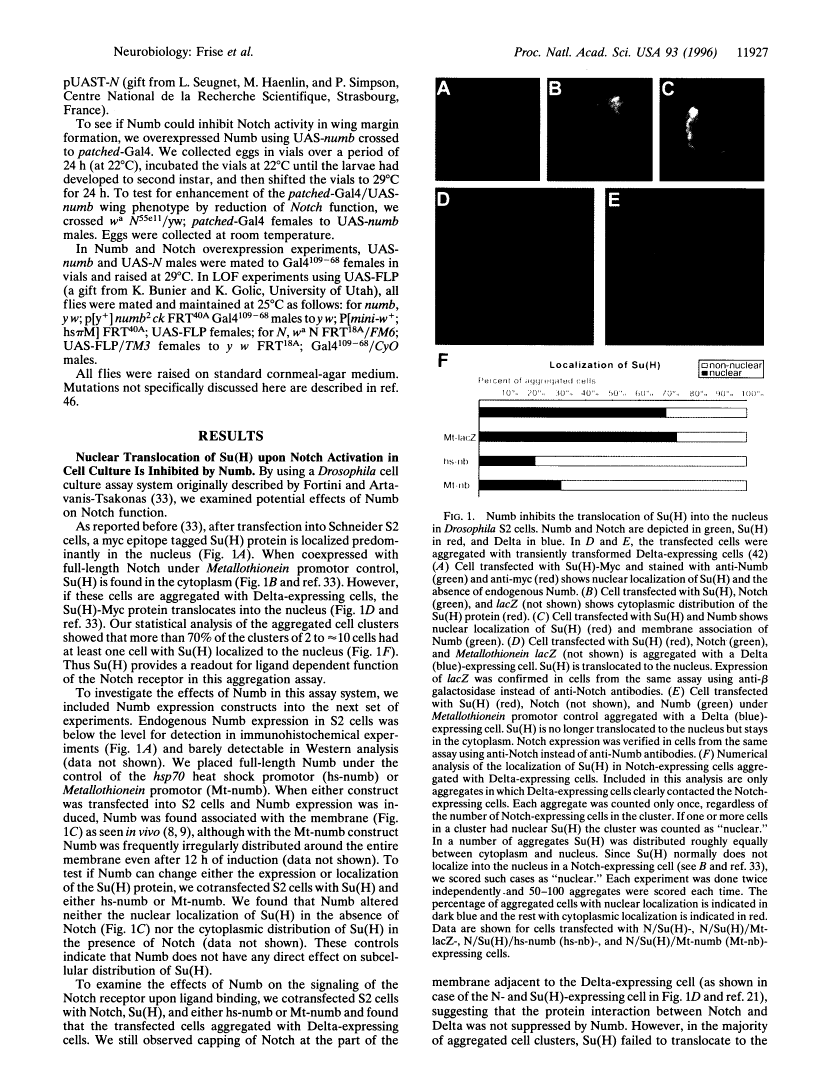
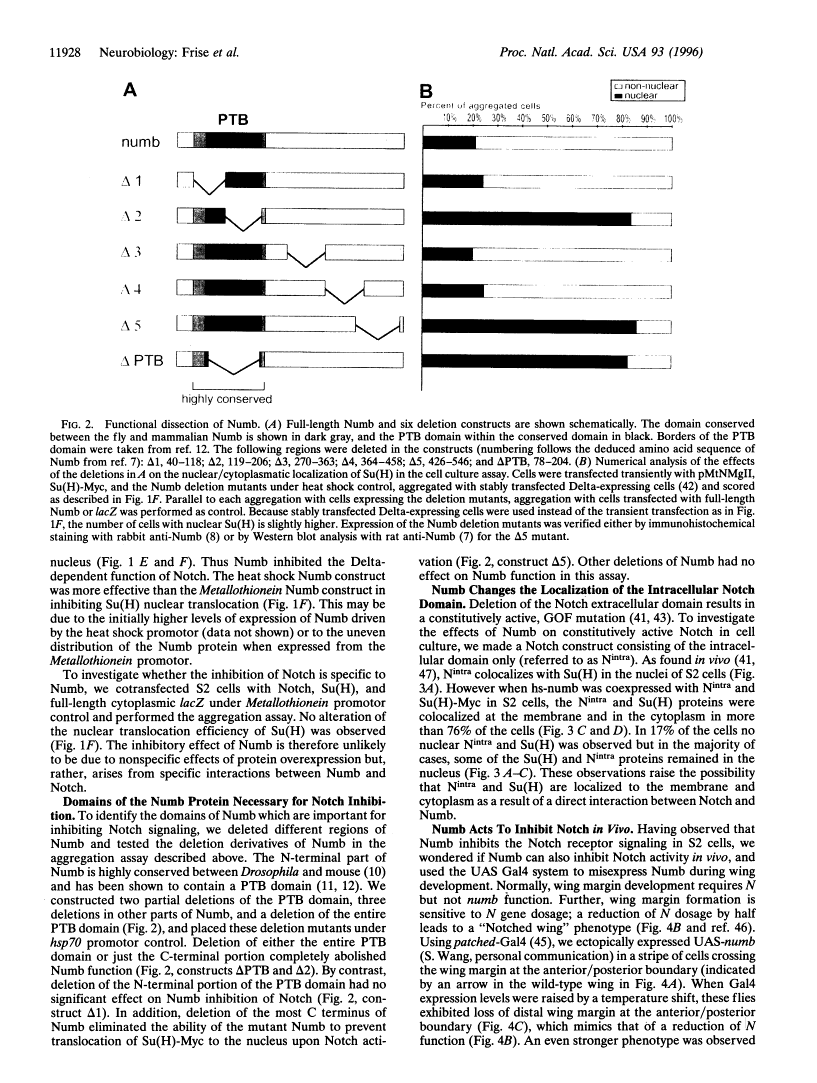
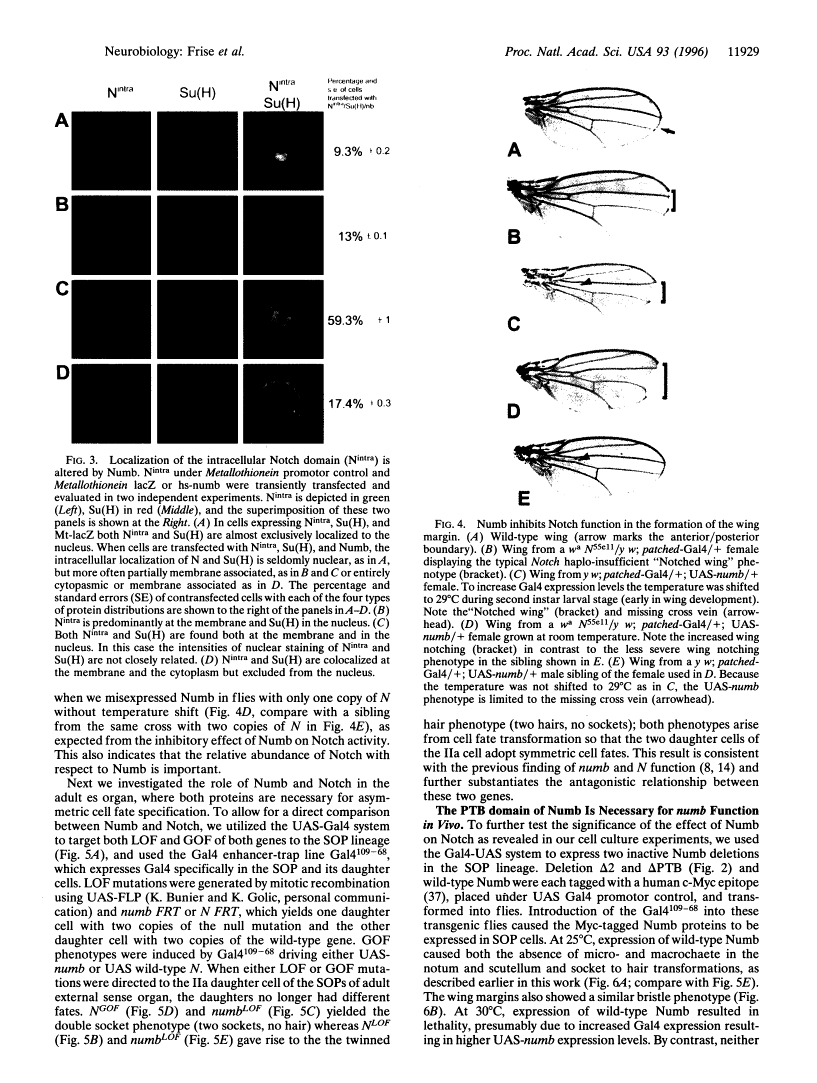
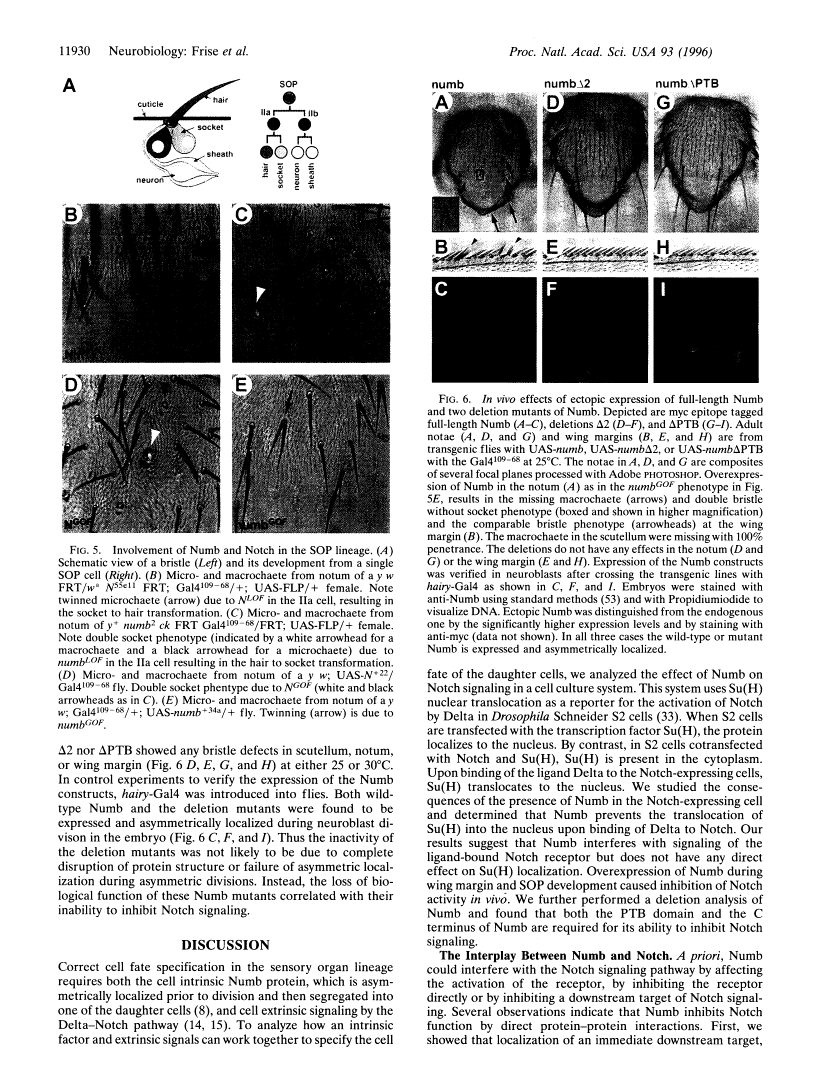
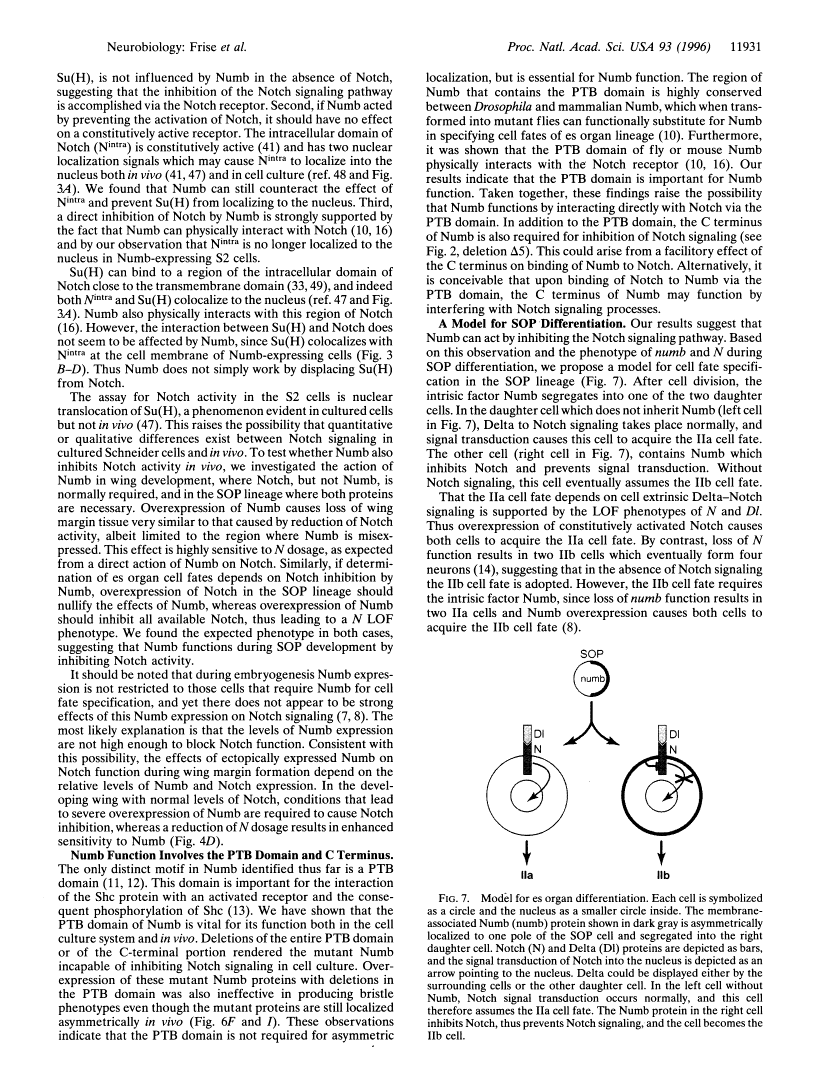
Abstract
Free full text

The Drosophila Numb protein inhibits signaling of the Notch receptor during cell-cell interaction in sensory organ lineage.
Abstract
Specification of unequal daughter cell fates in the Drosophila external sense organ lineage requires asymmetric localization of the intrinsic determinant Numb as well as cell-cell interactions mediated by the Delta ligand and Notch receptor. Previous genetic studies indicated that numb acts upstream of Notch, and biochemical studies revealed that Numb can bind Notch. For a functional assay of the action of Numb on Notch signaling, we expressed these proteins in cultured Drosophila cells and used nuclear translocation of Suppressor of Hairless [Su(H)] as a reporter for Notch activity. We found that Numb interfered with the ability of Notch to cause nuclear translocation of Su(H); both the C-terminal half of the phosphotyrosine binding domain and the C terminus of Numb are required to inhibit Notch. Overexpression of Numb during wing development, which is sensitive to Notch dosage, revealed that Numb is also able to inhibit the Notch receptor in vivo. In the external sense organ lineage, the phosphotyrosine binding domain of Numb was found to be essential for the function but not for asymmetric localization of Numb. Our results suggest that Numb determines daughter cell fates in the external sense organ lineage by inhibiting Notch signaling.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (2.8M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Click on the image to see a larger version.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Horvitz HR, Herskowitz I. Mechanisms of asymmetric cell division: two Bs or not two Bs, that is the question. Cell. 1992 Jan 24;68(2):237–255. [Abstract] [Google Scholar]
