Abstract
Background
Hepatocellular carcinoma is the third leading cause of cancer-related deaths worldwide. In the heterogeneous group of hepatocellular carcinomas, those with characteristics of embryonic stem-cell and progenitor-cell gene expression are associated with the worst prognosis. The oncofetal gene SALL4, a marker of a subtype of hepatocellular carcinoma with progenitor-like features, is associated with a poor prognosis and is a potential target for treatment.Methods
We screened specimens obtained from patients with primary hepatocellular carcinoma for the expression of SALL4 and carried out a clinicopathological analysis. Loss-of-function studies were then performed to evaluate the role of SALL4 in hepatocarcinogenesis and its potential as a molecular target for therapy. To assess the therapeutic effects of a peptide that targets SALL4, we used in vitro functional and in vivo xenograft assays.Results
SALL4 is an oncofetal protein that is expressed in the human fetal liver and silenced in the adult liver, but it is reexpressed in a subgroup of patients who have hepatocellular carcinoma and an unfavorable prognosis. Gene-expression analysis showed the enrichment of progenitor-like gene signatures with overexpression of proliferative and metastatic genes in SALL4-positive hepatocellular carcinomas. Loss-of-function studies confirmed the critical role of SALL4 in cell survival and tumorigenicity. Blocking SALL4-corepressor interactions released suppression of PTEN (the phosphatase and tensin homologue protein) and inhibited tumor formation in xenograft models in vivo.Conclusions
SALL4 is a marker for a progenitor subclass of hepatocellular carcinoma with an aggressive phenotype. The absence of SALL4 expression in the healthy adult liver enhances the potential of SALL4 as a treatment target in hepatocellular carcinoma. (Funded by the Singapore National Medical Research Council and others.).Free full text

Oncofetal Gene SALL4 in Aggressive Hepatocellular Carcinoma
Abstract
BACKGROUND
Hepatocellular carcinoma is the third leading cause of cancer-related deaths worldwide. In the heterogeneous group of hepatocellular carcinomas, those with characteristics of embryonic stem-cell and progenitor-cell gene expression are associated with the worst prognosis. The oncofetal gene SALL4, a marker of a subtype of hepatocellular carcinoma with progenitor-like features, is associated with a poor prognosis and is a potential target for treatment.
METHODS
We screened specimens obtained from patients with primary hepatocellular carcinoma for the expression of SALL4 and carried out a clinicopathological analysis. Loss-of-function studies were then performed to evaluate the role of SALL4 in hepatocarcinogenesis and its potential as a molecular target for therapy. To assess the therapeutic effects of a peptide that targets SALL4, we used in vitro functional and in vivo xenograft assays.
RESULTS
SALL4 is an oncofetal protein that is expressed in the human fetal liver and silenced in the adult liver, but it is reexpressed in a subgroup of patients who have hepatocellular carcinoma and an unfavorable prognosis. Gene-expression analysis showed the enrichment of progenitor-like gene signatures with overexpression of proliferative and metastatic genes in SALL4-positive hepatocellular carcinomas. Loss-of-function studies confirmed the critical role of SALL4 in cell survival and tumorigenicity. Blocking SALL4–corepressor interactions released suppression of PTEN (the phosphatase and tensin homologue protein) and inhibited tumor formation in xenograft models in vivo.
CONCLUSIONS
SALL4 is a marker for a progenitor subclass of hepatocellular carcinoma with an aggressive phenotype. The absence of SALL4 expression in the healthy adult liver enhances the potential of SALL4 as a treatment target in hepatocellular carcinoma. (Funded by the Singapore National Medical Research Council and others.)
Hepatocellular carcinoma is the third leading cause of cancer-related deaths globally. Although the epidemiologic risk factors for hepatocellular carcinoma are well known,1 the molecular mechanisms underlying hepatocarcinogenesis are not well characterized. Elucidation of these mechanisms may allow identification of new candidates for therapeutic targeting. Although surgery, liver transplantation, and radiologic intervention may be viable options for patients with early-stage disease, the prognosis associated with advanced-stage hepatocellular carcinoma remains bleak.2 Combination chemotherapy has not improved overall survival but has nonetheless been in wide use for many years because of its possible role in palliation. The need to understand the molecular pathogenesis of hepatocellular carcinoma and develop more effective targeted therapies remains urgent.
The human homologue of the Drosophila spalt homeotic gene, SALL4, encodes a C2H2 zinc-finger transcription factor.3 It is one of the key factors for maintenance of pluripotency and self-renewal of embryonic stem cells.4–6 In the murine liver, Sall4 expression diminishes gradually during development and is silenced by adulthood; this suggests a role for Sall4 during early-to-middle development of the fetal liver.7 SALL4 is an oncogene that was first described in leukemia. SALL4 was found to be constitutively expressed in human acute myeloid leukemia, and acute myeloid leukemia developed in SALL4B–transgenic mice.8 Subsequently, SALL4 expression has been reported in various types of cancers9,10 and is proposed to have diagnostic value in several of them.11–13
The progenitor-like subtype of hepatocellular carcinoma is known to be associated with a poor prognosis.14 The expression of Sall4 in murine fetal liver but not in adult liver led us to hypothesize that SALL4 might be an important marker for the progenitor-like subtype of hepatocellular carcinoma. We also hypothesized that SALL4 would contribute to the development and persistence of hepatocellular carcinoma when it was expressed in hepatocytes in adults.
