Abstract
Free full text

Reprogramming the Chromatin Landscape: interplay of the estrogen and glucocorticoid receptors at the genomic level
Abstract
Crosstalk between estrogen (ER) and glucocorticoid (GR) receptors has been shown to contribute to the development and progression of breast cancer. Importantly, the ER and GR status in breast cancer cells is a significant factor in determining the outcome of the disease. However, mechanistic details defining the cellular interactions between ER and GR are poorly understood. We investigated genome-wide binding profiles for ER and GR upon co-activation, and characterized status of the chromatin landscape. We describe a novel mechanism dictating the molecular interplay between ER and GR. Upon induction, GR modulates access of ER to specific sites in the genome by reorganization of the chromatin configuration for these elements. Binding to these newly accessible sites occurs either by direct recognition of ER response elements, or indirectly through interactions with other factors. The unveiling of this mechanism is important for understanding cellular interactions between ER and GR, and may represent a general mechanism for crosstalk between nuclear receptors in human disease.
Introduction
Steroid receptors (SRs), including the estrogen receptor (ER) progesterone receptor (PR), androgen receptor (AR), and glucocorticoid receptor (GR), are a class of ligand-inducible transcription factors that control a wide spectrum of physiological processes, including metabolism, development, and inflammatory responses. Importantly, steroid receptors have been shown to play prominent roles in many cancers. Both ER and PR have well established roles in breast cancer, however, increasing evidence suggests that GR is also an important factor in the patho-physiological progression of breast cancer (1,2). GR has been shown to be over expressed in grade 3 breast cancer when compared to normal breast tissue or grade 1–2 tumors (3). In addition, expression of GR, but not ER mRNA, is significantly increased in breast cancer stroma when compared to normal control tissue, suggesting that GR may play an important role in tumor-stroma interaction (4). Cross-talk between ER and GR may also be important in breast cancer development and treatment. The ER/GR status has been shown to be important for breast cancer outcome (5). In addition, lines of investigation have shown that co-treatment of cells with corticosteroids and estradiol, can have opposing effects on a cell’s response compared to single hormone treatments (6–9). Therefore, glucocorticoid treatment may offer a way to modulate estrogen response in breast cancer patients making it imperative that we understand the molecular interplay between ER and GR.
Members of the SR family interact with regulatory elements within the context of chromatin and mediate expression of genes involved in these pathways. Chromosomal architecture markedly restricts access to these sites, thereby contributing to cell-type specific activity of hormone response elements (HREs). SR have been shown to interact primarily with chromatin that is accessible prior to hormone induction (10–14), suggesting the chromatin landscape is primed by other factors in order to maintain a cell-type specific chromosomal environment for steroid receptor binding (15,16). However, even though the vast majority of GR binding sites are found at these pre-programmed elements, GR, in association with chromatin remodeling proteins, can cause alterations in chromatin structure upon induction (11). This allows for recruitment of secondary factors that have previously been excluded from these specific binding sites, a dynamic process recently designated as “assisted loading” (17). In addition, ER has been shown to bind to inaccessible chromatin through a FOXA1 dependent mechanism (12). Although genome-wide investigations on steroid receptors have provided invaluable information about their function, these studies were conducted in controlled environments where only one receptor is activated. In the physiological context, cells are contained in an environment with complex mixtures of hormones, allowing for multiple receptors to be activated at once. Crosstalk between steroid receptors can occur in these physiological environments and plays a role in controlling cellular processes (18–21).
Here, we provide evidence that co-activation of ER and GR re-programs the chromatin landscape causing global re-arrangement of steroid receptor binding in mouse mammary cells. Induction of GR facilitates selective access of ER to specific sites in the genome by maintaining an accessible configuration at these response elements. In addition, activation of ER can affect chromatin structure at estrogen-dependent GR binding sites, resulting in a new class of GR binding elements. Co-activation of ER and GR also leads to a loss of specific binding sites for each steroid receptor. These findings reveal that crosstalk occurs at the genome level and that activation of multiple steroid receptors has a dramatic impact on controlling which regulatory elements are accessible to each receptor.
Materials and Methods
Cell culture conditions
For maintenance, cells were cultured in DMEM supplemented with 10% fetal calf serum, sodium pyruvate, non-essential amino acids, 2 mM glutamine, and 53g/ml tetracycline to repress expression of the tet-regulated fusion proteins (22). Cells were maintained in a humidifier at 37° C and 5% CO2. Cells were plated for experiments in DMEM growth medium supplemented with 10% charcoal-dextran treated serum with tetracycline 48 hours prior to conducting the experiment. Twenty hours before hormone treatment, cells were washed three times with phosphate buffered-saline and fresh DMEM with 10% charcoal-dextran serum (no tetracycline) was added to the cells to induce expression of GFP-GR and Ch-ER.
