Abstract
Free full text

The Imprinted H19 LncRNA Antagonizes Let-7 MicroRNAs
SUMMARY
Abundantly expressed in fetal tissues and adult muscle, the developmentally regulated H19 long noncoding RNA (lncRNA) has been implicated in human genetic disorders and cancer. However, how H19 acts to regulate gene function has remained enigmatic, despite the recent implication of its encoded miR675 in limiting placental growth. We noted that vertebrate H19 harbors both canonical and noncanonical binding sites for the let-7 family of microRNAs, which plays important roles in development, cancer, and metabolism. Using H19 knockdown and overexpression, combined with in vivo crosslinking and genome-wide transcriptome analysis, we demonstrate that H19 modulates let-7 availability by acting as a molecular sponge. The physiological significance of this interaction is highlighted in culture where H19 depletion causes precocious muscle differentiation, a phenotype recapitulated by let-7 overexpression. Our results reveal an unexpected mode of action of H19 and identify this lncRNA as an important regulator of the major let-7 family of microRNAs.
INTRODUCTION
The H19 gene belongs to a highly conserved imprinted gene cluster that plays important roles in embryonal development and growth control (Gabory et al., 2010). The cluster also contains the nearby reciprocally imprinted gene for insulin-like growth factor 2 (Igf2) that is coordinately regulated by an intergenic control region and a common enhancer region. In human and mouse, H19 is expressed from the maternal allele, while Igf2 is paternally expressed. H19 expression is strongly induced during embryogenesis and down-regulated after birth, except in adult skeletal muscle and heart (Gabory et al., 2010; Onyango and Feinberg, 2011). Two human genetic disorders have been linked to the H19-Igf2 locus: Silver-Russell Syndrome and Beckwith-Wiedemann Syndrome, with the latter also displaying higher susceptibility to the development of embryonal tumors (Gabory et al., 2010). Further, a role of H19 acting either as a tumor suppressor (Yoshimizu et al., 2008) or an oncogene (Matouk et al., 2007) has been suggested. However, how H19 functions to impact these various processes remains poorly understood.
The human H19 encodes a predominantly cytoplasmic ~2.3-kilobase long capped, spliced, and polyadenylated RNA that produces no known protein product (Brannan et al., 1990). In the past two decades, extensive investigations using both deletion and transgenic mouse models have yielded important insights into the functional role of H19 (Gabory et al., 2010). It has been reported that mice with targeted H19 deletion (H19Δ3) exhibit an overgrowth phenotype (an increase in postnatal growth by 8% compared to wt littermates), which was restored to wt growth by transgenic expression of H19 (Gabory et al., 2009). Importantly, overexpression of several genes of the imprinted gene network (IGN) (Varrault et al., 2006), including Igf2 in the H19Δ3 mice was concomitantly restored to the wt level by such transgenic expression. This suggests that the H19 lncRNA acts in trans to regulate the IGN and control growth in mice (Gabory et al., 2009). However, the mechanism by which H19 does so is unclear. Recently, a role of H19 in limiting placental growth through its encoded miR675 has been reported (Keniry et al., 2012). This microRNA (miRNA) is produced from full-length H19 through Drosha processing and the miRNA is detected only in the mouse placenta and at a time window where placental growth normally ceases (Keniry et al., 2012). Finally, high H19 expression has been detected in adult skeletal muscle of both human and mouse (Gabory et al., 2010; Onyango and Feinberg, 2011), but the physiological significance of this expression remains to be investigated. In the current study, we demonstrate that the H19 lncRNA acts as a molecular sponge for the major let-7 family of miRNAs, which are known to play important roles in diverse physiological and pathological processes.
