Abstract
Free full text
Genome sequence of the human malaria parasite Plasmodium falciparum
Abstract
The parasite Plasmodium falciparum is responsible for hundreds of millions of cases of malaria, and kills more than one million African children annually. Here we report an analysis of the genome sequence of P. falciparum clone 3D7. The 23-megabase nuclear genome consists of 14 chromosomes, encodes about 5,300 genes, and is the most (A + T)-rich genome sequenced to date. Genes involved in antigenic variation are concentrated in the subtelomeric regions of the chromosomes. Compared to the genomes of free-living eukaryotic microbes, the genome of this intracellular parasite encodes fewer enzymes and transporters, but a large proportion of genes are devoted to immune evasion and host–parasite interactions. Many nuclear-encoded proteins are targeted to the apicoplast, an organelle involved in fatty-acid and isoprenoid metabolism. The genome sequence provides the foundation for future studies of this organism, and is being exploited in the search for new drugs and vaccines to fight malaria.
Despite more than a century of efforts to eradicate or control malaria, the disease remains a major and growing threat to the public health and economic development of countries in the tropical and subtropical regions of the world. Approximately 40% of the world’s population lives in areas where malaria is transmitted. There are an estimated 300–500 million cases and up to 2.7 million deaths from malaria each year. The mortality levels are greatest in sub-Saharan Africa, where children under 5 years of age account for 90% of all deaths due to malaria1. Human malaria is caused by infection with intracellular parasites of the genus Plasmodium that are transmitted by Anopheles mosquitoes. Of the four species of Plasmodium that infect humans, Plasmodium falciparum is the most lethal. Resistance to anti-malarial drugs and insecticides, the decay of public health infrastructure, population movements, political unrest, and environmental changes are contributing to the spread of malaria2. In countries with endemic malaria, the annual economic growth rates over a 25-year period were 1.5% lower than in other countries. This implies that the cumulative effect of the lower annual economic output in a malaria-endemic country was a 50% reduction in the per capita GDP compared to a non-malarious country3. Recent studies suggest that the number of malaria cases may double in 20 years if new methods of control are not devised and implemented1.
An international effort4 was launched in 1996 to sequence the P. falciparum genome with the expectation that the genome sequence would open new avenues for research. The sequences of two of the 14 chromosomes, representing 8% of the nuclear genome, were published previously5,6 and the accompanying Letters in this issue describe the sequences of chromosomes 1, 3-9 and 13 (ref. 7), 2, 10, 11 and 14 (ref. 8), and 12 (ref. 9). Here we report an analysis of the genome sequence of P. falciparum clone 3D7, including descriptions of chromosome structure, gene content, functional classification of proteins, metabolism and transport, and other features of parasite biology.
Sequencing strategy
A whole chromosome shotgun sequencing strategy was used to determine the genome sequence of P. falciparum clone 3D7. This approach was taken because a whole genome shotgun strategy was not feasible or cost-effective with the technology that was available at the beginning of the project. Also, high-quality large insert libraries of (A + T)-rich P. falciparum DNA have never been constructed in Escherichia coli, which ruled out a clone-by-clone sequencing strategy. The chromosomes were separated on pulsed field gels, and chromosomal DNA was extracted and used to construct shotgun libraries of 1–3-kilobase (kb) fragments of sheared DNA. Eleven of the fourteen chromosomes could be resolved on the gels, but chromosomes 6, 7 and 8 could not be resolved and were sequenced as a group. The shotgun sequences were assembled into contiguous DNA sequences (contigs), in some cases with low coverage shotgun sequences of yeast artificial chromosome (YAC) clones to assist in the ordering of contigs for closure. Sequence tagged sites (STSs)10, microsatellite markers11,12 and HAPPY mapping7 were also used to place and orient contigs during the gap closure process. The high (A + T) content of the genome made gap closure extremely difficult7-9. The predicted restriction enzyme maps of the chromosome sequences were compared to optical restriction maps to verify that the chromosomes had been assembled correctly13. Chromosomes 1–5, 9 and 12 were closed, whereas chromosomes 6–8, 10, 11, 13 and 14 contained 3–37 gaps (most <2.5 kb) per chromosome at the beginning of genome annotation. Efforts to close the remaining gaps are continuing.
Genome structure and content
The P. falciparum 3D7 nuclear genome is composed of 22.8 megabases (Mb) distributed among 14 chromosomes ranging in size from approximately 0.643 to 3.29 Mb (Fig. 1, and Supplementary Figs A-N). Thus the P. falciparum genome is almost twice the size of the genome of the fission yeast Schizosaccharomyces pombe. The overall (A + T) composition is 80.6%, and rises to ~90% in introns and intergenic regions. The structures of protein-encoding genes were predicted using several gene-finding programs and manually curated. Approximately 5,300 protein-encoding genes were identified, about the same as in S. pombe (Table 1, and Supplementary Table A). This suggests an average gene density in P. falciparum of 1 gene per 4,338 base pairs (bp), slightly higher than was found previously with chromosomes 2 and 3 (1 per 4,500 bp and 1 per 4,800 bp, respectively). The higher gene density reported here is probably the result of improved gene-finding software and larger training sets that enabled the detection of genes overlooked previously8. Introns were predicted in 54% of P. falciparum genes, a proportion roughly similar to that in S. pombe and Dictyostelium discoideum, but much higher than observed in Saccharomyces cerevisiae where only 5% of genes contain introns. Excluding introns, the mean length of P. falciparum genes was 2.3 kb, substantially larger than in the other organisms in which the average gene lengths range from 1.3 to 1.6 kb. Plasmodium falciparum genes showed a markedly greater proportion of genes (15.5%) longer than 4 kb compared to S. pombe and S. cerevisiae (3.0% and 3.6%, respectively). The explanation for the increased gene length in P. falciparum is not clear. Many of these large genes encode uncharacterized proteins that may be cytosolic proteins, as they do not possess recognizable signal peptides. No transposable elements or retrotransposons were identified.

Schematic representation of the P. falciparum 3D7 genome. Q Protein-encoding genes are indicated by open diamonds. All genes are depicted at the same scale regardless of their size or structure. The labels indicate the name for each gene. The rows of coloured rectangles represent, from top to bottom for each chromosome, the high-level Gene Ontology assignment for each gene in the ‘biological process’, ‘molecular function’, and ‘cellular component’ ontologies42; the life-cycle stage(s) at which each predicted gene product has been detected by proteomics techniques14,15; and Plasmodium yoelii yoelii genes that exhibit conserved sequence and organization with genes in P. falciparum, as shown by a position effect analysis. Rectangles surrounding clusters of P. yoelii genes indicate genes shown to be linked in the P. y. yoelii genome165. Boxes containing coloured arrowheads at the ends of each chromosome indicate subtelomeric blocks (SBs; see text and Fig. 2).
Table 1
| Feature | Value | ||||
|---|---|---|---|---|---|
| P. falciparum | S. pombe | S. cerevisiae | D. discoideum | A. thaliana | |
| Size (bp) | 22,853,764 | 12,462,637 | 12,495,682 | 8,100,000 | 115,409,949 |
| (G + C) content (%) | 19.4 | 36.0 | 38.3 | 22.2 | 34.9 |
| No. of genes | 5,268* | 4,929 | 5,770 | 2,799 | 25,498 |
| Mean gene length† (bp) | 2,283 | 1,426 | 1,424 | 1,626 | 1,310 |
| Gene density (bp per gene) | 4,338 | 2,528 | 2,088 | 2,600 | 4,526 |
| Per cent coding | 52.6 | 57.5 | 70.5 | 56.3 | 28.8 |
| Genes with introns (%) | 53.9 | 43 | 5.0 | 68 | 79 |
| Exons | |||||
 Number Number | 12,674 | ND | ND | 6,398 | 132,982 |
 No. per gene No. per gene | 2.39 | ND | NA | 2.29 | 5.18 |
 (G + C) content (%) (G + C) content (%) | 23.7 | 39.6 | 28.0 | 28.0 | ND |
 Mean length (bp) Mean length (bp) | 949 | ND | ND | 711 | 170 |
 Total length (bp) Total length (bp) | 12,028,350 | ND | ND | 4,548,978 | 33,249,250 |
| Introns | |||||
 Number Number | 7,406 | 4,730 | 272 | 3,587 | 107,784 |
 (G + C) content (%) (G + C) content (%) | 13.5 | ND | NA | 13.0 | ND |
 Mean length (bp) Mean length (bp) | 178.7 | 81 | NA | 177 | 170 |
 Total length (bp) Total length (bp) | 1,323,509 | 383,130 | ND | 643,899 | 18,055,421 |
| Intergenic regions | |||||
 (G + C) content (%) (G + C) content (%) | 13.6 | ND | ND | 14.0 | ND |
 Mean length (bp) Mean length (bp) | 1,694 | 952 | 515 | 786 | ND |
| RNAs | |||||
 No. of tRNA genes No. of tRNA genes | 43 | 174 | ND | 73 | ND |
 No. of 5S rRNA genes No. of 5S rRNA genes | 3 | 30 | ND | NA | ND |
 No. of 5.8S, 18S and 28S rRNA units No. of 5.8S, 18S and 28S rRNA units | 7 | 200–400 | ND | NA | 700–800 |
ND, not determined; NA, not applicable. ‘No. of genes’ for D. discoideum are for chromosome 2 (ref. 155) and in some cases represent extrapolations to the entire genome. Sources of data for the other organisms: S. pombe65, S. cerevisiae156, D. discoideum155 and A. thaiiana157.