Cell line validation
3617 and 7438 murine mammary epithelial cell lines have been described previously (17) and were constructed originally in our laboratory. All other cell lines were constructed from these two cell lines. Cells are routinely tested for GR and ER expression by western blots and fluorescent microscopy and for dex and estradiol responses.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation was conducted via standard protocols (23), with minor modifications using the following antibodies: GR cocktail (PA-510A and PA-511A, Affinity BioReagents; sc-1004 (M-20), Santa Cruz Biotechnology) and ER cocktail (Ab-10, Labvision; sc-543, Santa Cruz Biotechnology). Cells were untreated or induced with 100 nM dexamethasone (Sigma), 100 nM estradiol (Sigma), or 100 nM dexamethasone plus 100 nM estradiol for 30 mins. Three biological replicates were pooled as a single replicate before generating sequencing libraries. Two replicates per treatment were sequenced.
DNaseI digestion of chromatin and size fractionation
DNaseI digestion was conducted as previously described (11,24). Cells were untreated or induced with 100 nM dexamethasone (Sigma), 100 nM estradiol (Sigma), or 100 nM dexamethasone plus 100 nM estradiol for 30 mins. Nuclei were isolated and digested with 100–120 U/ml of DNaseI (Sigma) for 3 mins at 37°C. Digested chromatin was then incubated with 10μg/mL RNaseA (Roche) overnight at 55°C. Proteinase K (Ambion) was then added to the samples and incubated for 4 h at 55°C. DNA was purified by phenol-chloroform extraction and fractionated using sucrose gradient centrifugation in order to isolate 100–500bp fragments. These fragments were pooled, precipitated, and then assembled into libraries for sequencing
ChIP-Seq Data Analysis
Illumina Solexa genome analyzer platform was used to generate sequence reads (36-mer) and unique tags were aligned to the mouse reference genome (UCSC mm9 assembly). Hotspots, regions of enriched tags, were called using previously described methods with minor modifications (25). (See Supplemental Experimental Procedures for details of sequencing data analysis)
Results
Redistribution of ER and GR binding to the Genome upon co-activation
Crosstalk between ER and GR plays an important role in controlling many cellular processes (18–21). However, the mechanism used by these SRs to influence each other’s actions and control specific cellular outcomes is poorly understood. It has previously been shown that ER can bind to nucleosome-rich regions using FAIRE analysis (12). However, we show here, that when we overlay previously generated ChIP-Seq data for ER binding in MCF7 cells with DNaseI hypersensitivity data we generated in these cells, 83% of ER binding events are at pre-accessible sites (Supplementary Fig. S1A) (12). This suggests that other factors prime the chromatin landscape for ER binding genome-wide. Therefore, we wanted to test if GR could dictate ER binding genome-wide. We mapped ER and GR binding events by ChIP-Seq in cells treated with either the corticoid steroid dexamethasone (Dex), estradiol (E2), or both Dex and E2. Since PR itself can respond to corticoid steroids and bind to glucocorticoid response elements, it can be a challenge to study ER and GR crosstalk in cell lines containing all three steroid receptors (26,27). Therefore, we used a previously engineered mouse mammary cell line, which expresses GR and ER but not PR (17), to look at specific interactions between GR and ER.
We mapped 2916 ER binding events in the presence of E2 and 3949 ER-bound elements upon co-treatment of cells with Dex and E2 (Fig. 1; select examples in Fig. 2 and Supplementary Fig. S1B–D). Of these binding sites, 2863 are found to overlap in both treatments (Fig. 1A; blue dots). Interestingly, 28% of ER binding sites observed upon co-treatment of cells with Dex and E2 are Dex-dependent (Fig. 1A; green dots). In addition, 2% of ER binding events are lost upon co-treatment with Dex and E2 (Fig. 1A; red dots). These sites are unique binding sites observed only with the single hormone (E2) treatment. Together, these results demonstrate that there is a global redistribution of ER binding to the genome when both ER and GR are induced.