METHODS
PATIENTS AND SAMPLES
We obtained specimens of hepatocellular carcinoma from patients at the National University Hospital in Singapore (hereafter referred to as the Singapore specimens) to construct hepatocellular-carcinoma tissue microarrays. We also analyzed a second batch of specimens obtained from patients at the Queen Mary Hospital in Hong Kong (hereafter referred to as the Hong Kong specimens). The patients received various treatments and were not participants in any clinical trials evaluating treatment approaches. The institutional review board of the National University of Singapore approved the study, and patients provided written informed consent for the use of all surgically removed tissue specimens used in this study.
STUDY CONDUCT
The study had no commercial sponsors. The authors vouch for the accuracy and completeness of the data and analysis. No one who is not listed as an author contributed to the manuscript. Four of the authors hold a patent application for intellectual property derived from this study.
STATISTICAL ANALYSIS
All experiments were performed in triplicate unless otherwise stated. Statistical analyses were performed with the use of SPSS software, versions 15.0 and 16.0 (SPSS). To examine the association between levels of SALL4 expression and clinicopathological characteristics, Fisher’s exact test was used for analysis of the Singapore specimens of primary hepatocellular carcinomas and the chi-square test was used for analysis of the Hong Kong specimens. In the Hong Kong specimens, levels of SALL4 expression were divided into two groups. The group with a high level of SALL4 expression (high-SALL4 group) had SALL4 expression intensity that was equal to or exceeded the median level of expression detected on microarray analysis, whereas the group with a low level of SALL4 expression (low-SALL4 group) had SALL4 expression intensity that was lower than the median level of expression. Univariate analyses of cumulative overall survival and tumor recurrence were performed with the use of a Cox proportional-hazards regression model in the Singapore cohort and with the use of the Kaplan–Meier method followed by the log-rank test in the Hong Kong cohort. Factors with prognostic significance were included in the subsequent multivariate analyses with the use of the Cox proportional-hazards regression model in both cohorts. For statistical analysis of the Singapore specimens of primary hepatocellular carcinomas, 79 of the 179 specimens for which full clinicopathological data were available were analyzed. The correlation significance was determined by means of Spearman and Pearson correlation analyses. The chi-square test and Student’s t-test were used for comparisons between the high-SALL4 and low-SALL4 groups. A P value of 0.05 or less was considered to indicate statistical significance.
RESULTS
SALL4 EXPRESSION IN HEPATOCELLULAR CARCINOMA
To test our hypothesis that SALL4 is reexpressed in a subtype of hepatocellular carcinoma, we examined the expression of SALL4 in liver specimens from patients with and those without hepatocellular carcinomas. With the use of immunohistochemical analysis, we detected SALL4 expression in fetal liver specimens but not in adult liver specimens (Fig. 1A).
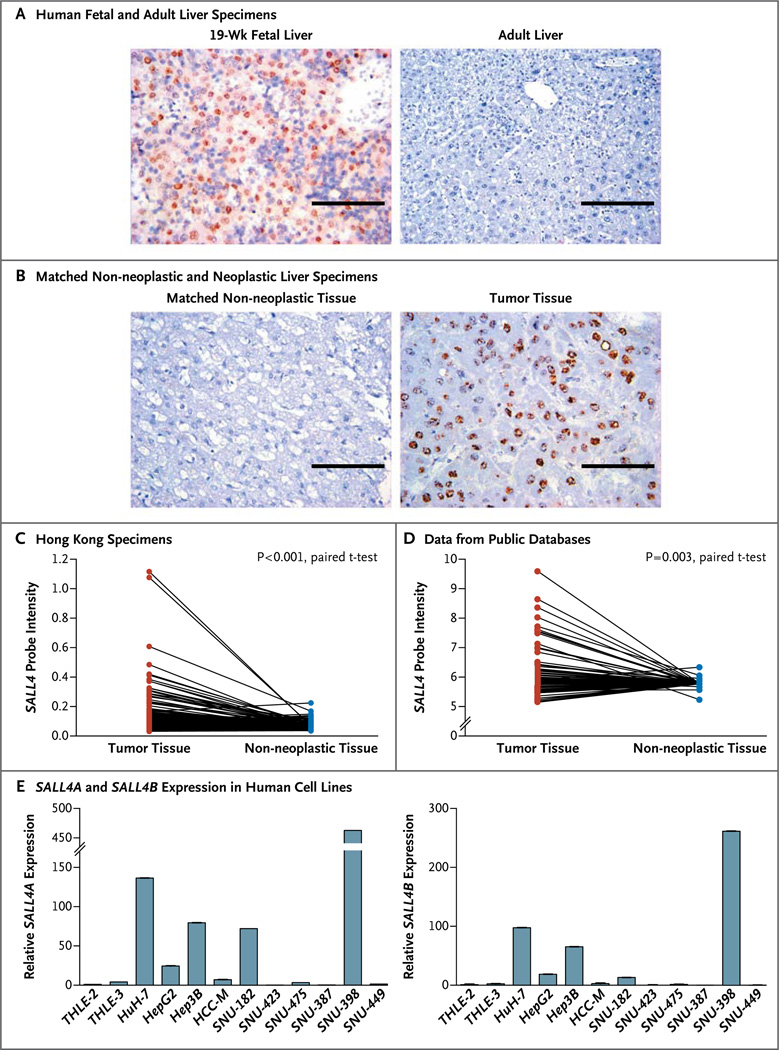
Panel A shows immunohistochemical staining of formalin-fixed, paraffin-embedded human fetal liver and human adult liver. SALL4 expression is indicated by brown staining. Scale bars indicate 100 µm. Panel B shows immunohistochemical staining of a hepatocellular carcinoma specimen and the matched non-neo-plastic liver specimen on tissue microarray analysis. SALL4 expression was detected in the hepatocellular carcinoma tissue and localized in the nucleus, as indicated by the brown staining. Scale bars indicate 100 µm. Panel C shows differential SALL4 expression in 228 matched specimens of primary hepatocellular carcinoma and non-neoplastic liver tissue from the Hong Kong cohort, as determined by means of microarray analysis. Panel D shows microarray SALL4 expression in primary hepatocellular carcinoma and non-neoplastic liver tissue on the basis of pooled data from public databases. Panel E shows the results of quantitative reverse-transcriptase–polymerase-chain-reaction analysis of SALL4A (left) and SALL4B (right) expression in 10 human hepatocellular-carcinoma cell lines and two immortalized nontransformed hepatocyte cell lines, THLE-2 and THLE-3. All values were normalized to ACTB and plotted relative to the expression of the THLE-2 cell line.