Fifty-two per cent of the predicted gene products (2,731) were detected in cell lysates prepared from several stages of the parasite life cycle by high-resolution liquid chromatography and tandem mass spectrometry14,15, including many predicted proteins with no similarity to proteins in other organisms. In addition, 49% of the genes overlapped (97% identity over at least 100 nucleotides) with expressed sequence tags (ESTs) derived from several life-cycle stages. As the proteomics and EST studies performed to date may not represent a complete sampling of all genes expressed during the complex life cycle of the parasite, this suggests that the annotation process identified substantial portions of most genes. However, in the absence of supporting EST or protein evidence, correct prediction of the 5′ ends of genes and genes with multiple small exons is challenging, and the gene models should be regarded as preliminary. Additional ESTs and full-length complementary DNA sequences16 are required for the development of better training sets for gene-finding programs and the verification of the predicted genes.
The nuclear genome contains a full set of transfer RNA (tRNA) ligase genes, and 43 tRNAs were identified to bind all codons except TGT and TGC, coding for Cys; it is possible that these tRNAs are located within the currently unsequenced regions. All codons ending in C and Tappear to be read by single tRNAs with a G in the first position, which is likely to read both codons via G:U wobble. Each anticodon occurs only once except for methionine (CAT), for which there are two copies, one for translation initiation and one for internal methionines, and the glycine (CCT) anticodon, which occurs twice. An unusual tRNA resembling a selenocysteinyl-tRNA was also found. A putative selenocysteine lyase was identified, which may provide selenium for synthesis of selenoproteins. Increased growth has been observed in selenium-supplemented Plasmodium culture17.
In almost all other eukaryotic organisms sequenced to date, the tRNA genes exhibit extensive redundancy, the only exception being the intracellular parasite Encephalitozoon cuniculi which contains 44 tRNAs18. Often, the abundance of specific anticodons is correlated with the codon usage of the organism19,20. This is not the case in P. falciparum, which exhibits minimal redundancy of tRNAs. The mitochondrial genome of Plasmodium is small (about 6 kb) and encodes no tRNAs, so the mitochondrion must import tRNAs21,22. Through their import, cytoplasmic tRNAs may serve mitochondrial protein synthesis in a manner seen with other organisms23,24. The apicoplast genome appears to encode sufficient tRNAs for protein synthesis within the organelle25.
Unlike many other eukaryotes, the malaria parasite genome does not contain long tandemly repeated arrays of ribosomal RNA (rRNA) genes. Instead, Plasmodium parasites contain several single 18S-5.8S-28S rRNA units distributed on different chromosomes. The sequence encoded by a rRNA gene in one unit differs from the sequence of the corresponding rRNA in the other units. Furthermore, the expression of each rRNA unit is developmentally regulated, resulting in the expression of a different set of rRNAs at different stages of the parasite life cycle26,27. It is likely that by changing the properties of its ribosomes the parasite is able to alter the rate of translation, either globally or of specific messenger RNAs (mRNAs), thereby changing the rate of cell growth or altering patterns of cell development. The two types of rRNA genes previously described in P. falciparum are the S-type, expressed primarily in the mosquito vector, and the A-type, expressed primarily in the human host. Seven loci encoding rRNAs were identified in the genome sequence (Fig. 1). Two copies of the S-type rRNA genes are located on chromosomes 11 and 13, and two copies of the A-type genes are located on chromosomes 5 and 7. In addition, chromosome 1 contains a third, previously uncharacterized, rRNA unit that encodes 18S and 5.8S rRNAs that are almost identical to the S-type genes on chromosomes 11 and 13, but has a significantly divergent 28S rRNA gene (65% identity to the A-type and 75% identity to the S-type). The expression profiles of these genes are unknown. Chromosome 8 also contains two unusual rRNA gene units that contain 5.8S and 28S rRNA genes but do not encode 18S rRNAs; it is not known whether these genes are functional. The sequences of the 18S and 28S rRNA genes on chromosome 7 and the 28S rRNA gene on chromosome 8 are incomplete as they reside at contig ends. The 5S rRNA is encoded by three identical tandemly arrayed genes on chromosome 14.
Chromosome structure
Plasmodium falciparum chromosomes vary considerably in length, with most of the variation occurring in the subtelomeric regions. Field isolates, even those from individuals residing in a single village28, exhibit extensive size polymorphism that is thought to be due to recombination events between different parasite clones during meiosis in the mosquito29. Chromosome size variation is also observed in cultures of erythrocytic parasites, but is due to chromosome breakage and healing events and not to meiotic recombination30,31. Subtelomeric deletions often extend well into the chromosome, and in some cases alter the cell adhesion properties of the parasite owing to the loss of the gene(s) encoding adhesion molecules32,33. Because many genes involved in antigenic variation are located in the subtelomeric regions, an understanding of subtelomere structure and functional properties is essential for the elucidation of the mechanisms underlying the generation of antigenic diversity.
The subtelomeric regions of the chromosomes display a striking degree of conservation within the genome that is probably due to promiscuous inter-chromosomal exchange of subtelomeric regions. Subtelomeric exchanges occur in other eukaryotes34-36, but the regions involved are much smaller (2.5–3.0 kb) in S. cerevisiae (data not shown). Previous studies of P. falciparum telomeres37,38 suggested that they contained six blocks of repetitive sequences that were designated telomere-associated repetitive elements (TAREs 1–6).
Whole genome analysis reveals a larger (up to 120 kb), more complex, subtelomeric repeat structure than was observed previously. The conserved regions fall into five large subtelomeric blocks (SBs; Fig. 2). The sequences within blocks 2, 4 and 5 include many tandem repeats in addition to those described previously, as well as non-repetitive regions. Subtelomeric block 1 (SB-1, equivalent to TARE-1), contains the 7-bp telomeric repeat in a variable number of near-exact copies39. SB-2 contains several sub-blocks of repeats of different sizes, including TAREs 2–5 and other sequences. The beginning of SB-2 consists of about 1,000–1,300 bp of nonrepetitive sequence, followed on some chromosomes by 2.5 copies of a 164-bp repeat. This is followed by another 300 bp of nonrepetitive sequence, and then 10 copies of a 135-bp repeat, the main element of TARE-2. TARE-2 is followed by 200 bp of non-repetitive sequence, and then two copies of a highly conserved 63-bp repeat. SB-2 extends for another 6 kb that contains non-repetitive sequence as well as other tandem repeats. Only four of the 28 telomeres are missing SB-2, which always occurs immediately adjacent to SB-1. A notable feature of SB-2 is the conserved order and orientation of each repeat variant as well as the sequence homology extending throughout the block. For almost any two chromosomes that were examined, a consistently ordered series of unique, identical sequences of >30 bp that are distributed across SB-2 were identified, suggesting that SB-2 is a repeat with a complex internal structure occurring once per telomere.
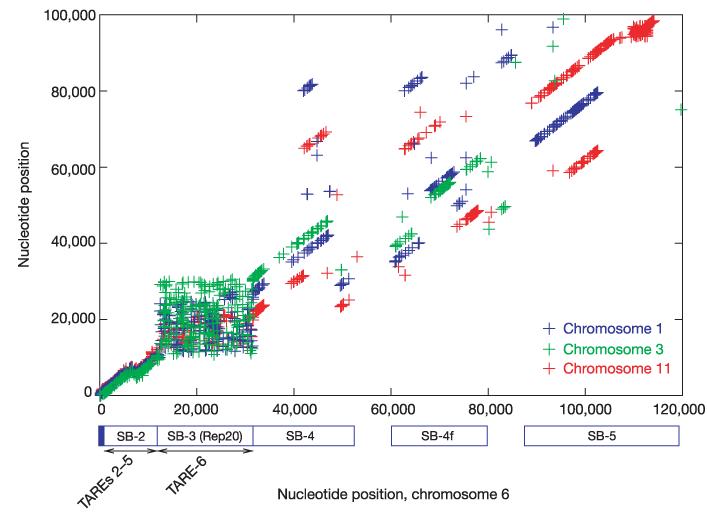
Alignment of subtelomeric regions of chromosomes 1, 3, 6 and 11. MUMmer2152 alignments showing exact matches between the left subtelomeric regions of chromosome 6 (horizontal axis) and chromosomes 11 (red), 1 (blue) and 3 (green), illustrating the conserved synteny between all telomeres. Each point represents an exact match of 40 bp or longer that is shared by two chromosomes and is not found anywhere else on either chromosome. Each collinear series of points along a diagonal represents an aligned region. SB, subtelomeric block; TARE, telomere-associated repetitive element.