- Bodmer R, Carretto R, Jan YN. Neurogenesis of the peripheral nervous system in Drosophila embryos: DNA replication patterns and cell lineages. Neuron. 1989 Jul;3(1):21–32. [Abstract] [Google Scholar]
- Hartenstein V, Posakony JW. Development of adult sensilla on the wing and notum of Drosophila melanogaster. Development. 1989 Oct;107(2):389–405. [Abstract] [Google Scholar]
- Campos-Ortega JA. Cellular interactions during early neurogenesis of Drosophila melanogaster. Trends Neurosci. 1988 Sep;11(9):400–405. [Abstract] [Google Scholar]
- Ghysen A, Dambly-Chaudière C, Jan LY, Jan YN. Cell interactions and gene interactions in peripheral neurogenesis. Genes Dev. 1993 May;7(5):723–733. [Abstract] [Google Scholar]
- Uemura T, Shepherd S, Ackerman L, Jan LY, Jan YN. numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell. 1989 Jul 28;58(2):349–360. [Abstract] [Google Scholar]
- Rhyu MS, Jan LY, Jan YN. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell. 1994 Feb 11;76(3):477–491. [Abstract] [Google Scholar]
- Knoblich JA, Jan LY, Jan YN. Asymmetric segregation of Numb and Prospero during cell division. Nature. 1995 Oct 19;377(6550):624–627. [Abstract] [Google Scholar]
- Zhong W, Feder JN, Jiang MM, Jan LY, Jan YN. Asymmetric localization of a mammalian numb homolog during mouse cortical neurogenesis. Neuron. 1996 Jul;17(1):43–53. [Abstract] [Google Scholar]
- Bork P, Margolis B. A phosphotyrosine interaction domain. Cell. 1995 Mar 10;80(5):693–694. [Abstract] [Google Scholar]
- Zhou MM, Ravichandran KS, Olejniczak EF, Petros AM, Meadows RP, Sattler M, Harlan JE, Wade WS, Burakoff SJ, Fesik SW. Structure and ligand recognition of the phosphotyrosine binding domain of Shc. Nature. 1995 Dec 7;378(6557):584–592. [Abstract] [Google Scholar]
- Kavanaugh WM, Williams LT. An alternative to SH2 domains for binding tyrosine-phosphorylated proteins. Science. 1994 Dec 16;266(5192):1862–1865. [Abstract] [Google Scholar]