We then investigated the level of expression of SALL4 in patients with hepatocellular carcinoma. We constructed a panel of tissue microarrays consisting of 179 surgically resected specimens of primary hepatocellular carcinomas and matched, archived, non-neoplastic liver specimens obtained from the National University Hospital in Singapore. The patients in the Singapore cohort were approximately 56 years of age, 81% were men, and 87.3% had grade I or II tumors. Detailed demographic and clinicopathological characteristics of these patients are described in Table S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org. With the use of immunohistochemical analysis, we observed differential expression of SALL4 in matched tissue specimens, with more SALL4-expressing cells in the specimens with hepatocellular carcinoma than in the matched non-neoplastic specimens (P<0.001) (Fig. 1B). Data from the immunohistochemical analysis showed SALL4 positivity in 55.6% (95 of 171) of the hepatocellular carcinoma specimens analyzed, albeit at various levels of expression (Table S2 in the Supplementary Appendix).
We further confirmed SALL4 up-regulation by means of gene-expression microarray analysis in various independent groups of specimens of primary hepatocellular carcinoma. In a group of 228 matched neoplastic and non-neoplastic liver specimens obtained from patients in Hong Kong, we detected differential SALL4 expression (P<0.001) (Fig. 1C), which was similar to the pattern of expression observed in the Singapore specimens. Table S3 in the Supplementary Appendix lists the demographic and clinicopathological characteristics of the patients with hepatocellular carcinoma in the Hong Kong cohort. Furthermore, we pooled various global gene-expression data from public databases and observed differential SALL4 expression in the hepatocellular-carcinoma and non-neoplastic liver-tissue specimens (P = 0.003) (Fig. 1D). From these sizable independent groups of adult liver specimens, we confirmed that SALL4 was up-regulated in the subgroup of specimens with hepatocellular carcinoma but remained silenced in the matched non-neoplastic specimens.
Analysis of endogenous SALL4 expression across a panel of 10 human hepatocellular carcinoma cell lines by means of a quantitative polymerase-chain-reaction assay showed the expression of SALL4 at various levels, reflecting the expression pattern observed in the hepatocellular carcinoma tissue specimens described above (Fig. 1E). Moreover, SALL4 expression was not detected in the two immortalized, nontransformed liver-cell lines, THLE-2 and THLE-3. These data suggest that these cell lines are appropriate models for further testing of our hypothesis and confirm our finding that SALL4 is reexpressed in a subgroup of hepatocellular carcinomas.
In summary, the results obtained from a number of different assays and groups of clinical specimens showed that SALL4 was expressed in fetal liver tissue, silenced in normal adult liver tissue, and reexpressed in many of the tissue specimens from adults with hepatocellular carcinoma (Fig. S2 in the Supplementary Appendix).
SALL4 AS A PROGNOSTIC FACTOR FOR HEPATOCELLULAR CARCINOMA
To analyze the clinical relevance of SALL4 reactivation in hepatocellular carcinoma, we carried out a clinicopathological analysis of the Singapore and Hong Kong specimens of primary hepatocellular carcinoma. This analysis showed that patients with a high level of SALL4 expression had a worse prognosis than patients with a low level of SALL4 expression. A univariate analysis of survival among patients in the Singapore cohort showed that the absence of SALL4 protein (an immunohistochemical score of 0 on a scale of 0 to 4, with higher scores indicating a greater proportion of positive cells [see the Supplementary Methods section in the Supplementary Appendix]), conferred a significant survival advantage (P = 0.02) (Fig. 2A). Similarly, SALL4 expression was associated with poor survival among the patients in the Hong Kong cohort (P = 0.002) (Fig. 2B). Our univariate analysis also showed that SALL4 was associated with disease recurrence in both the Singapore cohort (Table S4 in the Supplementary Appendix) and the Hong Kong cohort (Table S5 in the Supplementary Appendix).