SB-3 consists of the Rep20 element40, a large block of highly variable copies of a 21-bp repeat. The tandem repeats in SB-3 occur in a random order (Fig. 2). SB-4 has not been described previously, although it does contain the previously described R-FA3 sequence41. SB-4 also includes a complex mix of short (<28-bp) tandem repeats, and a 105-bp repeat that occurs once in each subtelomere. Many telomeres contain one or more var (variant antigen) gene exons within this block, which appear as gaps in the alignment. In five subtelomeres, fragments of 2–4 kb from SB-4 are duplicated and inverted. SB-5 is found in half of the subtelomeres, does not contain tandem repeats, and extends up to 120 kb into some chromosomes. The arrangement and composition of the subtelomeric blocks suggests frequent recombination between the telomeres.
Centromeres have not been identified experimentally in malaria parasites. However, putative centromeres were identified by comparison of the sequences of chromosomes 2 and 3 (ref. 6). Eleven of the 14 chromosomes contained a single region of 2–3 kb with extremely high (A + T) content (>97%) and imperfect short tandem repeats, features resembling the regional S. pombe centromeres; the 3 chromosomes lacking such regions were incomplete.
The proteome
Of the 5,268 predicted proteins, about 60% (3,208 hypothetical proteins) did not have sufficient similarity to proteins in other organisms to justify provision of functional assignments (Table 2). This is similar to what was found previously with chromosomes 2 and 3 (refs 5, 6). Thus, almost two-thirds of the proteins appear to be unique to this organism, a proportion much higher than observed in other eukaryotes. This may be a reflection of the greater evolutionary distance between Plasmodium and other eukaryotes that have been sequenced, exacerbated by the reduction of sequence similarity due to the (A + T) richness of the genome. Another 257 proteins (5%) had significant similarity to hypothetical proteins in other organisms. Thirty-one per cent (1,631) of the predicted proteins had one or more transmembrane domains, and 17.3% (911) of the proteins possessed putative signal peptides or signal anchors.
Table 2
| Feature | Number | Per cent |
|---|---|---|
| Total predicted proteins | 5,268 | |
| Hypothetical proteins | 3,208 | 60.9 |
| InterPro matches | 2,650 | 52.8 |
| Pfam matches | 1,746 | 33.1 |
| Gene Ontology | ||
 Process Process | 1,301 | 24.7 |
 Function Function | 1,244 | 23.6 |
 Component Component | 2,412 | 45.8 |
| Targeted to apicoplast | 551 | 10.4 |
| Targeted to mitochondrion | 246 | 4.7 |
| Structural features | ||
 Transmembrane domain(s) Transmembrane domain(s) | 1,631 | 31.0 |
 Signal peptide Signal peptide | 544 | 10.3 |
 Signal anchor Signal anchor | 367 | 7.0 |
 Non-secretory protein Non-secretory protein | 4,357 | 82.7 |
Of the apicoplast-targeted proteins, 126 were judged on the basis of experimental evidence or the predictions of multiple programs61,158 to be localized to the apicoplast with high confidence. Predicted apicoplast localization for 425 other proteins is based on an analysis using only one method and is of lower confidence. Predicted mitochondrial localization was based upon BLASTP searches of S. cerevisiae mitochondrion-targeted proteins159 and TargetP158 and MitoProtll160 predictions; 148 genes were judged to be targeted to the mitochondrion with a high or medium confidence level, and an additional 98 genes with a lower confidence of mitochondrial targeting. Other specialized searches used the following programs and databases: InterPro161; Pfam162; Gene Ontology42; transmembrane domains, TMHMM163; signal peptides and signal anchors, SignalP-2.0164.
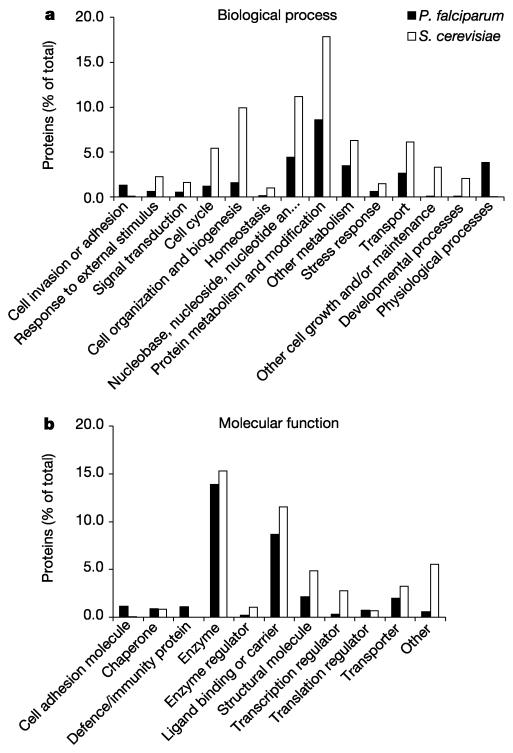
The Gene Ontology (GO)42 database is a controlled vocabulary that describes the roles of genes and gene products in organisms. GO terms were assigned manually to 2,134 gene products (40%) and a comparison of annotation with high-level GO terms for both S. cerevisiae and P. falciparum is shown in Fig. 3. In almost all categories, higher values can be seen for S. cerevisiae, reflecting the greater proportion of the genome that has been characterized compared to P. falciparum. There are two exceptions to this pattern that reflect processes specifically connected with the parasite life cycle. At least 1.3% of P. falciparum genes are involved in cell-to-cell adhesion or the invasion of host cells. As discussed below (see ‘Immune evasion’), P. falciparum has 208 genes (3.9%) known to be involved in the evasion of the host immune system. This is reflected in the assignment of many more gene products to the GO term ‘physiological processes’ in P. falciparum than in S. cerevisiae (Fig. 3). The comparison with S. cerevisiae also reveals that particular categories in P. falciparum appear to be under-represented. Sporulation and cell budding are obvious examples (they are included in the category ‘other cell growth and/or maintenance’), but very few genes in P. falciparum were associated with the ‘cell organization and biogenesis’, the ‘cell cycle’, or ‘transcription factor’ categories compared to S. cerevisiae (Fig. 3). These differences do not necessarily imply that fewer malaria genes are involved in these processes, but highlight areas of malaria biology where knowledge is limited.
The apicoplast
Malaria parasites and other members of the phylum apicomplexa harbour a relict plastid, homologous to the chloroplasts of plants and algae25,43,44. The ‘apicoplast’ is essential for parasite survival45,46, but its exact role is unclear. The apicoplast is known to function in the anabolic synthesis of fatty acids5,47,48, isoprenoids49 and haeme50,51, suggesting that one or more of these compounds could be exported from the apicoplast, as is known to occur in plant plastids. The apicoplast arose through a process of secondary endosymbiosis52-55, in which the ancestor of all apicomplexan parasites engulfed a eukaryotic alga, and retained the algal plastid, itself the product of a prior endosymbiotic event56. The 35-kb apicoplast genome encodes only 30 proteins25, but as in mitochondria and chloroplasts, the apicoplast proteome is supplemented by proteins encoded in the nuclear genome and post-translationally targeted into the organelle by the use of a bipartite targeting signal, consisting of an amino-terminal secretory signal sequence, followed by a plastid transit peptide55,57-60.
In total, 551 nuclear-encoded proteins (~10% of the predicted nuclear encoded proteins) that may be targeted to the apicoplast were identified using bioinformatic61 and laboratory-based methods. Apicoplast targeting of a few proteins has been verified by antibody localization and by the targeting of fluorescent fusion proteins to the apicoplast in transgenic P. falciparum or Toxoplasma gondii47 parasites. Some proteins may be targeted to both the apicoplast and mitochondrion, as suggested by the observation that the total number of tRNA ligases is inadequate for independentprotein synthesis in the cytoplasm, mitochondrion and apicoplast. In plants, some proteins lack a transit peptide but are targeted to plastids via an unknown process. Proteins that use an alternative targeting pathway in P. falciparum would have escaped detection with the methods used.
Nuclear-encoded apicoplast proteins include housekeeping enzymes involved in DNA replication and repair, transcription, translation and post-translational modifications, cofactor synthesis, protein import, protein turnover, and specific metabolic and transport activities. No genes for photosynthesis or light perception are apparent, although ferredoxin and ferredoxin-NADP reductase are present as vestiges of photosystem I, and probably serve to recycle reducing equivalents62. About 60% of the putative apicoplast-targeted proteins are of unknown function. Several metabolic pathways in the organelle are distinct from host pathways and offer potential parasite-specific targets for drug therapy63 (see ‘Metabolism’ and ‘Transport’ sections).
Evolution
Comparative genome analysis with other eukaryotes for which the complete genome is available (excluding the parasite E. cuniculi) revealed that, in terms of overall genome content, P. falciparum is slightly more similar to Arabidopsis thaliana than to other taxa. Although this is consistent with phylogenetic studies64, it could also be due to the presence in the P. falciparum nuclear genome of genes derived from plastids or from the nuclear genome of the secondary endosymbiont. Thus the apparent affinity of Plasmodium and Arabidopsis might not reflect the true phylogenetic history of the P. falciparum lineage. Comparative genomic analysis was also used to identify genes apparently duplicated in the P. falciparum lineage since it split from the lineages represented by the other completed genomes (Supplementary Table B).