- Hartenstein V, Posakony JW. A dual function of the Notch gene in Drosophila sensillum development. Dev Biol. 1990 Nov;142(1):13–30. [Abstract] [Google Scholar]
- Parks AL, Muskavitch MA. Delta function is required for bristle organ determination and morphogenesis in Drosophila. Dev Biol. 1993 Jun;157(2):484–496. [Abstract] [Google Scholar]
- Guo M, Jan LY, Jan YN. Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron. 1996 Jul;17(1):27–41. [Abstract] [Google Scholar]
- Wharton KA, Johansen KM, Xu T, Artavanis-Tsakonas S. Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell. 1985 Dec;43(3 Pt 2):567–581. [Abstract] [Google Scholar]
- Kidd S, Kelley MR, Young MW. Sequence of the notch locus of Drosophila melanogaster: relationship of the encoded protein to mammalian clotting and growth factors. Mol Cell Biol. 1986 Sep;6(9):3094–3108. [Europe PMC free article] [Abstract] [Google Scholar]
- Vässin H, Bremer KA, Knust E, Campos-Ortega JA. The neurogenic gene Delta of Drosophila melanogaster is expressed in neurogenic territories and encodes a putative transmembrane protein with EGF-like repeats. EMBO J. 1987 Nov;6(11):3431–3440. [Europe PMC free article] [Abstract] [Google Scholar]
- Kopczynski CC, Alton AK, Fechtel K, Kooh PJ, Muskavitch MA. Delta, a Drosophila neurogenic gene, is transcriptionally complex and encodes a protein related to blood coagulation factors and epidermal growth factor of vertebrates. Genes Dev. 1988 Dec;2(12B):1723–1735. [Abstract] [Google Scholar]
- Fehon RG, Kooh PJ, Rebay I, Regan CL, Xu T, Muskavitch MA, Artavanis-Tsakonas S. Molecular interactions between the protein products of the neurogenic loci Notch and Delta, two EGF-homologous genes in Drosophila. Cell. 1990 May 4;61(3):523–534. [Abstract] [Google Scholar]
- Cagan RL, Ready DF. Notch is required for successive cell decisions in the developing Drosophila retina. Genes Dev. 1989 Aug;3(8):1099–1112. [Abstract] [Google Scholar]