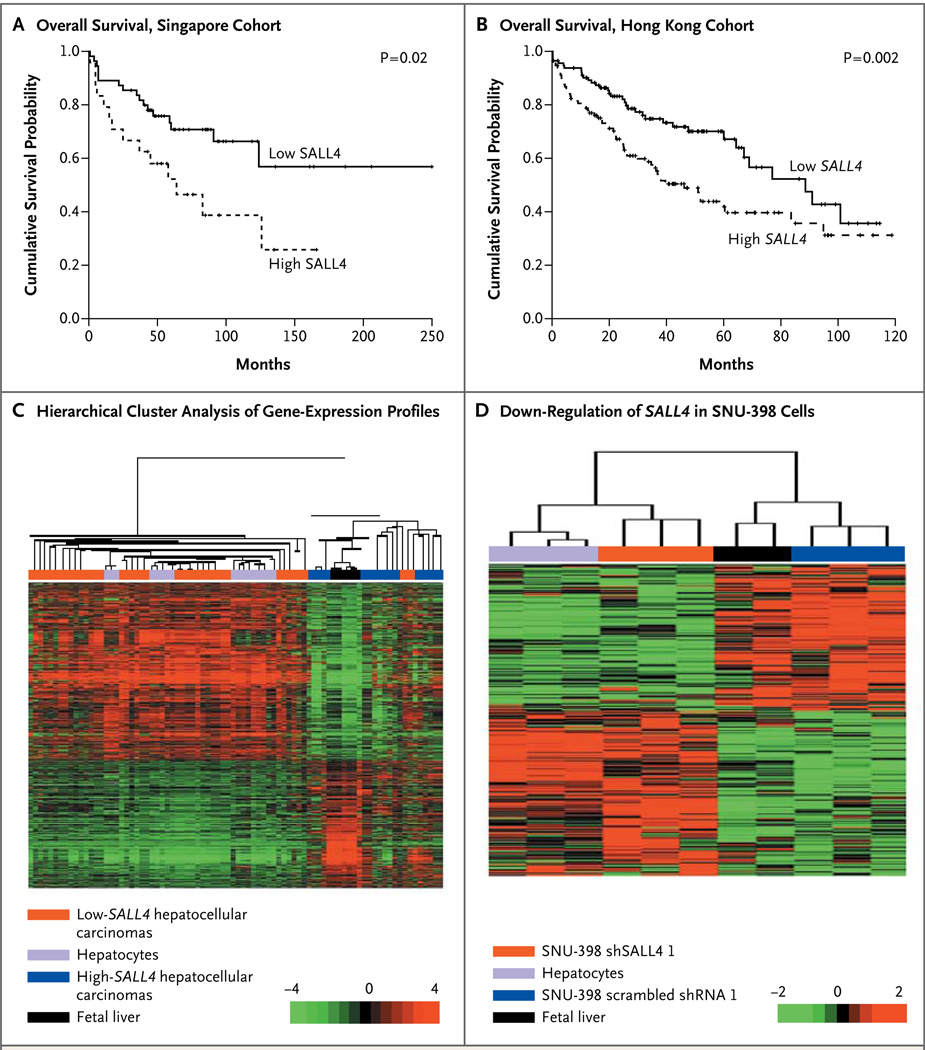
In Panel A, Kaplan–Meier curves show a lower rate of overall survival with SALL4-postive hepatocellular carcinomas than with SALL4-negative hepatocellular carcinomas in the Singapore cohort. In Panel B, Kaplan–Meier curves show a lower probability of overall survival with high-SALL4 hepatocellular carcinomas in the Hong Kong cohort. In Panel C, the dendrogram and heat map show the hierarchical cluster analysis of gene-expression data from human specimens of primary hepatocytes, fetal liver tissue, and hepatocellular carcinoma; gene-expression profiles were obtained from public databases. Columns represent individual samples, and rows represent individual genes. Each cell in the matrix represents the expression level of a gene in an individual sample. The scale bar indicates the level of expression; red indicates a high level of expression, and green a low level of expression. In Panel D, the dendrogram and heat map show the hierarchical cluster analysis of gene-expression data from human specimens of primary hepatocytes, fetal liver tissue, and SNU-398 cells transduced with scrambled short hairpin RNA 1 or SALL4-specific shRNA (shSALL4 1).
In addition, our association studies confirmed that SALL4 expression status was not associated with baseline liver function or the tumor-node–metastasis and Barcelona Clinic Liver Cancer stages in either the Singapore cohort (Table S1 in the Supplementary Appendix) or the Hong Kong cohort (Table S3 in the Supplementary Appendix). Moreover, SALL4 expression status in the Singapore cohort did not differ significantly according to whether the patients received or did not receive preoperative treatment. In a multivariate Cox regression model, SALL4 was an independent prognostic factor for overall survival (hazard ratio for death, 2.87; 95% confidence interval [CI], 1.09 to 7.52; P = 0.03) in the Singapore cohort (Table S6 in the Supplementary Appendix) and an independent predictor of both overall survival (hazard ratio for death, 1.52; 95% CI, 1.00 to 2.32; P = 0.05) and early recurrence (hazard ratio, 1.67; 95% CI, 1.11 to 2.51; P = 0.01) in the Hong Kong cohort (Table S7 in the Supplementary Appendix), after adjustment for other clinicopathological features that have conventionally been accepted as having prognostic value in hepatocellular carcinoma.