There are 237 P. falciparum proteins with strong matches to proteins in all completed eukaryotic genomes but no matches to proteins, even at low stringency, in any complete prokaryotic proteome (Supplementary Table C). These proteins help to define the differences between eukaryotes and prokaryotes. Proteins in this list include those with roles in cytoskeleton construction and maintenance, chromatin packaging and modification, cell cycle regulation, intracellular signalling, transcription, translation, replication, and many proteins of unknown function. This list overlaps with, but is somewhat larger than, the list generated by an analysis of the S. pombe genome65. The differences are probably due in part to the different stringencies used to identify the presence or absence of homologues in the two studies.
A large number of nuclear-encoded genes in most eukaryotic species trace their evolutionary origins to genes from organelles that have been transferred to the nucleus during the course of eukaryotic evolution. Similarity searches against other complete genomes were used to identify P. falciparum nuclear-encoded genes that may be derived from organellar genomes. Because similarity searches are not an ideal method for inferring evolutionary relatedness66, phylogenetic analysis was used to gain a more accurate picture of the evolutionary history of these genes. Out of 200 candidates examined, 60 genes were identified as being of probable mitochondrial origin. The proteins encoded by these genes include many with known or expected mitochondrial functions (for example, the tricarboxylic acid (TCA) cycle, protein translation, oxidative damage protection, the synthesis of haem, ubiquinone and pyrimidines), as well as proteins of unknown function. Out of 300 candidates examined, 30 were identified as being of probable plastid origin, including genes with predicted roles in transcription and translation, protein cleavage and degradation, the synthesis of isoprenoids and fatty acids, and those encoding four subunits of the pyruvate dehydrogenase complex. The origin of many candidate organelle-derived genes could not be conclusively determined, in part due to the problems inherent in analysing genes of very high (A + T) content. Nevertheless, it appears likely that the total number of plastid-derived genes in P. falciparum will be significantly lower than that in the plant A. thaliana (estimated to be over 1,000). Phylogenetic analysis reveals that, as with the A. thaliana plastid, many of the genes predicted to be targeted to the apicoplast are apparently not of plastid origin. Of 333 putative apicoplast-targeted genes for which trees were constructed, only 26 could be assigned a probable plastid origin. In contrast, 35 were assigned a probable mitochondrial origin and another 85 might be of mitochondrial origin but are probably not of plastid origin (they group with eukaryotes that have not had plastids in their history, such as humans and fungi, but the relationship to mitochondrial ancestors is not clear). The apparent non-plastid origin of these genes could either be due to inaccuracies in the targeting predictions or to the co-option of genes derived from the mitochondria or the nucleus to function in the plastid, as has been shown to occur in some plant species67.
Metabolism
Biochemical studies of the malaria parasite have been restricted primarily to the intra-erythrocytic stage of the life cycle, owing to the difficulty of obtaining suitable quantities of material from the other life-cycle stages. Analysis of the genome sequence provides a global view of the metabolic potential of P. falciparum irrespective of the life-cycle stage (Fig. 4). Of the 5,268 predicted proteins, 733 (~14%) were identified as enzymes, of which 435 (,8%) were assigned Enzyme Commission (EC) numbers. This is considerably fewer than the roughly one-quarter to one-third of the genes in bacterial and archaeal genomes that can be mapped to Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway diagrams68, or the 17% of S. cerevisiae open reading frames that can be assigned EC numbers. This suggests either that P. falciparum has a smaller proportion of its genome devoted to enzymes, or that enzymes are more difficult to identify in P. falciparum by sequence similarity methods. (This difficulty can be attributed either to the great evolutionary distance between P. falciparum and other well-studied organisms, or to the high (A + T) content of the genome.) A few genes might have escaped detection because they were located in the small regions of the genome that remain to be sequenced (Table 1). However, many biochemical pathways could be reconstructed in their entirety, suggesting that the similarity-searching approach was for the most part successful, and that the relative paucity of enzymes in P. falciparum may be related to its parasitic life-style. A similar picture has emerged in the analysis of transporters (see ‘Transport’). In erythrocytic stages, P. falciparum relies principally on anaerobic glycolysis for energy production, with regeneration of NAD+ by conversion of pyruvate to lactate69. Genes encoding all of the enzymes necessary for a functional glycolytic pathway were identified, including a phosphofructokinase (PFK) that has sequence similarity to the pyrophosphate-dependent class of enzymes but which is probably ATP-dependent on the basis of the characterization of the homologous enzyme in Plasmodium berghei70,71. A second putative pyrophosphate-dependent PFK was also identified which possessed N- and carboxy-terminal extensions that could represent targeting sequences.
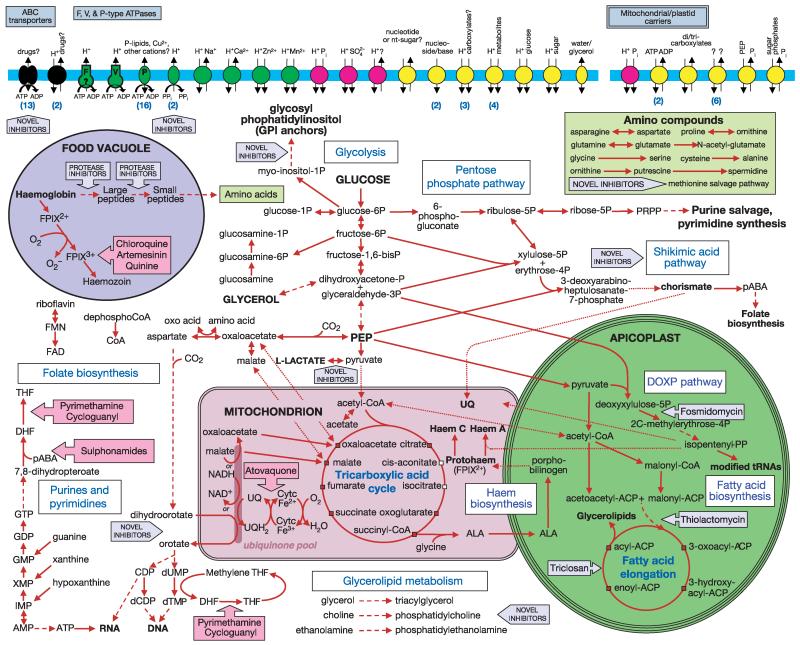
Overview of metabolism and transport in P. falciparum. Glucose and glycerol provide the major carbon sources for malaria parasites. Metabolic steps are indicated by arrows, with broken lines indicating multiple intervening steps not shown; dotted arrows indicate incomplete, unknown or questionable pathways. Known or potential organellar localization is shown for pathways associated with the food vacuole, mitochondrion and apicoplast. Small white squares indicate TCA (tricarboxylic acid) cycle metabolites that may be derived from outside the mitochondrion. Fuschia block arrows indicate the steps inhibited by antimalarials; grey block arrows highlight potential drug targets. Transporters are grouped by substrate specificity: inorganic cations (green), inorganic anions (magenta), organic nutrients (yellow), drug efflux and other (black). Arrows indicate direction of transport for substrates (and coupling ions, where appropriate). Numbers in parentheses indicate the presence of multiple transporter genes with similar substrate predictions. Membrane transporters of unknown or putative subcellular localization are shown in a generic membrane (blue bar). Abbreviations: ACP, acyl carrier protein; ALA, aminolevulinic acid; CoA, coenzyme A; DHF, dihydrofolate; DOXP, deoxyxylulose phosphate; FPIX2+ and FPIX3+, ferro- and ferriprotoporphyrin IX, respectively; pABA, para-aminobenzoic acid; PEP, phosphoenolpyruvate; Pi, phosphate; PPi, pyrophosphate; PRPP, phosphoribosyl pyrophosphate; THF, tetrahydrofolate; UQ, ubiquinone.
A gene encoding fructose bisphosphatase could not be detected, suggesting that gluconeogenesis is absent, as are enzymes for synthesis of trehalose, glycogen or other carbohydrate stores. Candidate genes for all but one enzyme of the conventional pentosephosphate pathway were found. These include a bifunctional glucose-6-phosphate dehydrogenase/6-phosphogluconate dehydrogenase required to generate NADPH and ribose 5-phosphate for other biosynthetic pathways72,73. Transaldolase appears to be absent, but erythrose 4-phosphate required for the chorismate pathway could probably be generated from the glycolytic intermediates fructose 6-phosphate and glyceraldehyde 3-phosphate via a putative transketolase (Fig. 4).
The genes necessary for a complete TCA cycle, including a complete pyruvate dehydrogenase complex, were identified. However, it remains unclear whether the TCA cycle is used for the full oxidation of products of glycolysis, or whether it is used to supply intermediates for other biosynthetic pathways. The pyruvate dehydrogenase complex seems to be localized in the apicoplast, and the only protein with significant similarity to aconitases has been reported to be a cytosolic iron-response element binding protein that did not possess aconitase activity74. Also, malate dehydrogenase appears to be cytosolic rather than mitochondrial, even though it seems to have originated from the mitochondrial genome75. Genes encoding malate-quinone oxidoreductase and type I fumarate dehydratase are present. Malate-quinone oxidoreductase, which is probably targeted to the mitochondrion, may well replace malate dehydrogenase in the TCA cycle, as it does in Helicobacter pylori. A gene encoding phosphoenolpyruvate carboxylase (PEPC) was also found. Like bacteria and plants, P. falciparum may cope with a drain of TCA cycle intermediates by using phosphoenolpyruvate (PEP) to replenish oxaloacetate (Fig. 4). This would seem to be supported by reports of CO2-incorporating activity in asexual stage parasite cultures76. Thus, the TCA cycle appears to be unconventional in erythrocytic stages, and may serve mainly to synthesize succinyl-CoA, which in turn can be used in the haem biosynthesis pathway.