- Corbin V, Michelson AM, Abmayr SM, Neel V, Alcamo E, Maniatis T, Young MW. A role for the Drosophila neurogenic genes in mesoderm differentiation. Cell. 1991 Oct 18;67(2):311–323. [Abstract] [Google Scholar]
- Ruohola H, Bremer KA, Baker D, Swedlow JR, Jan LY, Jan YN. Role of neurogenic genes in establishment of follicle cell fate and oocyte polarity during oogenesis in Drosophila. Cell. 1991 Aug 9;66(3):433–449. [Abstract] [Google Scholar]
- Rulifson EJ, Blair SS. Notch regulates wingless expression and is not required for reception of the paracrine wingless signal during wing margin neurogenesis in Drosophila. Development. 1995 Sep;121(9):2813–2824. [Abstract] [Google Scholar]
- Doherty D, Feger G, Younger-Shepherd S, Jan LY, Jan YN. Delta is a ventral to dorsal signal complementary to Serrate, another Notch ligand, in Drosophila wing formation. Genes Dev. 1996 Feb 15;10(4):421–434. [Abstract] [Google Scholar]
- Artavanis-Tsakonas S, Matsuno K, Fortini ME. Notch signaling. Science. 1995 Apr 14;268(5208):225–232. [Abstract] [Google Scholar]
- Heitzler P, Simpson P. The choice of cell fate in the epidermis of Drosophila. Cell. 1991 Mar 22;64(6):1083–1092. [Abstract] [Google Scholar]
- Spana EP, Doe CQ. Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron. 1996 Jul;17(1):21–26. [Abstract] [Google Scholar]
- Schweisguth F, Posakony JW. Suppressor of Hairless, the Drosophila homolog of the mouse recombination signal-binding protein gene, controls sensory organ cell fates. Cell. 1992 Jun 26;69(7):1199–1212. [Abstract] [Google Scholar]
- Furukawa T, Maruyama S, Kawaichi M, Honjo T. The Drosophila homolog of the immunoglobulin recombination signal-binding protein regulates peripheral nervous system development. Cell. 1992 Jun 26;69(7):1191–1197. [Abstract] [Google Scholar]
- Fortini ME, Artavanis-Tsakonas S. The suppressor of hairless protein participates in notch receptor signaling. Cell. 1994 Oct 21;79(2):273–282. [Abstract] [Google Scholar]