RESULTS
H19 contains potential let-7 binding sites
While in the process of defining the molecular function of H19, we unexpectedly noticed increased levels of the miRNA-processing enzyme Dicer in response to ectopic H19 expression in multiple cell types. Given that Dicer is a known target of let-7 (Forman et al., 2008; Tokumaru et al., 2008) and that lncRNAs can act as “sponges” to bind specific miRNAs and regulate their function (Ebert and Sharp, 2010; Salmena et al., 2011), we suspected that H19 might bind let-7 and interfere with its function. Let-7 regulates target gene expression by binding to imperfectly complementary sequences in mRNAs, leading to translational repression and/or mRNA destabilization (Fabian and Sonenberg, 2012). Subsequent bioinformatic analysis revealed putative complementary sequences for let-7 in human H19 (Figure 1A). Among the four predicted let-7 sites in human H19, the site with starting nucleotide at position 1617 is an offset 6-mer seed site with strong compensatory base-pairing for the 3′-end of let-7 (Bartel, 2009). The other three sites are noncanonical (seedless) sites. However, the site at position 1244 has strong base-paring in the seed region despite involving a G:U pair. From conservation analysis (based on sequence alignments of 33 mammalian genomes), we found that the conservation for human H19 varies greatly from region to region. After defining a conservation score threshold of 0.57 (Siepel et al., 2005), the sites at positions 1244 and 750 are well and moderately conserved with average conservation scores of 0.75 and 0.64, respectively. The sites at positions 1617 and 2102 are poorly conserved with average conservation scores of 0.03 and 0.02, respectively (Figure 1B). The experimental results below for the site at position 1244 are consistent with previous reports that noncanonical sites can be functional (Didiano and Hobert, 2006; Lal et al., 2009; Landthaler et al., 2008; Tay et al., 2008; Vella et al., 2004). This is further supported by the observation of large numbers of noncanonical sites identified by genome-wide RIP-Chip and CLIP-seq studies (Chi et al., 2009; Hafner et al., 2010; Landthaler et al., 2008). The findings on the seed site at position 1617 suggest that there may be many more functional nonconserved seed sites than are currently known, as nonconserved seed sites outnumber conserved seed sites by about an order of magnitude (Bartel, 2009). Experiments described in Figure 2 suggest that both canonical and noncanonical sites can function in target regulation.
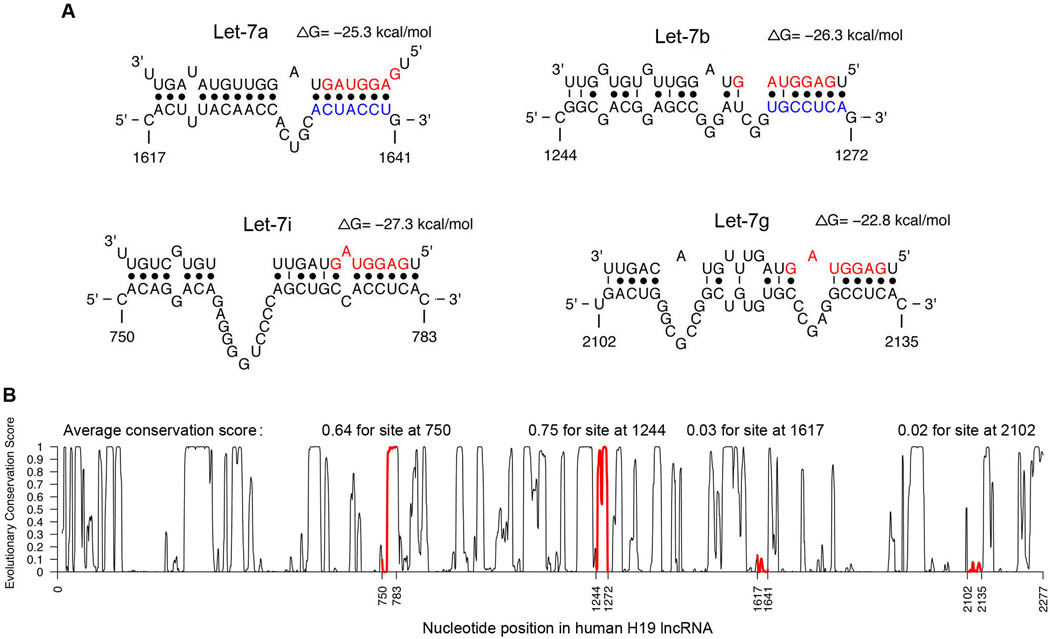
(A) Partial sequences of H19 and sequences of four let-7 subtypes are shown. Nucleotides of the miRNA seed region (position 2 to 8) are in red. Numbers are in nucleotides relative to the transcriptional start site of H19. The deleted nucleotides in psiCHECK2-H19D are marked in blue. See also Figure S2A. (B) Profile of evolutionary conservation scores for human H19 lncRNA. The portions of the profile for four let-7 binding sites are in red, with average conservation score given on top.