Genes encoding all subunits of the catalytic F1 portion of ATP synthase, the protein that confers oligomycin sensitivity, and the gene that encodes the proteolipid subunit c for the F0 portion of ATP synthase, were detected in the parasite genome. The F0 a and b subunits could not be detected, raising the question as to whether the ATP synthase is functional. Because parts of the genome sequence are incomplete, the presence of the a and b subunits could not be ruled out. Erythrocytic parasites derive ATP through glycolysis and the mitochondrial contribution to the ATP pool in these stages appears to be minimal77,78. It is possible that the ATP synthase functions in the insect or sexual stages of the parasite. However, in the absence of the F0 a and b subunits, an ATP synthase cannot use the proton gradient79.
A functional mitochondrion requires the generation of an electrochemical gradient across the inner membrane. But the P. falciparum genome seems to lack genes encoding components of a conventional NADH dehydrogenase complex I. Instead, a single subunit NADH dehydrogenase gene specifies an enzyme that can accomplish ubiquinone reduction without proton pumping, thus constituting a non-electrogenic step. Other dehydrogenases targeted to the mitochondrion also serve to reduce ubiquinone in P. falciparum, including dihydroorotate dehydrogenase, a critical enzyme in the essential pyrimidine biosynthesis pathway80. The parasite genome contains some genes specifying ubiquinone synthesis enzymes, in agreement with recent metabolic labelling studies81. Re-oxidation of ubiquinol is carried out by the cytochrome bc1 complex that transfers electrons to cytochrome c, and is accompanied by proton translocation82. Apocytochrome b of this complex is encoded by the mitochondrial genome21,22, but the rest of the components are encoded by nuclear genes. Ubiquinol cycling is a critical step in mitochondrial physiology, and its selective inhibition by hydroxynaphthoquinones is the basis for their antimalarial action83. The final step in electron transport is carried out by the proton-pumping cytochrome c oxidase complex, of which only two subunits are encoded in the mitochondrial DNA (mtDNA). In most eukaryotes, subunit II of cytochrome c oxidase is encoded by a gene on the mitochondrial genome. In P. falciparum, however, the coxII gene is divided such that the N-terminal portion is encoded on chromosome 13 and the C-terminal portion on chromosome 14. A similar division of the coxII gene is also seen in the unicellular alga, Chlamydomonas reinhardtii84. An alternative oxidase that transfers electrons directly from ubiquinol to oxygen has been seen in plants as well in many protists, and an earlier biochemical study suggested its presence in P. falciparum85. The genome sequence, however, fails to reveal such an oxidase gene.
Biochemical, genetic and chemotherapeutic data suggest that malaria and other apicomplexan parasites synthesize chorismate from erythrose 4-phosphate and phosphoenolpyruvate via the shikimate pathway86-89. It was initially suggested that the pathway was located in the apicoplast88, but chorismate synthase is phylogenetically unrelated to plastid isoforms90 and has subsequently been localized to the cytosol91. The genes for the preceding enzymes in the pathway could not be identified with certainty, but a BLASTP search with the S. cerevisiae arom polypeptide92, which catalyses 5 of the preceding steps, identified a protein with a low level of similarity (E value 7.9 × 10−8).
In many organisms, chorismate is the pivotal precursor to several pathways, including the biosynthesis of aromatic amino acids and ubiquinone. We found no evidence, on the basis of similarity searches, for a role of chorismate in the synthesis of tryptophan, tyrosine or phenylalanine, although para-aminobenzoate (pABA) synthase does have a high degree of similarity to anthranilate (2-amino benzoate) synthase, the enzyme catalysing the first step in tryptophan synthesis from chorismate. In accordance with the supposition that the malaria parasite obtains all of its amino acids either by salvage from the host or by globin digestion, we found no enzymes required for the synthesis of other amino acids with the exception of enzymes required for glycine–serine, cysteine–alanine, aspartate–asparagine, proline–ornithine and glutamine–glutamate interconversions. In addition to pABA synthase, all but one of the enzymes (dihydroneopterin aldolase) required for de novo synthesis of folate from GTP were identified.
Several studies have shown that the erythrocytic stages of P. falciparum are incapable of de novo purine synthesis (reviewed in ref. 80). This statement can now be extended to all life-cycle stages, as only adenylsuccinate lyase, one of the 10 enzymes required to make inosine monophosphate (IMP) from phosphoribosyl pyrophosphate, was identified. This enzyme also plays a role in purine salvage by converting IMP to AMP. Purine transporters and enzymes for the interconversion of purine bases and nucleosides are also present. The parasite can synthesize pyrimidines de novo from glutamine, bicarbonate and aspartate, and the genes for each step are present. Deoxyribonucleotides are formed via an aerobic ribonucleoside diphosphate reductase93,94, which is linked via thioredoxin to thioredoxin reductase. Gene knockout experiments have recently shown that thioredoxin reductase is essential for parasite survival95.
The intraerythrocytic stages of the malaria parasite uses haemoglobin from the erythrocyte cytoplasm as a food source, hydrolysing globin to small peptides, and releasing haem that is detoxified in the form of haemazoin. Although large amounts of haem are toxic to the parasite, de novo haem biosynthesis has been reported96 and presumably provides a mechanism by which the parasite can segregate host-derived haem from haem required for synthesis of its own iron-containing proteins. However, it has been unclear whether de novo synthesis occurs using imported host enzymes97 or parasite-derived enzymes. Genes encoding the first two enzymes in the haem biosynthetic pathway, aminolevulinate synthase98 and aminolevulinate dehydratase99, were cloned previously, and genes encoding every other enzyme in the pathway except for uroporphyrinogen-III synthase were found (Fig. 4).
Haem and iron–sulphur clusters form redox prosthetic groups for a wide range of proteins, many of which are localized to the mitochondrion and apicoplast. The parasite genome appears to encode enzymes required for the synthesis of these molecules. There are two putative cysteine desulphurase genes, one which also has homology to selenocysteine lyase and may be targeted to the mitochondrion, and the second which may be targeted to the apicoplast, suggesting organelle specific generation of elemental sulphur to be used in Fe–S cluster proteins. The subcellular localization of the enzymes involved in haem synthesis is uncertain. Ferrochelatase and two haem lyases are likely to be localized in the mitochondrion.
The role of the apicoplast in type II fatty-acid biosynthesis was described previously5,47. The genes encoding all enzymes in the pathway have now been elucidated, except for a thioesterase required for chain termination. No evidence was found for the associative (type I) pathway for fatty-acid biosynthesis common to most eukaryotes. The apicoplast also houses the machinery for mevalonate-independent isoprenoid synthesis. Because it is not present in mammals, the biosynthesis of isopentyl diphosphate from pyruvate and glyceraldehyde-3-phosphate provides several attractive targets for chemotherapy. Three enzymes in the pathway have been identified, including 1-deoxy-d-xylulose-5-phosphate synthase, 1-deoxy-d-xylulose-5-phosphate reductoisomerase49, and 2C-methyl-d-erythritol 2,4-cyclodiphosphate synthase100,101. One predicted protein was similar to the fourth enzyme, 2C-methyl-d-erythritol-4-phosphate cytidyltransferase (BLASTP E value 9.6 × 10−15).
Transport
On the basis of genome analysis, P. falciparum possesses a very limited repertoire of membrane transporters, particularly for uptake of organic nutrients, compared to other sequenced eukaryotes (Fig. 5). For instance, there are only six P. falciparum members of the major facilitator superfamily (MFS) and one member of the amino acid/polyamine/choline APC family, less than 10% of the numbers seen in S. cerevisiae, S. pombe or Caenorhabditis elegans (Fig. 5). The apparent lack of solute transporters in P. falciparum correlates with the lower percentage of multispanning membrane proteins compared with other eukaryotic organisms (Fig. 5). The predicted transport capabilities of P. falciparum resemble those of obligate intracellular prokaryotic parasites, which also possess a limited complement of transporters for organic solutes102.
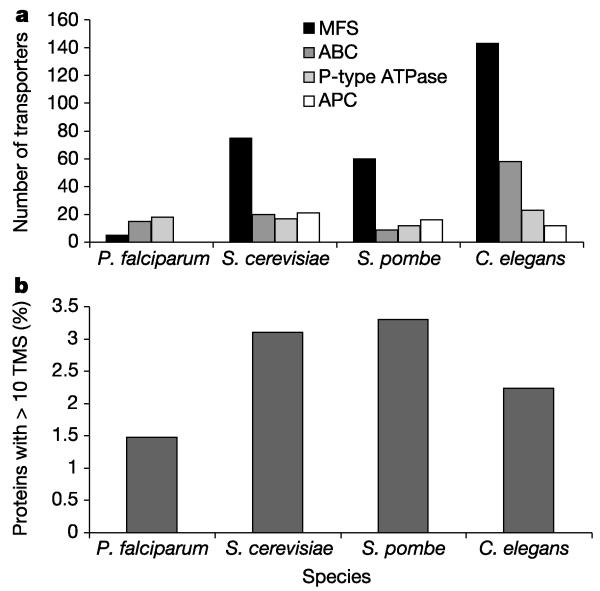
Analysis of transporters in P. falciparum. a, Comparison of the numbers of transporters belonging to the major facilitator superfamily (MFS), ATP-binding cassette (ABC) family, P-type ATPase family and the amino acid/polyamine/choline (APC) family in P. falciparum and other eukaryotes. Analyses were performed as previously described102. b, Comparison of the numbers of proteins with ten or more predicted transmembrane segments163 (TMS) in P. falciparum and other eukaryotes. Prediction of membrane spanning segments was performed using TMHMM.