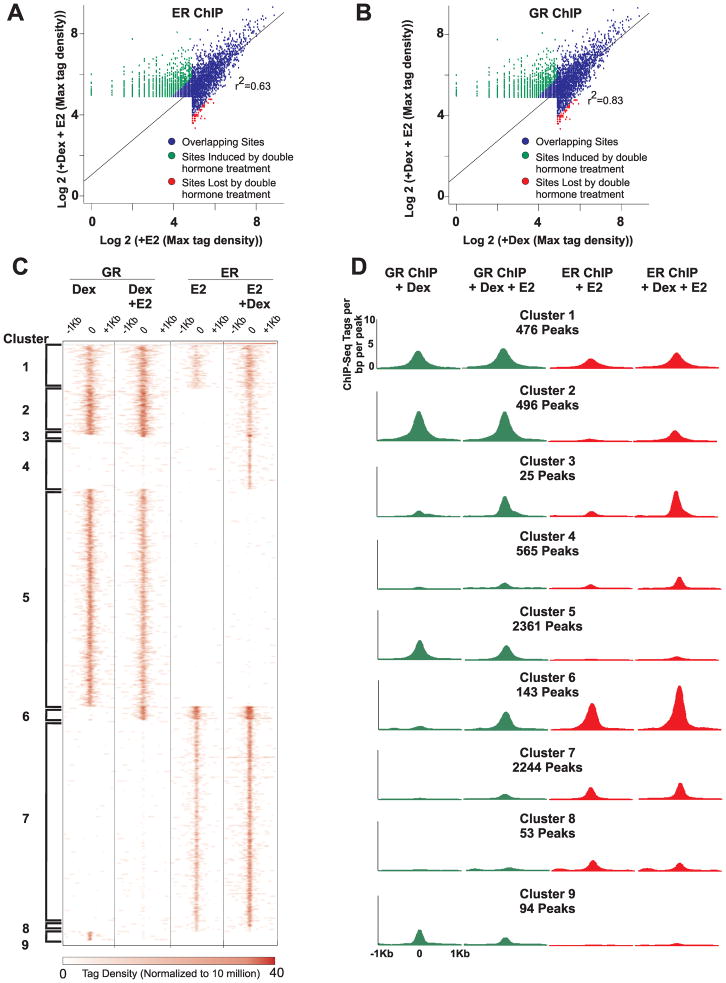
A. Global changes in ER binding upon co-treatment of cells with Dex and E2. Binding profiles of ER were determined by ChIP-Seq after treatment of cells with E2 or with both Dex and E2. Scatter plot summarizes global changes in ER binding between the two treatments. Sites induced or lost by double hormone treatments have at least a 2-fold difference in tag density over the background of the dual hormone treatment.
B. Global changes in GR binding upon co-treatment of cells with Dex and E2. Binding profiles of GR were determined by ChIP-Seq after treatment of cells with Dex or with both Dex and E2. Scatter plot illustrates genome-wide changes in GR binding between the two treatments. Sites induced or lost by double hormone treatments have at least a 2-fold difference in tag density over the background of the dual hormone treatment.
C. Different ER and GR binding modules. Clustering analysis of 6487 unique peaks identified for GR and/or ER by ChIP-Seq. Cells were treated with either Dex, E2, or both. Heat map portrays the number of reads per 106 sequences as well as the positions of the reads within 2kb of the ChIP-Seq peak. Nine major modules identified by the analysis are highlighted by brackets.
D. ER and GR binding profiles for each identified cluster. Distribution graphs of GR and ER binding sites within a 2kb interval of the ChIP-Seq peak for each cluster. Graphs are shown for GR binding in the presence of Dex or Dex and E2 and for ER binding in the presence of E2 or Dex and E2.
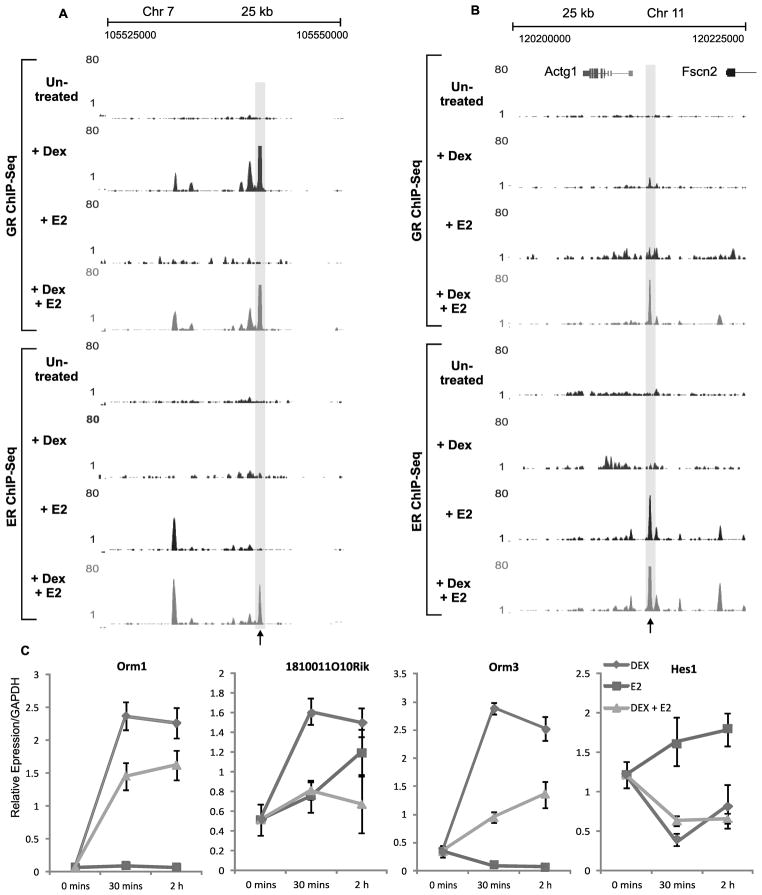
A. Genomic region illustrating assisted loading of ER by GR. Example [UCSC browser shot; (40)] of ER and GR ChIP-Seq in the absence of hormone or from cells treated with Dex, E2, or Dex and E2. ER and GR ChIP experiments were done after 30 mins of hormone treatment. Black arrow denotes ER assisted loading site.