Gene-profiling studies suggest that patients with cancers that have embryonic stem-cell or hepatic progenitor cell–like gene-expression signatures have a poor prognosis.14–16 To investigate whether SALL4-positive hepatocellular carcinomas share a gene-expression pattern with fetal hepatoblasts, we extracted global gene-expression data on hepatocytes, fetal liver tissue, and hepatocellular carcinomas (all from humans) from the Gene Expression Omnibus database for hierarchical cluster analysis. Our analysis showed that high-SALL4 hepatocellular carcinomas clustered tightly with fetal livers, whereas the low-SALL4 hepatocellular carcinomas clustered with normal hepatocytes (Fig. 2C). These findings suggest that hepatocellular carcinomas that express SALL4 have a gene-expression pattern that is similar to the pattern in hepatic progenitor cells and thus are poorly differentiated, aggressive, and associated with a poor prognosis.
We next examined the importance of SALL4 in hepatocellular carcinoma by performing loss-of-function studies. Knocking down SALL4 by means of short hairpin RNA (shRNA) caused a decrease in cell viability, an increase in apoptosis, and a decrease in tumorigenicity of hepatocellular carcinoma cells (Fig. S1 in the Supplementary Appendix). To investigate whether loss of SALL4 in hepatocellular carcinoma cells reverses the aggressive progenitor-like phenotype, we carried out microarray analysis of gene-expression profiles of SNU-398 cells treated with control scrambled shRNA (Scr shRNA 1) or SALL4-specific shRNA (shSALL4 1) and then performed hierarchical clustering of these profiles along with those derived from normal human hepatocytes and fetal liver tissues. As expected, SNU-398 cells treated with scrambled shRNA 1 clustered closely with human fetal liver tissue, since both express high levels of SALL4. SALL4–knocked down SNU-398 cells clustered with human hepatocytes; this suggested that down-regulation of SALL4 can render hepatocellular carcinoma more hepatocyte-like with respect to gene expression (Fig. 2D).
To correlate the subgroup of SALL4-positive hepatocellular carcinomas with oncogenic pathways in primary hepatocellular carcinoma specimens, we carried out gene-set enrichment analysis to investigate the enrichment of pathways that have prognostic value in SALL4-high and SALL4-low hepatocellular carcinomas. We found that genes that were up-regulated in hepatocellular carcinoma associated with poor survival, genes in an embryonic stem-cell signature, and genes that were up-regulated in metastasis, hepatoblastoma, and a proliferation subclass of hepatocellular carcinoma17 were significantly enriched in the high-SALL4 hepatocellular-carcinoma subgroup (Fig. 3A through 3E). Taken together, these data suggest that the expression of oncofetal protein SALL4 can be used as a molecular marker to identify an aggressive progenitor cell–like subtype of hepatocellular carcinoma and that SALL4 expression has prognostic value for patients with hepatocellular carcinoma.
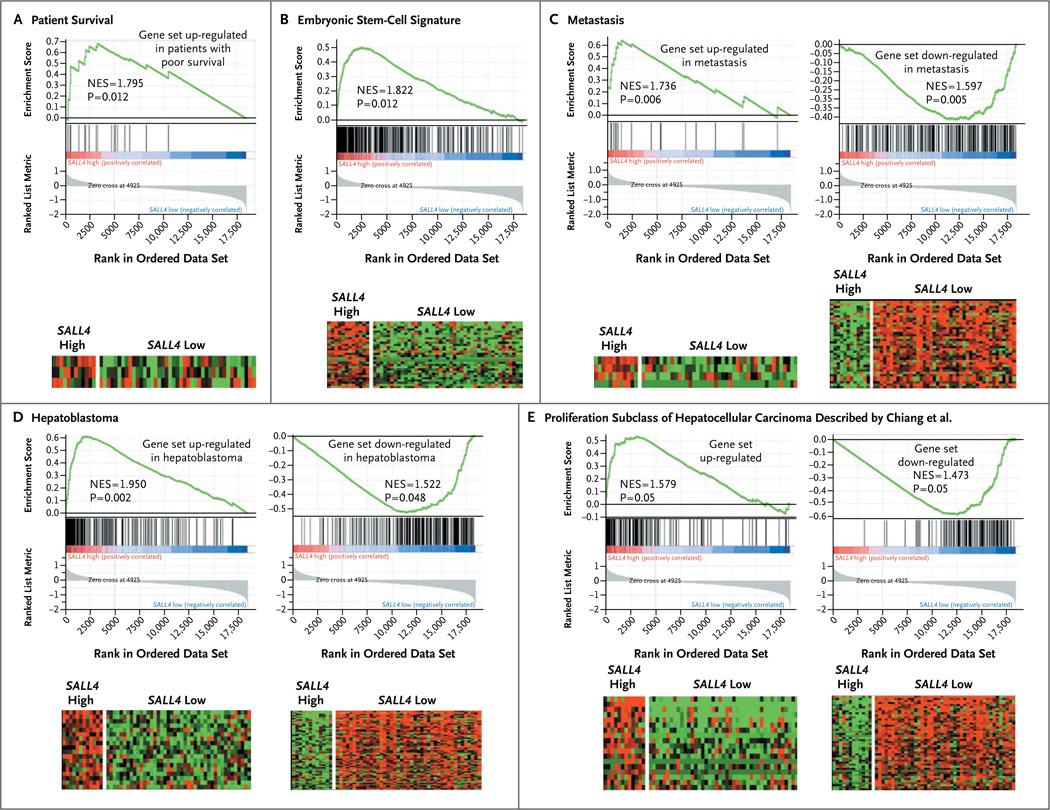
Enrichment of genes associated with poor survival (Panel A), embryonic stem cells (Panel B), metastasis (Panel C), hepatoblastoma (Panel D), and a proliferation subclass of hepatocellular carcinoma was described by Chiang et al.17 (Panel E) in high-SALL4 hepatocellular carcinoma. The heat maps show the enrichment of genes implicated in the gene sets. Columns represent individual samples, and rows represent each gene. Each cell in the matrix represents the expression level of a gene in an individual sample. Red indicates a high level of expression, and green a low level of expression. NES denotes normalized enrichment score in gene-set enrichment analysis. An NES score of more than 1 indicates enrichment of the gene set in high-SALL4 hepatocellular carcinoma. The ranked list metric was generated by calculating the signal-to-noise ratio,18 which is based on the difference of means scaled according to the standard deviation. The larger the signal-to-noise ratio, the more distinct the gene expression is in each phenotype and the more the gene acts as a “class marker.” The Broad Institute Gene Set Enrichment Analysis website (www.broad.mit.edu/gsea) provides detailed information about the computational method.