A complete catalogue of the identified transporters is presented in Fig. 4. In addition to the glucose/proton symporter103 and the water/glycerol channel104, one other probable sugar transporter and three carboxylate transporters were identified; one or more of the latter are probably responsible for the lactate and pyruvate/proton symport activity of P. falciparum105. Two nucleoside/nucleobase transporters are encoded on the P. falciparum genome, one of which has been localized to the parasite plasma membrane106. No obvious amino-acid transporters were detected, which emphasizes the importance of haemoglobin digestion within the food vacuole as an important source of amino acids for the erythrocytic stages of the parasite. How the insect stages of the parasite acquire amino acids and other important nutrients is unknown, but four metabolic uptake systems were identified whose substrate specificity could not be predicted with confidence. The parasite may also possess novel proteins that mediate these activities. Nine members of the mitochondrial carrier family are present in P. falciparum, including an ATP/ADP exchanger107 and a di/tri-carboxylate exchanger, probably involved in transport of TCA cycle intermediates across the mitochondrial membrane. Probable phosphoenolpyruvate/phosphate and sugar phosphate/phosphate antiporters most similar to those of plant chloroplasts were identified, suggesting that these transporters are targeted to the apicoplast membrane. The former may enable uptake of phosphoenolpyruvate as a precursor of fatty-acid biosynthesis.
A more extensive set of transporters could be identified for the transport of inorganic ions and for export of drugs and hydrophobic compounds. Sodium/proton and calcium/proton exchangers were identified, as well as other metal cation transporters, including a substantial set of 16 P-type ATPases. An Nramp divalent cation transporter was identified which may be specific for manganese or iron. Plasmodium falciparum contains all subunits of V-type ATPases as well as two proton translocating pyrophosphatases108, which could be used to generate a proton motive force, possibly across the parasite plasma membrane as well as across a vacuolar membrane. The proton pumping pyrophosphatases are not present in mammals, and could form attractive antimalarial targets. Only a single copy of the P. falciparum chloroquineresistance gene crt is present, but multiple homologues of the multidrug resistance pump mdr1 and other predicted multidrug transporters were identified (Fig. 3). Mutations in crt seem to have a central role in the development of chloroquine resistance109.
Plasmodium falciparum infection of erythrocytes causes a variety of pleiotropic changes in host membrane transport. Patch clamp analysis has described a novel broad-specificity channel activated or inserted in the red blood cell membrane by P. falciparum infection that allows uptake of various nutrients110. If this channel is encoded by the parasite, it is not obvious from genome analysis, because no clear homologues of eukaryotic sodium, potassium or chloride ion channels could be identified. This suggests that P. falciparum may use one or more novel membrane channels for this activity.
DNA replication, repair and recombination
DNA repair processes are involved in maintenance of genomic integrity in response to DNA damaging agents such as irradiation, chemicals and oxygen radicals, as well as errors in DNA metabolism such as misincorporation during DNA replication. The P. falciparum genome encodes at least some components of the major DNA repair processes that have been found in other eukaryotes111,112. The core of eukaryotic nucleotide excision repair is present (XPB/Rad25, XPG/Rad2, XPF/Rad1, XPD/Rad3, ERCC1) although some highly conserved proteins with more accessory roles could not be found (for example, XPA/Rad4, XPC). The same is true for homologous recombinational repair with core proteins such as MRE11, DMC1, Rad50 and Rad51 present but accessory proteins such as NBS1 and XRS2 not yet found. These accessory proteins tend to be poorly conserved and have not been found outside of animals or yeast, respectively, and thus may be either absent or difficult to identify in P. falciparum. However, it is interesting that Archaea possess many of the core proteins but not the accessory proteins for these repair processes, suggesting that many of the accessory eukaryotic repair proteins evolved after P. falciparum diverged from other eukaryotes.
The presence of MutL and MutS homologues including possible orthologues of MSH2, MSH6, MLH1 and PMS1 suggests that P. falciparum can perform post-replication mismatch repair. Orthologues of MSH4 and MSH5, which are involved in meiotic crossing over in other eukaryotes, are apparently absent in P. falciparum. The repair of at least some damaged bases may be performed by the combined action of the four base excision repair glycosylase homologues and one of the apurinic/apyrimidinic (AP) endonucleases (homologues of Xth and Nfo are present). Experimental evidence suggests that this is done by the long-patch pathway113.
The presence of a class II photolyase homologue is intriguing, because it is not clear whether P. falciparum is exposed to significant amounts of ultraviolet irradiation during its life cycle. It is possible that this protein functions as a blue-light receptor instead of a photolyase, as do members of this gene family in some organisms such as humans. Perhaps most interesting is the apparent absence of homologues of any of the genes encoding enzymes known to be involved in non-homologous end joining (NHEJ) in eukaryotes (for example, Ku70, Ku86, Ligase IVand XRCC1)112. NHEJ is involved in the repair of double strand breaks induced by irradiation and chemicals in other eukaryotes (such as yeast and humans), and is also involved in a few cellular processes that create double strand breaks (for example, VDJ recombination in the immune system in humans). The role of NHEJ in repairing radiation-induced double strand breaks varies between species114. For example, in humans, cells with defects in NHEJ are highly sensitive to γ-irradiation while yeast mutants are not. Double strand breaks in yeast are repaired primarily by homologous recombination. As NHEJ is involved in regulating telomere stability in other organisms, its apparent absence in P. falciparum may explain some of the unusual properties of the telomeres in this species115.
Secretory pathway
Plasmodium falciparum contains genes encoding proteins that are important in protein transport in other eukaryotic organisms, but the organelles associated with a classical secretory pathway and protein transport are difficult to discern at an ultra-structural level116. In order to identify additional proteins that may have a role in protein translocation and secretion, the P. falciparum protein database was searched with S. cerevisiae proteins with GO assignments for involvement in protein export. We identified potential homologues of important components of the signal recognition particle, the translocon, the signal peptidase complex and many components that allow vesicle assembly, docking and fusion, such as COPI and COPII, clathrin, adaptin, v- and t-SNARE and GTP binding proteins. The presence of Sec62 and Sec63 orthologues raises the possibility of post-translational translocation of proteins, as found in S. cerevisiae.
Although P. falciparum contains many of the components associated with a classical secretory system and vesicular transport of proteins, the parasite secretory pathway has unusual features. The parasite develops within a parasitophorous vacuole that is formed during the invasion of the host cell, and the parasite modifies the host erythrocyte by the export of parasite-encoded proteins117. The mechanism(s) by which these proteins, some of which lack signal peptide sequences, are transported through and targeted beyond the membrane of the parasitophorous vacuole remains unknown. But these mechanisms are of particular importance because many of the proteins that contribute to the development of severe disease are exported to the cytoplasm and plasma membrane of infected erythrocytes.
Attempts to resolve these observations resulted in the proposal of a secondary secretory pathway118. More recent studies suggest export of COPII vesicle coat proteins, Sar1 and Sec31, to the erythrocyte cytoplasm as a mechanism of inducing vesicle formation in the host cell, thereby targeting parasite proteins beyond the parasitophorous vacuole, a new model in cell biology119,120. A homologue of N-ethylmaleimide-sensitive factor (NSF), a component of vesicular transport, has also been located to the erythrocyte cytoplasm121. The 41-2 antigen of P. falciparum, which is also found in the erythrocyte cytoplasm and plasma membrane122, is homologous with BET3, a subunit of the S. cerevisiae transport protein particle (TRAPP) that mediates endoplasmic reticulum to Golgi vesicle docking and fusion123. It is not clear how these proteins are targeted to the cytoplasm, as they lack an obvious signal peptide. Nevertheless, the expanded list of protein-transport-associated genes identified in the P. falciparum genome should facilitate the development of specific probes to further elucidate the intra- and extracellular compartments of its protein transport system.
Immune evasion
In common with other organisms, highly variable gene families are clustered towards the telomeres. Plasmodium falciparum contains three such families termed var, rif and stevor, which code for proteins known as P. falciparum erythrocyte membrane protein 1 (PfEMP1), repetitive interspersed family (rifin) and sub-telomeric variable open reading frame (stevor), respectively5,124-130. The 3D7 genome contains 59 var, 149 rif and 28 stevor genes, but for each family there are also a number of pseudogenes and gene truncations present.