B. Genomic region illustrating assisted loading of GR by ER. Example (UCSC browser shot) of ER and GR ChIP-Seq in the absence of hormone or from cells treated with Dex, E2, or Dex and E2. ER and GR ChIP experiments were done after 30 mins of hormone treatment. Black arrow denotes GR assisted loading site.
C. Co-activation of ER and GR effect gene expression. Expression analysis of Orm1, Orm3, 1810011010Rik, and Hes1, from cells treated with Dex (blue), E2 (red), or Dex and E2 (green).
To determine if the redistribution of binding upon co-treatment of cells with Dex and E2 is unique to ER or is also observed with GR, we compared the binding events of GR in the presence of Dex and Dex plus E2 (Fig. 1; select examples in Fig. 2 and Supplementary Fig. S1B–D). We observe 3427 GR binding sites for cells treated with Dex and 3501 GR binding elements for cells co-treated with Dex and E2. There are 3334 peaks that overlap between the treatments (Fig. 1B; blue dots). Similar to ER, we observe a global re-arrangement of GR binding events when cells are simultaneously treated with both hormones. Five percent of GR binding sites in cells co-treated with Dex and E2 are estrogen-dependent and 3% of GR binding events observed in cells treated with Dex are unique to the single hormone treatment (Fig. 1B; green and red dots respectively).
To illustrate the overall changes in ER and GR binding across the genome upon dual hormone treatment, the ChIP-Seq data was combined and 6487 unique chromosomal positions were identified. Sequence-read densities for each hormone treatment were obtained over a 2kb interval for each unique peak. Supervised clustering was conducted to extract specific binding modules for ER and GR, which resulted in the identification of 9 major clusters (Fig. 1C–D; Supplementary Fig. S2A). Analysis of the binding pattern at genomic elements for each cluster was found to be similar, with majority of binding sites occurring at intron and intergenic regions (Supplementary Fig. S2B). Clusters 5 and 7 represent the basic GR and ER binding patterns respectively, where GR and ER can bind to these sites upon activation with their corresponding hormone. Three different clusters (2–4) represent the unique ER binding sites observed upon co-treatment of cells with Dex and E2 in Figure 1A, whereas the unique GR binding sites observed in the dual hormone treatment has two identified binding modules (cluster 3, 6). Of most interest to us is the binding of ER at sites in cluster 2 (496 binding sites) and of GR at sites in cluster 6 (143 binding sites). In these modules, binding of the receptor is dependent on having the other receptor activated, in addition to the other receptor also binding at these sites. This suggests that ER and GR binding to these sites are modulated by an assisted-loading mechanism. A similar mechanism has been observed for pBox and AP1 binding. In both examples pBox and AP1 are dependent on the activation of GR for binding at specific sites (15,17).
In addition to global re-arrangement of receptor binding, co-treatment of cells with Dex and E2 can greatly affect expression levels at a subset of genes as shown by expression microarray analysis (Supplementary Fig. S3). A representative set of genes were confirmed by quantitative PCR. (Fig. 2C). In addition, we found that many of the genes whose expression changed upon dual hormone treatment were associated with changes in either ER or GR binding within 20 kb of their transcriptional start sites (Supplementary Fig. S3C). The observation that gene expression changes upon dual hormone treatments is in agreement with previously published studies which show that crosstalk between ER and GR regulates a subset of proinflammatory genes and that corticoid steroids can reverse the effect of estradiol on a small subset of genes in human leiomyoma cells (8,9).