- Schweisguth F, Posakony JW. Antagonistic activities of Suppressor of Hairless and Hairless control alternative cell fates in the Drosophila adult epidermis. Development. 1994 Jun;120(6):1433–1441. [Abstract] [Google Scholar]
- Bailey AM, Posakony JW. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 1995 Nov 1;9(21):2609–2622. [Abstract] [Google Scholar]
- Lecourtois M, Schweisguth F. The neurogenic suppressor of hairless DNA-binding protein mediates the transcriptional activation of the enhancer of split complex genes triggered by Notch signaling. Genes Dev. 1995 Nov 1;9(21):2598–2608. [Abstract] [Google Scholar]
- Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985 Dec;5(12):3610–3616. [Europe PMC free article] [Abstract] [Google Scholar]
- Bunch TA, Grinblat Y, Goldstein LS. Characterization and use of the Drosophila metallothionein promoter in cultured Drosophila melanogaster cells. Nucleic Acids Res. 1988 Feb 11;16(3):1043–1061. [Europe PMC free article] [Abstract] [Google Scholar]
- Thummel CS, Boulet AM, Lipshitz HD. Vectors for Drosophila P-element-mediated transformation and tissue culture transfection. Gene. 1988 Dec 30;74(2):445–456. [Abstract] [Google Scholar]
- Struhl G, Fitzgerald K, Greenwald I. Intrinsic activity of the Lin-12 and Notch intracellular domains in vivo. Cell. 1993 Jul 30;74(2):331–345. [Abstract] [Google Scholar]
- Diederich RJ, Matsuno K, Hing H, Artavanis-Tsakonas S. Cytosolic interaction between deltex and Notch ankyrin repeats implicates deltex in the Notch signaling pathway. Development. 1994 Mar;120(3):473–481. [Abstract] [Google Scholar]
- Rebay I, Fehon RG, Artavanis-Tsakonas S. Specific truncations of Drosophila Notch define dominant activated and dominant negative forms of the receptor. Cell. 1993 Jul 30;74(2):319–329. [Abstract] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993 Jun;118(2):401–415. [Abstract] [Google Scholar]