TARGETING SALL4 BY A NOVEL PEPTIDE
Our loss-of-function studies suggested that SALL4 is a potential therapeutic target for hepatocellular carcinoma and that down-regulation of SALL4 might reverse the aggressive phenotype of hepatocellular carcinomas. We next sought to develop targeted therapy to antagonize the oncogenic role of SALL4. It is known that SALL4 functions as a transcription repressor by recruiting a histone deacetylase (HDAC)–containing nucleosome remodeling and HDAC (NuRD) complex, and that the tumor suppressor PTEN (phosphatase and tensin homologue) is among the target genes repressed by SALL4.19 Our SALL4 loss-of-function microarray data provided direct evidence that in hepatocellular carcinoma, the PTEN–AKT pathway is involved in SALL4-induced hepatocarcinogenesis, since the class I phosphatidylinositol 3–kinase (PI3K) signaling events mediated by AKT were significantly affected by SALL4 down-regulation (P = 0.02).
We recently described a SALL4 12-AA peptide as a competitive inhibitor that blocks the interaction between SALL4 and NuRD and as a result blocks the NuRD-mediated SALL4-repression function.20 We tested whether this 12-AA SALL4 peptide was effective in targeting the SALL4–PTEN–AKT pathway and resulted in decreased viability of hepatocellular carcinoma cells. When 5 µM or 20 µM of nonmutant SALL4 peptide was added to SNU-398 cells, the number of viable cells was reduced, as compared with SNU-398 cells treated with control mutant peptide and scrambled peptide. Trichostatin A, a general HDAC inhibitor, was used as a positive control drug.21,22 When the PTEN inhibitor SF1670 was added to the cells treated with SALL4 peptide, it rescued the phenotype, as evidenced by the maintenance of cell viability (Fig. 4A). These results suggest that PTEN plays an important role in the SALL4 peptide–induced loss of tumor-cell viability in hepatocellular carcinoma. In contrast, SALL4 peptide had no effect on SNU-387 cells with undetectable endogenous SALL4 expression (Fig. 4B). This finding suggests that SALL4 peptide is a specific agent inhibiting SALL4-overexpressing hepatocellular carcinoma cells, with minimal toxic effects on SALL4-negative cells, as compared with the HDAC inhibitor trichostatin A.
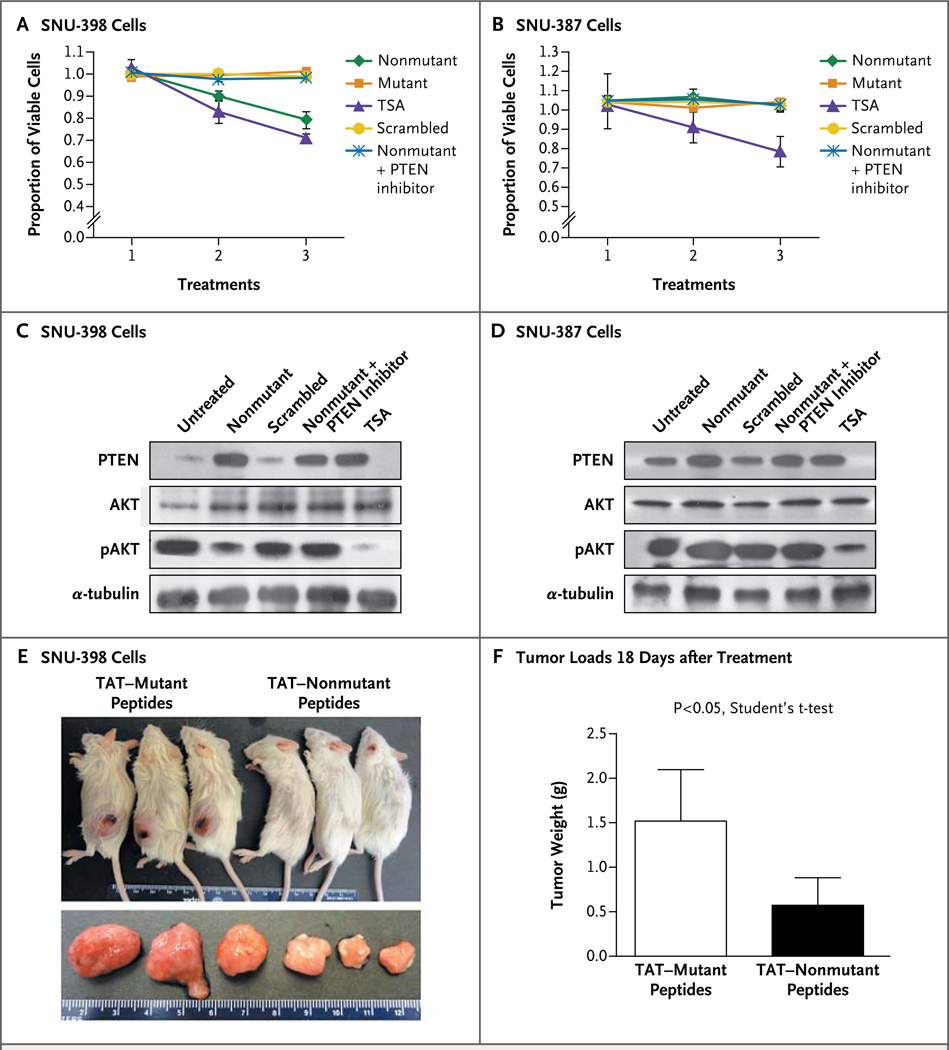
A 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium cell-viability assay shows the effects of SALL4 peptide on SNU-398 cells with high endogenous SALL4 expression (Panel A) and SNU-387 cells with undetectable SALL4 expression (Panel B). Cell viability was determined 72 hours after peptide treatments. Absorbance at 490 nm was directly correlated with cell viability. Nonmutant functional SALL4 peptide, mutant nonfunctional peptide, trichostatin A (TSA), scrambled control peptide, and nonmutant peptide plus PTEN (phosphatase and tensin homologue) inhibitor were added to the cells at the indicated concentration. The PTEN inhibitor concentration was 400 nM (Panels A through D). In Panels A and B, treatment 1 was a no-treatment control, treatment 2 was treatment of cells with 5 µM of peptides or 50 nM of TSA, and treatment 3 was treatment of cells with 20 µM of peptides or 100 nM of TSA. Western blot analyses show the effects of peptides on PTEN, AKT, and phosphorylated