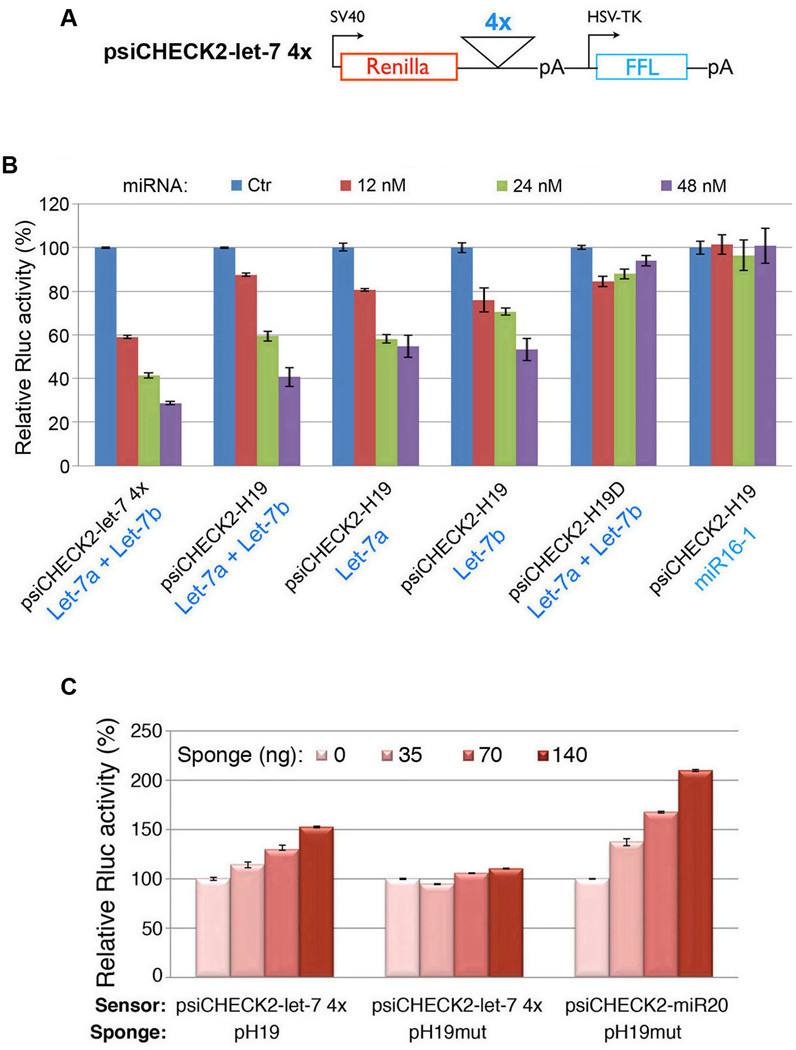
(A) Reporter constructs. (B) The indicated constructs were each transfected into HEK293 cells, together with control miRNA (Ctr), let-7 or miR16-1 at a final concentration of 12, 24, or 48 nM. In Let-7a + Let-7b, equal concentrations of let-7a and let-7b were combined to give the indicated final concentrations. Numbers are mean ± SD (n=3). See also Figure S1. (C) Let-7 sensor (psiCHECK2-let-7 4×) or miR20 sensor (psiCHECK2-miR20) were transfected into HEK293 cells, together with 0, 35, 70, or 140 ng of sponge plasmid pH19 or pH19mut. Numbers are mean ± SD (n=3). See also Figures S2B and S3A.