Changes in DNA Accessibility Occur at Assisted-Loading Sites
To begin to understand the molecular mechanisms used for ER and GR assisted-loading we wanted to determine if the assisted loading sites (cluster 2 and 6) we observe with dual hormone treatments result from changes in chromatin accessibility at these sites upon hormone induction. We therefore conducted DNaseI-seq to map changes in DNaseI hypersensitive sites (DHS) between single and dual hormone treatments (11,24).
Interestingly, ER binding events that are assisted by GR occur at genomic locations where DNaseI accessibility increases upon treatment of cells with Dex and E2 compared to E2 alone (Fig. 3A–B; select example in Fig. 3C). A similar increase in hypersensitivity is observed at these sites when untreated cells are compared to cells treated with Dex, indicating that these changes are Dex dependent (Fig. 3A; select example in Fig. 3C). This change is highly significant when compared to ER binding sites that are only dependent on estrogen (Fig. 3A). This observation suggests a possible model in which GR initially binds to and recruits chromatin remodelers to these sites. GR has previously been shown to recruit SWI/SNF complexes to de novo sites which leads to increases in accessibility at these response elements (10). In contrast, ER binding events that are unique to the treatment of cells with only E2 (cluster 8) occur at sites which have no change in accessibility (Supplementary Fig. S4A; select example in Supplementary Fig. S4B). In clusters 1, 3, and 4 we also observe slight increases in hypersensitivity at these sites upon co-treatment with Dex and E2 (Fig. 3A; Supplementary Fig. S4A), which could account for slight increases in ER binding at these sites.

A. Significant changes in hypersensitivity at ER assisted loading sites (cluster 2). Box plot comparing changes in DNaseI hypersensitivity upon treatment of cells with Dex compared to untreated cells (top) or dual hormone treatment compared to treatment with E2 (bottom) for ER binding modules 1, 2, and 7. Increases in hypersensitivity in ER assisted-loading sites (cluster 2) are statistically significant compared to sites that bind ER in the presence of E2.
B. Increases in accessibility at ER assisted loading sites highly correlate with ER binding. Scatter plot illustrates that ER binding sites unique to the double hormone treatment and overlapping GR binding sites are highly correlated with increases in DNaseI accessibility upon dual hormone treatment.
C. Genomic region illustrating changes in DHS at ER Assisted Loading site. Example (UCSC browser shot) of DHS in the absence of hormone or from cells treated with Dex, E2, or Dex and E2. Tracks are overlaid with ER and GR ChIP-Seq data from the same genomic site. Black arrows denote ER assisted loading sites.
D. Significant changes in accessibility at GR assisted loading sites (cluster 6). Box plot comparing changes in DNaseI hypersensitivity upon treatment of cells with E2 compared to untreated cells (top) or dual hormone treatment compared to treatment with Dex (bottom) for GR binding modules 1, 5, and 6. Increases in hypersensitivity in GR assisted-loading sites (cluster 6) are statistically significant compared to sites that bind GR in the presence of Dex.
E. Increases in hypersensitivity at GR assisted loading sites are highly correlated with ER binding. Scatter plot reveals that GR Binding sites unique to the double hormone treatment and overlaping ER binding sites highly correlate with increases in DNaseI hypersensitivity upon dual hormone treatment.
F. Genomic region illustrating changes in DHS at GR assisted loading site. Example (UCSC browser shot) of DHS in the absence of hormone or from cells treated with Dex, E2, or Dex and E2. Tracks are overlaid with ER and GR ChIP-Seq data from the same genomic site. Black arrow denotes GR assisted loading site.
Previous studies have shown that induction of ER can increase DNaseI hypersensitivity at response elements (13). We therefore analyzed changes in DNaseI accessibility at binding sites where GR loading is assisted by ER (cluster 6). We found that increase in chromatin accessibility at these sites is highly correlated with assisted loading of GR upon treatment of cells with Dex and E2 compared to cells treated with Dex alone (Fig. 3D–E; select example in Fig. 3F). A similar increase in hypersensitivity is also observed at these sites when untreated cells are compared to cells treated with E2, proving that these changes are E2 dependent (Fig. 3D; select example in Fig. 3F). However, GR binding events unique to treatment of cells with only Dex occur at sites which either have no change in accessibility or a slight decrease (Supplementary Fig. S4C; select example in Supplementary Fig S4D). We also analyzed changes in chromatin accessibility upon treatment of cells with Dex and E2 compared with cells treated with Dex at other GR binding modules. Whereas GR binding at cluster 3 is highly correlated with changes in DNaseI accessibility, other GR binding modules are not dependent on changes in chromatin accessibility (Fig. 3D; Supplementary Fig. S4C). These findings indicate that both GR and ER causes changes in chromatin structure at specific response elements allowing for recruitment of the other receptor.