AKT (pAKT) expression on SNU-398 cells (Panel C) and SNU-387 cells (Panel D) 72 hours after peptide treatments; α-tubulin was analyzed as a loading control. Twenty µM of nonmutant or scrambled peptide or 100 nM of TSA was used. The PTEN inhibitor concentration was 400 nM. Panel E shows nonobese diabetic mice with severe combined immunodeficiency that underwent transplantation with 3 million SNU-398 cells at the left flank and were treated intraperitoneally for 5 consecutive days with 56 mg of TAT-mutant peptide per kilogram of body weight or 60 mg of nonmutant peptide per kilogram (top panel). The dissected subcutaneous tumors are shown below. Panel F shows the mean weight of tumors dissected at day 18 after transplantation and treatment in five mice that received TAT fusion peptides intraperitoneally. T bars indicate standard deviations.
To confirm the involvement of the tumor suppressor PTEN in the SALL4 peptide–induced reduction of hepatocellular carcinoma cell viability, we analyzed PTEN expression by means of Western blotting. SALL4 peptide treatment induced an extensive increase in levels of PTEN protein in SNU-398 cells (Fig. 4C). In contrast, SALL4 peptide had a negligible effect on PTEN expression in SNU-387 cells (Fig. 4D). We next examined whether the increase in PTEN expression had an effect on PI3K signaling. PTEN functions as a phosphatase to dephosphorylate AKT. Western blot analysis showed a marked reduction of phosphorylated AKT protein levels with SALL4 peptide treatment in SNU-398 cells (Fig. 4C), but not in low-SALL4 SNU-387 cells (Fig. 4D); this suggests that the increase in the level of PTEN expression had a functional role in blocking the PI3K survival signaling by dephosphorylating AKT. Furthermore, this effect could be rescued by the PTEN inhibitor, which is consistent with the role of SALL4 in regulating this pathway.
To further test the therapeutic effect of this peptide for in vivo treatment, we conjugated mutant and nonmutant peptides with the transactivator of transcription (TAT) protein transduction domain and administered the peptides intraperitoneally in nonobese diabetic mice with severe combined immunodeficiency, in which SNU-398 cells had been transplanted subcutaneously. TAT fusion nonmutant peptides reduced the tumorigenicity of SNU-398 hepatocellular carcinoma cells (Fig. 4E). The tumor loads at 18 days after treatment in mice that received TAT-nonmutant peptides were significantly smaller than tumor loads in mice that received TAT–mutant peptides (Fig. 4F). These experiments confirm that the SALL4 peptide had biologic activity both in vitro and in vivo in a xenotransplant model.
Our in vitro and murine data suggest that the SALL4 peptide is a potential targeted therapy for a subgroup of hepatocellular carcinomas, since it effectively and specifically reduced viable SALL4-positive hepatocellular carcinoma cells.
DISCUSSION
Hepatocellular carcinoma is one of the deadliest cancers, with a ratio of mortality to incidence of almost 1.1 Our study shows the unique expression pattern of the stem-cell factor SALL4 in human livers at various stages — activated in fetal liver tissue, silenced in adult liver tissue, and reexpressed as an oncofetal protein in hepatocellular carcinoma.