- Hinz U, Giebel B, Campos-Ortega JA. The basic-helix-loop-helix domain of Drosophila lethal of scute protein is sufficient for proneural function and activates neurogenic genes. Cell. 1994 Jan 14;76(1):77–87. [Abstract] [Google Scholar]
- Gho M, Lecourtois M, Géraud G, Posakony JW, Schweisguth F. Subcellular localization of Suppressor of Hairless in Drosophila sense organ cells during Notch signalling. Development. 1996 Jun;122(6):1673–1682. [Abstract] [Google Scholar]
- Lieber T, Kidd S, Alcamo E, Corbin V, Young MW. Antineurogenic phenotypes induced by truncated Notch proteins indicate a role in signal transduction and may point to a novel function for Notch in nuclei. Genes Dev. 1993 Oct;7(10):1949–1965. [Abstract] [Google Scholar]
- Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, Honjo T. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J kappa/Su(H). Curr Biol. 1995 Dec 1;5(12):1416–1423. [Abstract] [Google Scholar]
- Bang AG, Bailey AM, Posakony JW. Hairless promotes stable commitment to the sensory organ precursor cell fate by negatively regulating the activity of the Notch signaling pathway. Dev Biol. 1995 Dec;172(2):479–494. [Abstract] [Google Scholar]
- Knust E, Schrons H, Grawe F, Campos-Ortega JA. Seven genes of the Enhancer of split complex of Drosophila melanogaster encode helix-loop-helix proteins. Genetics. 1992 Oct;132(2):505–518. [Europe PMC free article] [Abstract] [Google Scholar]
- Delidakis C, Artavanis-Tsakonas S. The Enhancer of split [E(spl)] locus of Drosophila encodes seven independent helix-loop-helix proteins. Proc Natl Acad Sci U S A. 1992 Sep 15;89(18):8731–8735. [Europe PMC free article] [Abstract] [Google Scholar]
- Guo M, Bier E, Jan LY, Jan YN. tramtrack acts downstream of numb to specify distinct daughter cell fates during asymmetric cell divisions in the Drosophila PNS. Neuron. 1995 May;14(5):913–925. [Abstract] [Google Scholar]
Associated Data
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.93.21.11925