DNA Sequence Specifies Assisted-Loading Sites
To determine if ER assisted loading sites contained unique sequences compared to other ER binding modules we performed de novo motif analysis. Surprisingly, we do not observe enrichment of estrogen response elements (EREs) at assisted ER loading sites (Fig. 4A). Instead there is a high prevalence for glucocorticoid response elements (GREs) and the activator protein 1 (AP-1) binding motif. AP-1 is a transcription factor that has been shown to interact with both ER and GR and plays a role in the recruitment of these receptors upon induction to specific sites within the genome (15,28,29). In comparison, motif and FIMO analysis of other ER binding modules show a higher prevalence for an ERE (Fig. 4B; Supplementary Fig. S5A) and a lower occurrence rate for GREs (Fig. 4C).

A. Motif analysis of ER assisted loading sites. De novo motif analysis was conducted on cluster 2 using ChIP-MEME. The identity of the top two enriched motifs (eval <10−2) were determined using a TOMTOM search against the Transfac database of characterized transcription factors.
B. Motif distribution for EREs within each binding module. Frequency of finding an ERE at a binding site within ER binding modules and GR only binding sites was determined using FIMO. Results are reported as the number of motif occurrences per number of binding sites within a cluster (pval<10−5).
C. Motif distribution for GREs within each cluster. Frequency of finding an GRE at a binding site within ER binding modules and GR only binding sites was determined using FIMO. Results are reported as the number of motifs occurrences per number of binding sites within a cluster ( (pval<10−5).
To confirm that ER binding at ER assisted-loading sites (cluster 2) is not dependent on direct binding to DNA at EREs, we constructed an ER DNA binding domain (DBD) mutant (Fig. 5A; Supplementary Fig. S5B–C; Supplementary Fig. S6), which has previously been shown to abolish binding of ER to EREs but still allow tethering of ER to AP1 (30). We repeated the ChIP-Seq analysis on cells that did not express wild-type ER but did express the DBD mutant. Mutation of the ER DBD domain did not affect ER binding at sites in clusters 1 and 2 but did affect ER binding in all other ER binding modules (Fig. 5B). The ER DBD mutant has a 72% overlap in binding with wild type ER at cluster 2 sites and 55% overlap at cluster 1 sites. Taken together, these results suggest that ER is recruited to cluster 2 assisted loading sites by other factors, after GR induces chromatin remodeling at these sites.
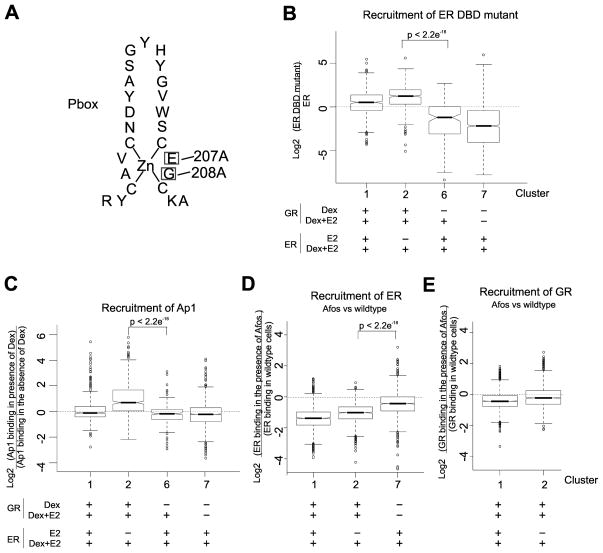
A. Mutation in the DNA binding domain of ER. Diagram showing the location of two mutations made in the DNA binding domain of ER. Mutations are E207A and G208A.
B. Mutations in the DNA binding domain of ER do not affect ER recruitment to binding sites found in cluster 2 (ER assisted-loading sites). Shown are box plots of the effect of the ER DNA binding mutant on recruitment of ER to binding sites within clusters 1, 2, 6, and 7. The ER mutant is incapable of binding directly to EREs but can still be recruited to binding sites through tethering with other factors.
C. AP1 binding at sites in cluster 2 is dependent upon activation of GR. Shown are box plots of the effect of Dex on AP1 binding at sites within clusters 1, 2, 6, and 7. AP1 binding at elements within cluster 2 (ER assisted loading sites) is dependent on the treatment of cells with Dex whereas AP1 binding at sites within clusters 1, 6, and 7 (ER binding sites in the presence of E2) are not dependent upon treatment with Dex.
D. Afos affects ER binding at sites in cluster 2.Shown are box plots illustrating the effect of Afos expression on the binding of ER in clusters 1, 2, and 7. Afos expression disrupts ER binding at assisted loading sites(cluster 2) whereas ER binding at cluster 7 is not affected.