Identification of reliable biomarkers that can be used to predict patient outcomes ensures more effective clinical management, and we propose that SALL4 is such a biomarker in hepatocellular carcinoma. The reexpression of SALL4 in hepatocellular carcinoma is clinically significant, since patients with the subtype that overexpresses SALL4 have enriched hepatic progenitor cell–like gene signatures and tend to have a poor prognosis. Given the strong prognostic value of SALL4 established in our retrospective study, we propose that the prognostic value of SALL4 should be further verified in prospective clinical trials.
The discovery of a role for SALL4 in hepatocellular carcinoma, its association with prognosis, and the antitumor effects of a newly identified peptide blocker targeting it have potential therapeutic significance. Testing for the presence of SALL4 at diagnosis may be helpful not only for determining the prognosis but also for identifying patients who are likely to have a response to treatment. Given the expression of SALL4 in hepatocellular carcinoma cells but not in normal adult hepatocytes, treatment with SALL4 peptide may have less tissue toxicity, which is especially beneficial in patients with underlying cirrhosis whose baseline liver function is already compromised. Future studies will determine whether this peptide has synergistic potential with other molecular therapies, such as sorafenib, which act on parallel nonoverlapping pathways.
In conclusion, our study shows that the oncofetal protein SALL4 plays an important role in the extensive network of heterogeneous cellular pathways underlying hepatocarcinogenesis. A 12–amino acid peptide can block the oncogenic role of SALL4 and hence has therapeutic potential in SALL4-positive hepatocellular carcinoma.
Acknowledgments
Supported by a Singapore Translational Research Award from the Singapore National Medical Research Council (NMRC/ STaR/0001/2008, to Dr. Tenen) and grants from the Singapore Ministry of Education and National Research Foundation (to Dr. Tenen) and the National Institutes of Health (PO1DK080665, to Drs. Tenen and Chai; and RO1HL092437 and PO1HL095489, to Dr. Chai).
We thank the members of the laboratories of Drs. Tenen and Chai for many helpful discussions and technical support; the Centre for Translational Research and Diagnostics for the generation of microarray data; and Dr. Hongbo R. Luo for discussion about studies of PTEN and AKT.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
REFERENCES
Full text links
Read article at publisher's site: https://doi.org/10.1056/nejmoa1300297
Read article for free, from open access legal sources, via Unpaywall:
https://www.nejm.org/doi/pdf/10.1056/NEJMoa1300297?articleTools=true
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Tumour stemness and poor clinical outcomes in haemochromatosis patients with hepatocellular carcinoma.
J Clin Pathol, 77(10):669-675, 19 Sep 2024
Cited by: 1 article | PMID: 37253536 | PMCID: PMC11503110
Oncofetal SNRPE promotes HCC tumorigenesis by regulating the FGFR4 expression through alternative splicing.
Br J Cancer, 131(1):77-89, 25 May 2024
Cited by: 3 articles | PMID: 38796598
SALL4 in gastrointestinal tract cancers: upstream and downstream regulatory mechanisms.
Mol Med, 30(1):46, 08 Apr 2024
Cited by: 0 articles | PMID: 38584262 | PMCID: PMC11000312
Review Free full text in Europe PMC
The association between tumour heterogeneity and immune evasion mechanisms in hepatocellular carcinoma and its clinical implications.
Br J Cancer, 131(3):420-429, 17 May 2024
Cited by: 2 articles | PMID: 38760445 | PMCID: PMC11300599
Review Free full text in Europe PMC
Mechanisms of angiogenesis in tumour.
Front Oncol, 14:1359069, 25 Mar 2024
Cited by: 3 articles | PMID: 38590656 | PMCID: PMC10999665
Review Free full text in Europe PMC
Go to all (144) article citations
Other citations
Wikipedia
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Sal-like protein 4 (SALL4), a stem cell biomarker in liver cancers.
Hepatology, 57(4):1469-1483, 06 Mar 2013
Cited by: 122 articles | PMID: 23175232 | PMCID: PMC6669886
Coexpression of SALL4 with HDAC1 and/or HDAC2 is associated with underexpression of PTEN and poor prognosis in patients with hepatocellular carcinoma.
Hum Pathol, 64:69-75, 12 Apr 2017
Cited by: 14 articles | PMID: 28411180
Oncofetal gene SALL4 in aggressive hepatocellular carcinoma.
N Engl J Med, 369(12):1171, 01 Sep 2013
Cited by: 7 articles | PMID: 24047071
SALL4 Oncogenic Function in Cancers: Mechanisms and Therapeutic Relevance.
Int J Mol Sci, 23(4):2053, 12 Feb 2022
Cited by: 19 articles | PMID: 35216168 | PMCID: PMC8876671
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NCI NIH HHS (2)
Grant ID: R01 CA118316
Grant ID: P01 CA066996
NHLBI NIH HHS (6)
Grant ID: P01 HL095489
Grant ID: P01HL095489
Grant ID: R01 HL056745
Grant ID: R01HL092437
Grant ID: R01 HL092437
Grant ID: R01 HL112719
NIDDK NIH HHS (2)
Grant ID: P01DK080665
Grant ID: P01 DK080665





