sensitivity and specificities of microscopy depend on the type of organism, its life cycle, and the pattern of egg or larval shedding. Although qPCR also relies on these factors, the ability to detect attograms of parasitic DNA overcomes many of these potential obstacles because of higher sensitivity. In the present study, we compared our qPCR to direct smear microscopy primarily to normalize the conditions between the two tests (Table 2). Although the Kato-Katz/fecal concentration method generally has a higher sensitivity than direct microscopy, and for A. lumbricoides and T. trichiura is semi-quantitative, it is more time-consuming and relies on collection of multiple stool samples to maximize sensitivity. Antigen detection assays with improved parasite detection ability are available (e.g., for G. lamblia and E. histolytica), but these assays are costly and not readily available in resource-limited areas.7,13
Another finding was the higher levels of parasitic co-infections detected by using qPCR than by direct microscopy in this study. Polyparasitism is an important factor in the decision for implementing mass drug administration and for selecting appropriate anti-parasitic drugs.14 Although polyparasitism is common among adults and adolescents in many parasite-endemic areas, using a highly sensitive qPCR, even with samples from 13-month-old children, we found an 8.5% prevalence of co-infection with two or more parasites. The implications are significant in that community-based mass drug administrations are often directed at soil-transmitted helminths, but not for other parasites such as G. lamblia, C. parvum/C. hominis, and E. histolytica.15 Moreover, identifying polyparasitized persons may provide insights into the pathologic consequences of the interaction between phylogenetically distinct parasites sharing the same anatomic niche.
A relatively low prevalence of A. duodenale and S. stercoralis was found by using qPCR that would have been missed if microscopy alone was used (Table 2). The median concentrations of DNA from these organisms were found to range from 0.089 to 37.5 fg/μL (Table 2). Because no eggs/larvae were seen by microscopy, we cannot say with any certainty that our failure to detect infection by microscopy was caused by low infection levels. A reasonable alternative and biologically plausible explanation is that qPCR can detect DNA from any lifecycle stage (e.g., larvae for hookworm), whereas microscopy typically is optimized for a single stage.
The major limitation of the present study was the absence of an internal control to assess potential inhibition of the DNA extraction method or the qPCR. Although an internal control is important for detecting inhibition and thereby false-negative samples, it was not available at the time the study was conducted. For the present study, PCR inhibition (although always a possibility) appeared to be of little consequence given the general increase in detection seen by the qPCR method. Furthermore, our methods of DNA extraction were chosen to prevent such inhibition.16,17 In a separate experiment, stools were processed with and without polyvinylpyrrolidone. No differences in DNA detection for A. lumbricoides were observed between the two groups. Future projects will include the use of limiting concentrations of a control plasmid in all reactions.18
The benefits of qPCR are the increased detection rates and significantly increased specificity. For example, the increased specificity of qPCR enabled us to distinguish between the two species of hookworm. Although treatment is the same for infection with either species, understanding the predominant species will help understand transmission sources and enable better characterization of the clinical differences between the two genera.19 Another important feature of the higher specificity of molecular diagnostics is the ability to distinguish between the non-pathogenic E. dispar and the human pathogen E. histolytica, a parasite that causes 100,000 deaths per year.20 Our assay could distinguish between the pathogenic and non-pathogenic Entamoebae, enabling pathogen-directed treatment. Our ability to identify Cryptosporidium without relying on another diagnostic approach (such as using the modified acid-fast or enzyme-linked immunosorbent assay) is also useful.21 Because our qPCR is genus specific, we could detect C. parvum and C. hominis. Although cryptosporidiosis is generally a self-limiting illness, it can cause severe illness and high mortality rates in immunocompromised patients.22 Because of these reasons, qPCR can replace a large range of species-specific microscopic methods.
Our assay was also useful in monitoring post-treatment efficacy for infection with A. lumbricoides and G. lamblia. Surprisingly for A. lumbricoides, two persons who were parasite negative before therapy showed parasite-positive results when tested by qPCR, suggesting that they acquired A. lumbricoides infection during the study period.
Results for G. lamblia were mixed; approximately 65% of the children had G. lamblia DNA in stool. This finding was expected because the infection in more than 50% of the patients with giardiasis will self-resolve, and five days of treatment with albendazole has been shown to be as effective as treatment with metronidazole.23,24 In the present study, patients received albendazole for three days, a dosing schedule that might eliminate G. lamblia in some situations. The ability to screen a large number of persons at one time and perform follow-up after treatment would be useful in the context of a public health setting.
To decrease the per-reaction cost, we were also able to show we could reduce the reagent costs by 67%, by merely lowering the volume of the reagent mixture, without compromising detection limits. In addition, we identified a method of DNA extraction that then enabled detection of T. trichiura DNA in stool. This relatively simple method overcomes the difficulty that has heretofore affected stool-based molecular diagnostics for helminths.25 Not taking into account the cost of the instrument needed to perform the qPCR, we have decreased the cost of analyzing all eight parasites in a patient's stool for less than $1.00 U.S. dollar per sample compared with recent published cost of $2.60 for microscopy.25 Although stool samples were stored in either −40°C or −80°C freezers, this step was necessary because the stool had been collected and stored for other analyses. For qPCR, the samples could be assayed immediately after collection, stored at room temperature in 70% ethanol, or stored in a 4°C refrigerator for extended periods. In the case of storage at 4°C, parasite cysts and ova may be stable for prolonged periods, thus avoiding the expense of a deep-freezer.