E. GR binding is not disrupted by Afos expression at ER assisted loading sites (cluster 2)Shown are box plots illustrating the effect of Afos expression on the binding of GR in clusters 1 and 2. Afos expression has not effect on GR binding at assisted loading sites(cluster 2).
AP1 binding at ER Assisted-Loading Sites
To determine if AP1 could be a potential factor for tethering ER to sites in Cluster 2, we overlaid previous AP1 ChIP-Seq data with ER binding sites in ER binding modules 1, 2, 6 and 7 (Fig. 5C). This previous data is from a cell line derived from the same parent cell line as the cell line used in the other studies in our paper (15). Interestingly, we found that there is an increase in AP1 at these ER assisted-loading sites (cluster 2) upon treatment of cells with Dex. In contrast, an increase in AP1 binding upon treatment with Dex is not observed in clusters 1, 6, or 7 in which ER binding is not dependent on GR at these sites (Fig. 5C). To access if AP1 is necessary for ER binding at cluster 2 sites we over-expressed Afos, a truncated Fos protein consisting of the leucine zipper domain and a substitution of the basic region with an acidic extension that maintains dimerization with Jun but inhibits its ability to bind DNA (15). ChIP-Seq of ER and GR binding in the presence of Afos shows that the dominant negative AP1 complex effects ER binding at cluster 2 sites, but has no effect on GR binding at these elements (Fig 5D–E). These results suggest that ER loading at the binding elements in cluster 2 requires GR activation and recruitment to these sites to induce changes in chromatin accessibility. For cluster 2 sites, ER, once activated, is then recruited through tethering with other factors such as AP1 (Fig. 6).
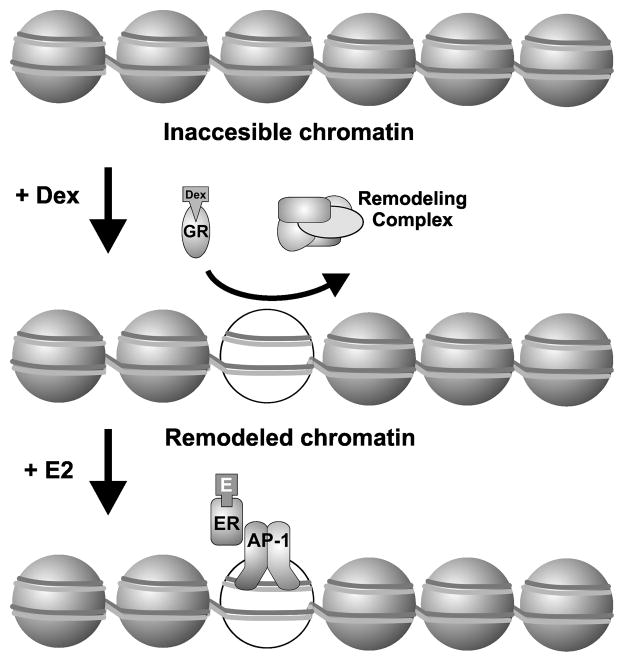
Upon treatment of cells with Dex, GR binds to chromatin which leads to increases in accessibility at these sites (possibly due to recruitment of chromatin remodelers). Now upon treatment with E2, ER can now bind to these previously inaccessible sites through tethering with AP1 or other factors.
Discussion
The molecular interplay between GR and ER in breast cancer is becoming more apparent. Studies have indicated that the interactions between ER and GR are functionally significant in breast cancer cells, and that the ER and GR status of cancer cells may be important in guiding the treatment of breast cancer patients (1–5). It is therefore important to understand the mechanisms that govern ER and GR crosstalk.
Genome-wide analysis has shown that transcription factors occupy only a small fraction of their consensus binding sites due to restrictive effects of chromatin structure. The chromatin landscape largely determines which binding elements are available for transcription factor binding. Here we show that ER and GR can dictate each other’s binding at specific recognition sites through an assisted-loading mechanism (17). Under this mechanism, a given factor with chromatin access at a specific site recruits chromatin remodeling systems to the site, thereby creating a transient window of access for secondary factors with binding elements within the remodeled region. The findings presented here demonstrate the generality of this model, and show that the mechanism can function in both directions. GR reprograms accessibility for ER, and ER can modulate the landscape for GR access. This suggests that the time of induction for each steroid receptor, either during differentiation or the initiation and development of cancer, can be critical for dictating binding patterns of other steroid receptors across the genome and cellular response to hormones. Our model also provides a mechanistic basis for the observation that co-activation of GR and NF-kB alters their binding sites and target genes (30).