The var genes code for proteins which are exported to the surface of infected red blood cells where they mediate adherence to host endothelial receptors131, resulting in the sequestration of infected cells in a variety of organs. These and other adherence properties132-135 are important virulence factors that contribute to the development of severe disease. Rifins, products of the rif genes, are also expressed on the surface of infected red cells and undergo antigenic variation131. Proteins encoded by stevor genes show sequence similarity to rifins, but they are less polymorphic than the rifins129. The function of rifins and stevors is unknown. PfEMP1 proteins are targets of the host protective antibody response136, but transcriptional switching between var genes permits antigenic variation and a means of immune evasion, facilitating chronic infection and transmission. Products of the var gene family are thus central to the pathogenesis of malaria and to the induction of protective immunity.
Figure 6 shows the genome-wide arrangement of these multigene families. In the 24 chromosomal ends that have a var gene as the first transcriptional unit, there are three basic types of gene arrangement. Eight have the general pattern var-rif var +/− (rif/stevor)n, ten can be described as var-(rif/stevor)n, three have a var gene alone and two have two or more adjacent var genes. This telomeric organization is consistent with exchange between chromosome ends, although the extent of this re-assortment may be limited by the varied gene combinations. The var, rif and stevor genes consist of two exons. The first var exon is between 3.5 and 9.0 kb in length, polymorphic and encodes an extracellular region of the protein. The second exon is between 1.0 and 1.5 kb, and encodes a conserved cytoplasmic tail that contains acidic amino-acid residues (ATS; ‘acidic terminal sequence’). The first rif and stevor exons are about 50–75 bp in length, and encode a putative signal sequence while the second exon is about 1 kb in length, with the rif exon being on average slightly larger than that for stevor. The rifin sequences fall into two major subgroups determined by the presence or absence of a consensus peptide sequence, KEL (X15) IPTCVCR, approximately 100 amino acids from the N terminus. The var genes are made up of three recognizable domains known as ‘Duffy binding like’ (DBL); ‘cysteine rich interdomain region’ (CIDR) and ‘constant2’ (C2)137-139. Alignment of sequences existing before the P. falciparum genome project had placed each of these domains into a number of sub-classes; α to ε for DBL domains, and α to γ for CIDR domains. Despite these recognizable signatures, there is a low level of sequence similarity even between domains of the same sub-type. Alignment and tree construction of the DBL domains identified here showed that a small number did not fit well into existing categories, and have been termed DBL-X. Similar analysis of all 3D7 CIDR sequences showed that with this data they were best described as CIDRa or CIDR non-α, as distinct tree branches for the other domain types were not observed. In terms of domain type and order, 16 types of var gene sequences were identified in this study.

Organization of multi-gene families in P. falciparum. a, Telomeric regions of all chromosomes showing the relative positions of members of the multi-gene families: rif (blue) stevor (yellow) and var (colour coded as indicated; see b and c). Grey boxes represent pseudogenes or gene fragments of any of these families. The left telomere is shown above the right. Scale: ~0.6 mm = 1 kb. b, c, var gene domain structure. var genes contain three domain types: DBL, of which there are six sequence classes; CIDR, of which there are two sequence classes; and conserved 2 (C2) domains (see text). The relative order of the domains in each gene is indicated (c). var genes with the same domain types in the same order have been colour coded as an identical class and given an arbitrary number for their type (b) and the total number of members of each class in the genome of P. falciparum clone 3D7. d, Internal multi-gene family clusters. Key as in a.
Type 1 var genes, consisting of DBLα, CIDRα, DBLδ, and CIDR non-α followed by the ATS, are the most common structures, with 38 genes in this category (Fig. 6b). A total of 58 var genes commence with a DBLa domain, and in 51 cases this is followed by CIDRα, and in 46 var genes the last domain of the first exon is CIDR non-α. Four var genes are atypical with the first exon consisting solely of DBL domains (type 3 and type 13). There is non-randomness in the ordering and pairing of DBL and CIDR sub-domains140, suggesting that some—for example, DBLδ–CIDR non-α and DBLβ–C2 (Table 3)—should either be considered as functional–structural combinations, or that recombination in these areas is not favoured, thereby preserving the arrangement. Eighteen of the 24 telomeric proximal var genes are of type 1. With two exceptions, type 4 on chromosome 7 and type 9 on chromosome 11, all of the telomeric var genes are transcribed towards the centromere. The inverted position of the two var genes may hinder homologous recombination at these loci in telomeric clusters that are formed during asexual multiplication115. A further 12 var genes are located near to telomeres, with the remaining var genes forming internal clusters on chromosomes 4, 7, 8 and 12 and a single internal gene being located on chromosome 6.
Table 3
| Domain type | Number of domains |
|---|---|
| DBLα | 58 |
| DBLβ-C2 | 18 |
| DBLγ | 13 |
| DBLδ | 44 |
| DBUε | 13 |
| DBL-X | 13 |
| CIDRα | 51 |
| CIDR non-α | 54 |
|
| |
| Preferred pairings | Frequency |
|
| |
| DBLα–CIDRα | 51/58 |
| DBLβ–C2 | 18/18 |
| DBLγ–CIDR non-α | 44/44 |
| CIDRα–DBLδ | 39/51 |
| CIDRα–DBLβ | 10/51 |
| DBILβ–C2–DBLγ | 10/18 |
| DBLγ–DBL-X | 8/13 |
Top, the total number of each DBL orCIDR domain type in intact var genes within the P. falciparum 3D7 genome. Bottom, the frequencies of the most common individual domain pairings found within intact var genes. The denominator refers to the total number of the first-named domains in intact var genes, and the numerator refers to the number of second-named domains found adjacent. See text for discussion of domain types.
Alignment of sequences 1.5 kb upstream of all of the var genes revealed three classes of sequences, upsA, upsB and upsC (of which there are 11, 35 and 13 members, respectively) that show preferential association with different var genes. Thus, upsB is associated with 22 out of 24 telomeric var genes, upsA is found with the two remaining telomeric var genes that are transcribed towards the telomere and with most telomere associated var genes (9 out of 12) which also point towards the telomere141. All 13 upsC sequences are associated with internal var clusters. Nearly all the telomeric var genes have an (A + T)-rich region approximately 2 kb upstream characterized by a number of poly(A) tracts as well as one or more copies of the consensus GGATCTAG. An analysis of the regions 1.0 kb downstream of var genes shows three sequence families, with members of one family being associated primarily with var genes next to the telomeric repeats. The intron sequences within the var genes have been associated with locus specific silencing142. They vary in length from 170 to ~1,200 bp and are ~89% A/T. On the coding strand, at the 5′ end the non-A/T bases are mainly G residues with 70% of sequences having the consensus TGTTTGGATATATA. The central regions are highly A-rich, and contain a number of semiconserved motifs. The 3′ region is comparably rich in C, with one or more copies in most genes of the sequence (TA)n CCCATAACTACA. The 3′ end has an extended and atypical splice consensus of ACANATATAGTTA(T)n TAG. Sequences upstream of rif and stevor genes also have distinguishable upstream sequences, but a proportion of rif genes have the stevor type of 5′ sequence. Because the majority of telomeric var genes share a similar structure and 5′ and 3′ sequences, they may form a unique group in terms of regulation of gene expression.
The most conserved var gene previously identified, which mediates adherence to chondroitin sulphate A in the placenta143, is incomplete in 3D7 because of deletion of part of exon 1 and all of exon 2. This gene is located on the right telomere of chromosome 5 (Fig 6). The majority of var genes sequenced previously had been identified as they mediated adhesion to particular receptors, and most of them had more than four domains in exon 1. The fact that type 1 var genes containing only 4 domains predominate in the 3D7 genome suggests that previous analyses had been based on a highly biased sample. The significance of this in terms of the function of type 1 var genes remains to be determined.
Immune-evasion mechanisms such as clonal antigenic variation of parasite-derived red cell surface proteins (PfEMP1s, rifins) and modulation of dendritic cell function have been documented in P. falciparum131,132. A putative homologue of human cytokine macrophage migration inhibitory factor (MIF) was identified in P. falciparum. In vertebrates, MIFs have been shown to function as immunomodulators and as growth factors144, and in the nematode Brugia malayi, recombinant MIF modulated macrophage migration and promoted parasite survival145. An MIF-type protein in P. falciparum may contribute to the parasite’s ability to modulate the immune response by molecular mimicry or participate in other host–parasite interactions.
Implications for vaccine development
An effective malaria vaccine must induce protective immune responses equivalent to, or better than, those provided by naturally acquired immunity or immunization with attenuated sporozoites146. To date, about 30 P. falciparum antigens that were identified via conventional techniques are being evaluated for use in vaccines, and several have been tested in clinical trials. Partial protection with one vaccine has recently been attained in a field setting147. The present genome sequence will stimulate vaccine development by the identification of hundreds of potential antigens that could be scanned for desired properties such as surface expression or limited antigenic diversity. This could be combined with data on stage-specific expression obtained by microarray and proteomics14,15 analyses to identify potential antigens that are expressed in one or more stages of the life cycle. However, highthroughput immunological assays to identify novel candidate vaccine antigens that are the targets of protective humoral and cellular immune responses in humans need to be developed if the genome sequence is to have an impact on vaccine development. In addition, new methods for maximizing the magnitude, quality and longevity of protective immune responses will be required in order to produce effective malaria vaccines.