Although multiplex PCR has made great strides in advancing the diagnosis of gastrointestinal parasites,2,3,25 we provide an alternative method that requires less sophisticated instrumentation. More importantly, this system was the most cost-effective method of transferring this technology to a relatively resource-limited area. This finding will enable more widespread use.
With more than two billion gastrointestinal parasite infections worldwide and an increasing body of evidence that long-term infections lead to decrease cognitive abilities, developmental delays, and stunted growth rates, we present a high throughput system that has been field tested in a resource-limited area.26 This method provides a sensitive and specific approach to gastrointestinal parasitism and polyparasitism, with broad implications for community based therapies and methods for assessing efficacy of treatment.
Footnotes
Authors' addresses: Rojelio Mejia, National School of Tropical Medicine, Baylor College of Medicine, Houston, TX, E-mail: [email protected]. Thomas B. Nutman, Helminth Immunology Section, Laboratory of Parasitic Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, E-mail: vog.hin.diain@namtunt. Yosselin Vicuña, Nely Broncano, Carlos Sandoval, Maritza Vaca, Martha Chico, and Philip J. Cooper, Colegio de Ciencias de la Salud, Universidad San Francisco de Quito, Fundación Ecuatoriana para la Investigación en Salud, Quito, Ecuador, E-mails: moc.liamg@ivssoy, se.liamtoh@seylen, moc.liamtoh@solraca_lavodnas, moc.oohay@qce_ramiram, se.oohay@6ocihcahtram, and [email protected].
References
Articles from The American Journal of Tropical Medicine and Hygiene are provided here courtesy of The American Society of Tropical Medicine and Hygiene
Full text links
Read article at publisher's site: https://doi.org/10.4269/ajtmh.12-0726
Read article for free, from open access legal sources, via Unpaywall:
http://www.ajtmh.org/deliver/fulltext/14761645/88/6/1041.pdf?itemId=%2Fcontent%2Fjournals%2F10.4269%2Fajtmh.12-0726&mimeType=pdf&containerItemId=content/journals/14761645
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.4269/ajtmh.12-0726
Article citations
Effective Laboratory Diagnosis of Parasitic Infections of the Gastrointestinal Tract: Where, When, How, and What Should We Look For?
Diagnostics (Basel), 14(19):2148, 27 Sep 2024
Cited by: 0 articles | PMID: 39410552 | PMCID: PMC11475984
Review Free full text in Europe PMC
Comparison of multi-parallel quantitative real-time PCRs targeting different DNA regions and detecting soil-transmitted helminths in stool.
Parasit Vectors, 17(1):390, 13 Sep 2024
Cited by: 0 articles | PMID: 39272159 | PMCID: PMC11397029
Molecular Testing of Environmental Samples as a Potential Source to Estimate Parasite Infection.
Trop Med Infect Dis, 9(10):226, 26 Sep 2024
Cited by: 0 articles | PMID: 39453253 | PMCID: PMC11511502
Soil surveillance for monitoring soil-transmitted helminths: Method development and field testing in three countries.
PLoS Negl Trop Dis, 18(9):e0012416, 06 Sep 2024
Cited by: 2 articles | PMID: 39241051 | PMCID: PMC11469484
Parasitic Contamination of Soil in the Southern United States.
Am J Trop Med Hyg, 111(3):506-514, 23 Jul 2024
Cited by: 3 articles | PMID: 39043177 | PMCID: PMC11376165
Go to all (160) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (Showing 7 of 7)
- (1 citation) ENA - AB571301
- (1 citation) ENA - AJ001599
- (1 citation) ENA - FM991956
- (1 citation) ENA - X75434
- (1 citation) ENA - AJ293299
- (1 citation) ENA - AF279916
- (1 citation) ENA - EU344797
Show less
RefSeq - NCBI Reference Sequence Database
- (1 citation) RefSeq - XM_625506.1
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Multi-parallel qPCR provides increased sensitivity and diagnostic breadth for gastrointestinal parasites of humans: field-based inferences on the impact of mass deworming.
Parasit Vectors, 9:38, 27 Jan 2016
Cited by: 106 articles | PMID: 26813411 | PMCID: PMC4729172
Identification of human intestinal parasites affecting an asymptomatic peri-urban Argentinian population using multi-parallel quantitative real-time polymerase chain reaction.
Parasit Vectors, 8:380, 17 Jul 2015
Cited by: 50 articles | PMID: 26183074 | PMCID: PMC4504406
High throughput multiplex PCR and probe-based detection with Luminex beads for seven intestinal parasites.
Am J Trop Med Hyg, 84(2):332-337, 01 Feb 2011
Cited by: 113 articles | PMID: 21292910 | PMCID: PMC3029193
Is real-time PCR-based diagnosis similar in performance to routine parasitological examination for the identification of Giardia intestinalis, Cryptosporidium parvum/Cryptosporidium hominis and Entamoeba histolytica from stool samples? Evaluation of a new commercial multiplex PCR assay and literature review.
Clin Microbiol Infect, 22(2):190.e1-190.e8, 06 Nov 2015
Cited by: 38 articles | PMID: 26548509
Review
Funding
Funders who supported this work.
Wellcome Trust (1)
Impact of early infectious and microbial exposures on the development of immunity and allergic inflammatory disease in children living in a tropical region of Ecuador.
Professor Philip Cooper, St George's University of London
Grant ID: 088862