While nuclear receptors are thought to interact with the genome primarily by binding to specific DNA recognition sequences, they can also be recruited to the genome at non-canonical sites through interactions with other transcription factors (15,31,32). In our studies, de novo motif analysis of ER sites that are reprogrammed during dual hormone treatments implicated involvement of AP-1 in mediating ER binding at these re-arranged binding sites. Suppression of AP-1 binding with the Afos mutant confirmed the action of AP-1 at these sites. A previous study has shown that ER and GR influence each other’s activity at an AP-1 response element. In this study dexamethasone was shown to inhibit estradiol stimulation of transcription through an AP-1 response element (33). Taken together, these observations suggest that AP-1 plays a prominent role in mediating ER and GR responses upon co-activation. It should be noted in this context that AP1 has also been shown to be important in breast cancer progression (34–36).
Multiple mechanisms have been proposed to describe crosstalk between ER and GR (37–39). One class of models suggests that ER regulates expression or degradation of GR. These mechanisms, however, are based on long-term treatment of cells with hormones and may be the result of secondary effects (37,38). Here we propose a molecular mechanism for direct crosstalk between ER and GR at the genome level, whereby co-activation of two receptors leads to rapid reprogramming of chromatin structure and receptor binding (Supplementary Fig. S2A). The unveiling of this mechanism is important for understanding the molecular interplay between ER and GR and may represent a general mechanism for crosstalk between transcription factors.
Acknowledgments
The authors would like to thank Stephanie Morris, Diego Presman, Mike Guertin, and Erin Swinstead for critical reading of the manuscript and the National Cancer Institute Advanced Technology Program Sequencing Facility for sequencing services. This research was supported, in part, by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. TBM was supported, in part, by a National Institute of General Medical Sciences Pharmacological Research and Training Fellowship. LG was supported by a research grant from the Lundbeck Foundation.
Footnotes
Conflict of Interest:
The authors declare that they have no conflict of interest.
All data is available through GEO (#GSE46124).
References
Full text links
Read article at publisher's site: https://doi.org/10.1158/0008-5472.can-13-0742
Read article for free, from open access legal sources, via Unpaywall:
https://cancerres.aacrjournals.org/content/canres/73/16/5130.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1158/0008-5472.can-13-0742
Article citations
Exploring the epigenetic profile of ID4 in breast cancer: bioinformatic insights into methylation patterns and chromatin accessibility dynamics.
Breast Cancer Res Treat, 207(1):91-101, 03 May 2024
Cited by: 0 articles | PMID: 38702584
Glucocorticoid receptor action in prostate cancer: the role of transcription factor crosstalk.
Front Endocrinol (Lausanne), 15:1437179, 04 Jul 2024
Cited by: 0 articles | PMID: 39027480 | PMCID: PMC11254642
Review Free full text in Europe PMC
Glucocorticoid receptors orchestrate a convergence of host and cellular stress signals in triple negative breast cancer.
J Steroid Biochem Mol Biol, 243:106575, 29 Jun 2024
Cited by: 0 articles | PMID: 38950871
Review
Mechanistic analysis of enhancer sequences in the estrogen receptor transcriptional program.
Commun Biol, 7(1):719, 11 Jun 2024
Cited by: 1 article | PMID: 38862711
Upregulation of coagulation factor V by glucocorticoid in the preovulatory follicles of zebrafish.
J Steroid Biochem Mol Biol, 241:106521, 16 Apr 2024
Cited by: 0 articles | PMID: 38631601
Go to all (84) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (1 citation) GEO - GSE46124
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Glucocorticoid receptor modulation decreases ER-positive breast cancer cell proliferation and suppresses wild-type and mutant ER chromatin association.
Breast Cancer Res, 21(1):82, 24 Jul 2019
Cited by: 31 articles | PMID: 31340854 | PMCID: PMC6651939
GR and ER Coactivation Alters the Expression of Differentiation Genes and Associates with Improved ER+ Breast Cancer Outcome.
Mol Cancer Res, 14(8):707-719, 02 May 2016
Cited by: 61 articles | PMID: 27141101 | PMCID: PMC5008962
Binding of glucocorticoid receptors to mammary chromatin acceptor sites.
J Steroid Biochem, 28(6):581-586, 01 Dec 1987
Cited by: 7 articles | PMID: 3695510
Genome-wide crosstalk between steroid receptors in breast and prostate cancers.
Endocr Relat Cancer, 28(9):R231-R250, 22 Jul 2021
Cited by: 13 articles | PMID: 34137734 | PMCID: PMC8345902
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Intramural NIH HHS (2)
Grant ID: Z01 BC005450
Grant ID: Z01 BC005450-25