Concluding remarks
The P. falciparum, Anopheles gambiae and Homo sapiens genome sequences have been completed in the past two years, and represent new starting points in the centuries-long search for solutions to the malaria problem. For the first time, a wealth of information is available for all three organisms that comprise the life cycle of the malaria parasite, providing abundant opportunities for the study of each species and their complex interactions that result in disease. The rapid pace of improvements in sequencing technology and the declining costs of sequencing have made it possible to begin genome sequencing efforts for Plasmodium vivax, the second major human malaria parasite, several malaria parasites of animals, and for many related parasites such as Theileria and Toxoplasma. These will be extremely useful for comparative purposes. Last, this technology will enable sampling of parasite, vector and host genomes in the field, providing information to support the development, deployment and monitoring of malaria control methods.
In the short term, however, the genome sequences alone provide little relief to those suffering from malaria. The work reported here and elsewhere needs to be accompanied by larger efforts to develop new methods of control, including new drugs and vaccines, improved diagnostics and effective vector control techniques. Much remains to be done. Clearly, research and investments to develop and implement new control measures are needed desperately if the social and economic impacts of malaria are to be relieved. The increased attention given to malaria (and to other infectious diseases affecting tropical countries) at the highest levels of government, and the initiation of programmes such as the Global Fund to Fight AIDS, Tuberculosis and Malaria148, the Multilateral Initiative on Malaria in Africa149, the Medicines for Malaria Venture150, and the Roll Back Malaria campaign151, provide some hope of progress in this area. It is our hope and expectation that researchers around the globe will use the information and biological insights provided by complete genome sequences to accelerate the search for solutions to diseases affecting the most vulnerable of the world’s population.
Methods
Sequencing, gap closure and annotation
The techniques used at each of the three participating centres for sequencing, closure and annotation are described in the accompanying Letters7-9. To ensure that each centres’ annotation procedures produced roughly equivalent results, the Wellcome Trust Sanger Institute (‘Sanger’) and the Institute for Genomic Research (‘TIGR’) annotated the same 100-kb segment of chromosome 14. The number of genes predicted in this sequence by the two centres was 22 and 23; the discrepancy being due to the merging of two single genes by one centre. Of the 74 exons predicted by the two centres, 50 (68%) were identical, 9 (2%) overlapped, 6 (8%) overlapped and shared one boundary, and the remainder were predicted by one centre but not the other. Thus 88% of the exons predicted by the two centres in the 100-kb fragment were identical or overlapped.
Finished sequence data and annotation were transferred in XML (extensible markup language) format from Sanger and the Stanford Genome Technology Center to TIGR, and made available to co-authors over the internet. Genes on finished chromosomes were assigned systematic names according the scheme described previously5. Genes on unfinished chromosomes were given temporary identifiers.
Analysis of subtelomeric regions
Subtelomeric regions were analysed by the alignment of all of the chromosomes to each other using MUMmer2152 with a minimum exact match length ranging from 30 to 50 bp. Tandem repeats were identified by extracting a 90-kb region from the ends of all chromosomes and using Tandem Repeat Finder153 with the following parameter settings: match = 2, mismatch = 7, indel = 7, pm = 75, pi = 10, minscore = 100, maxperiod = 500. Detailed pairwise alignments of internal telomeric blocks were computed with the ssearch program from the Fasta3 package154.
Evolutionary analyses
Plasmodium falciparum proteins were searched against a database of proteins from all complete genomes as well as from a set of organelle, plasmid and viral genomes. Putative recently duplicated genes were identified as those encoding proteins with better BLASTP matches (based on E value with a 10−15 cutoff) to other proteins in P. falciparum than to proteins in any other species. Proteins of possible organellar descent were identified as those for which one of the top six prokaryotic matches (based on E value) was to either a protein encoded by an organelle genome or by a species related to the organelle ancestors (members of the Rickettsia subgroup of the α-Proteobacteria or cyanobacteria). Because BLAST matches are not an ideal method of inferring evolutionary history, phylogenetic analysis was conducted for all these proteins. For phylogenetic analysis, all homologues of each protein were identified by BLASTP searches of complete genomes and of a nonredundant protein database. Sequences were aligned using CLUSTALW, and phylogenetic trees were inferred using the neighbour-joining algorithms of CLUSTALW and PHYLIP. For comparative analysis of eukaryotes, the proteomes of all eukaryotes for which complete genomes are available (except the highly reduced E. cuniculi) were searched against each other. The proportion of proteins in each eukaryotic species that had a BLASTP match in each of the other eukaryotic species was determined, and used to infer a ‘whole-genome tree’ using the neighbour-joining algorithm. Possible eukaryotic conserved and specific proteins were identified as those with matches to all the complete eukaryotic genomes (10−30 E-value cutoff) but without matches to any complete prokaryotic genome (10−15 cutoff).
Supplementary Material
Legends for figures and tables
Supplementary table A
Supplementary table B
Supplementary table C
Supplementary figure A
Supplementary figure B
Supplementary figure C
Supplementary figure D
Supplementary figure E
Supplementary figure F
Supplementary figure G
Supplementary figure H
Supplementary figure I
Supplementary figure J
Supplementary figure K
Supplementary figure L
Supplementary figure M
Supplementary figure N
Acknowledgements
We thank our colleagues at The Wellcome Trust Sanger Institute, The Institute for Genomic Research, the Stanford Genome Technology Center, and the Naval Medical Research Center for their support. We thank J. Foster for providing markers for chromosome 14; R. Huestis and K. Fischer for providing RT–PCR data for chromosomes 2 and 3 before publication; A. Waters for assistance with ribosomal RNAs; S. Cawley for assistance with phat; and M. Crawford and R. Wang for discussions. This work was supported by the Wellcome Trust, the Burroughs Wellcome Fund, the National Institute for Allergy and Infectious Diseases, the Naval Medical Research Center, and the US Army Medical Research and Materiel Command.
Footnotes
Supplementary Information accompanies the paper on Nature’s website (http://www.nature.com/nature).
Competing interests statement
The authors declare that they have no competing financial interests.
Sequences and annotation are available at the following websites: PlasmoDB (http://plasmodb.org), The Institute for Genomic Research (http://www.tigr.org), the Wellcome Trust Sanger Institute (http://www.sanger.ac.uk/Projects/Protozoa/), and the Stanford Genome Technology Center (http://www-sequence.stanford.edu/group/malaria). Chromosome sequences were submitted to EMBL or GenBank with accession numbers AL844501-AL844509 (chromosomes 1, 3-9 and 13), AE001362.2 (chromosome 2), AE014185-AE014187 (chromosomes 10, 11 and 14) and AE014188 (chromosome 12).
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nature01097
Read article for free, from open access legal sources, via Unpaywall:
https://www.nature.com/articles/nature01097.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/101890776
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/nature01097
Article citations
Plasmodium falciparum genetic diversity and multiplicity of infection among asymptomatic and symptomatic malaria-infected individuals in Uganda.
Trop Med Health, 52(1):86, 14 Nov 2024
Cited by: 0 articles | PMID: 39543779 | PMCID: PMC11562702
A unique symbiosome in an anaerobic single-celled eukaryote.
Nat Commun, 15(1):9726, 09 Nov 2024
Cited by: 0 articles | PMID: 39521804 | PMCID: PMC11550330
Systematic identification of interchromosomal interaction networks supports the existence of specialized RNA factories.
Genome Res, 34(10):1610-1623, 29 Oct 2024
Cited by: 0 articles | PMID: 39322282
A <i>Plasmodium falciparum</i> MORC protein complex modulates epigenetic control of gene expression through interaction with heterochromatin.
Elife, 12:RP92201, 16 Oct 2024
Cited by: 2 articles | PMID: 39412522 | PMCID: PMC11483127
Host 5-HT affects Plasmodium transmission in mosquitoes via modulating mosquito mitochondrial homeostasis.
PLoS Pathog, 20(10):e1012638, 15 Oct 2024
Cited by: 0 articles | PMID: 39405338 | PMCID: PMC11508672
Go to all (2,659) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (2)
- (1 citation) ENA - AL844501
- (1 citation) ENA - AL844509
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Sequence of Plasmodium falciparum chromosome 12.
Nature, 419(6906):534-537, 01 Oct 2002
Cited by: 33 articles | PMID: 12368869
Sequence of Plasmodium falciparum chromosomes 2, 10, 11 and 14.
Nature, 419(6906):531-534, 01 Oct 2002
Cited by: 104 articles | PMID: 12368868
Malaria research. Parasite genome sequenced, scrutinized.
Science, 298(5591):33-34, 01 Oct 2002
Cited by: 2 articles | PMID: 12364754
Functional genomic technologies applied to the control of the human malaria parasite, Plasmodium falciparum.
Pharmacogenomics, 2(2):137-142, 01 May 2001
Cited by: 5 articles | PMID: 11368752
Review
Funding
Funders who supported this work.
NIAID NIH HHS (1)
Grant ID: R01 AI028398
Wellcome Trust (1)
Molecular and immunological studies of proteins expressed on plasmodium falciparum infected erythrocytes.
Prof Chris Newbold, University of Oxford
Grant ID: 061524