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc38160?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1073/pnas.93.21.11925
Article citations
Numb Suppresses Notch-Dependent Activation of Enhancer of split during Lateral Inhibition in the Drosophila Embryonic Nervous System.
Biomolecules, 14(9):1062, 26 Aug 2024
Cited by: 0 articles | PMID: 39334829 | PMCID: PMC11429637
Spatio-Temporal Regulation of Notch Activation in Asymmetrically Dividing Sensory Organ Precursor Cells in Drosophila melanogaster Epithelium.
Cells, 13(13):1133, 30 Jun 2024
Cited by: 0 articles | PMID: 38994985 | PMCID: PMC11240559
Review Free full text in Europe PMC
Par3/bazooka binds NICD and promotes notch signaling during Drosophila development.
Dev Biol, 514:37-49, 15 Jun 2024
Cited by: 1 article | PMID: 38885804
Low-level repressive histone marks fine-tune gene transcription in neural stem cells.
Elife, 12:e86127, 14 Jun 2023
Cited by: 1 article | PMID: 37314324 | PMCID: PMC10344426
Zika Virus Induces Degradation of the Numb Protein Required through Embryonic Neurogenesis.
Viruses, 15(6):1258, 27 May 2023
Cited by: 1 article | PMID: 37376558 | PMCID: PMC10301363
Go to all (194) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Control of daughter cell fates during asymmetric division: interaction of Numb and Notch.
Neuron, 17(1):27-41, 01 Jul 1996
Cited by: 417 articles | PMID: 8755476
Lethal giant larvae acts together with numb in notch inhibition and cell fate specification in the Drosophila adult sensory organ precursor lineage.
Curr Biol, 13(9):778-783, 01 Apr 2003
Cited by: 31 articles | PMID: 12725738
Only a subset of the binary cell fate decisions mediated by Numb/Notch signaling in Drosophila sensory organ lineage requires Suppressor of Hairless.
Development, 124(22):4435-4446, 01 Nov 1997
Cited by: 61 articles | PMID: 9409662
Numb: "Adapting" notch for endocytosis.
Dev Cell, 3(2):155-156, 01 Aug 2002
Cited by: 22 articles | PMID: 12194846
Review