The let-7-binding sites are functional
Experiments using let-7 biosensors were carried out to determine the functionality of two predicted let-7 binding sites: positions 1244 and 1617. psiCHECK2-let-7 4× (Iwasaki et al., 2009) harbors four copies of let-7 binding sites (called 4× hereafter) in the 3′-UTR of a Renilla luciferase (Rluc) gene (Figure 2A). This plasmid also contains a downstream constitutively expressed firefly luciferase gene as an internal control for normalization. As expected, co-transfected let-7 inhibited Rluc expression in a dose-dependent manner (Figure 2B, 1st column from left). A dose-dependent inhibition was also observed with psiCHECK2-H19 containing a human H19 fragment at the 3′-UTR of the Rluc in place of 4× (Figure 2B, 2nd column from left). This H19 fragment was predicted to bind both let-7a and let-7b at positions 1244 and 1617, respectively (Figure 1A). The extent of inhibition with psiCHECK2-H19 was slightly lower than that with psiCHECK2-let-7 4×, likely due to fewer let-7 binding sites (2 vs. 4). In metazoans multiple sites for the same or different miRNAs are generally required for effective repression (Bartel, 2009; Filipowicz et al., 2008). When let-7a and let-7b were tested individually, dose-dependent inhibitions were evident as well (Figure 2B, 3rd and 4th columns from left). To confirm that the observed inhibition was dependent on the predicted let-7 sites, we tested psiCHECK2-H19D with deleted sequences complementary to the “seed” region of both let-7a and let-7b (Figure 1A, nucleotides marked in blue). This mutant no longer responded to let-7 inhibition in a dose-dependent manner (Figure 2B, 2nd column from right). The slight repression seen at 12 nM and 24 nM might be results of let-7 3′-end interaction with H19 sequences. When miR16-1 (not predicted to bind H19) was tested on psiCHECK2-H19, a dose-dependent inhibition was not observed (Figure 2B, 1st column from right). Together, these results suggest that positions 1244 and 1617 contain functional let-7 binding sites.
H19 acts as a molecular sponge for let-7
To determine whether H19 might act as a “sponge” to sequester let-7, we transfected psiCHECK2-let-7 4× (Sensor) into HEK293 cells, which do not express endogenous H19, but express appreciable levels of let-7 miRNAs (Supplemental Figure S1A), together with increasing amounts of pH19 (Sponge) that expresses full-length human H19. The Rluc activity increased in response to pH19 in a dose-dependent manner (Figure 2C, left column), suggesting that ectopically expressed H19 specifically sequestered endogenous let-7, thereby preventing it from inhibiting Rluc expression. To confirm whether let-7 binding sites are required for this effect, a mutant H19 (pH19mut) in which all four predicted let-7 interaction sites were mutated to sequences complementary to miR20 (Supplemental Figure S2A, B) was tested. Remarkably, this mutant no longer elicited such an effect (Figure 2C, middle column); rather, it gained an ability to derepress a miR20 sensor (right column). Consistent with absence of miR20 sites in H19, dose-dependent derepression of psiCHECK2-miR20 by pH19 was not observed (Supplemental Figure S3A). Derepression of psiCHECK2-miR20 by pH19mut was more efficient than that of psiCHECK2-let-7 4× by pH19, likely due to the fact that the miR20 sites in pH19mut are much more complementary to miR20 than the let-7 sites in pH19 to let-7. Together, these results strongly point to a role of H19 as a molecular “sponge” for let-7.
H19 associates with miRNPs
miRNAs are known to be present in the cytoplasm in the form of miRNA ribonucleoprotein complexes (miRNPs) that also contain Ago2, the core component of the RNA-induced silencing complex (RISC) (Filipowicz et al., 2008; Izaurralde, 2012). To test whether H19 associates with miRNPs, coimmunoprecipitation (co-IP) experiments using antibodies against Ago2 on extracts of differentiating human embryonal carcinoma (EC) cell line PA-1 were carried out. H19 is expressed at low levels in undifferentiated embryonic stem (ES) cells and EC cells, but its expression is strongly up-regulated during differentiation (Brannan et al., 1990; McKarney et al., 1996). H19 was preferentially enriched (~4.5-fold) in Ago2-containing miRNPs relative to control IgG immunoprecipitates (Figure 3A, top panel, left column, blue bar vs. yellow bar), whereas beta-actin mRNA, expressed at a level ~1.5-fold that of H19 (bottom panel), did not detectably associate with the miRNPs (top panel, right column). The specificity of the Ago2 antibody was confirmed by IP and immunoblotting (Figure 3B). Selective H19 enrichment in miRNPs was also detected in HEK293 cells ectopically expressing H19 (data not shown). Thus, H19 is present in Ago2-containing miRNPs, likely through association with let-7 and other miRNAs, as our bioinformatic analysis suggests that H19 contains putative binding sites for additional miRNAs (Supplemental Table S1).
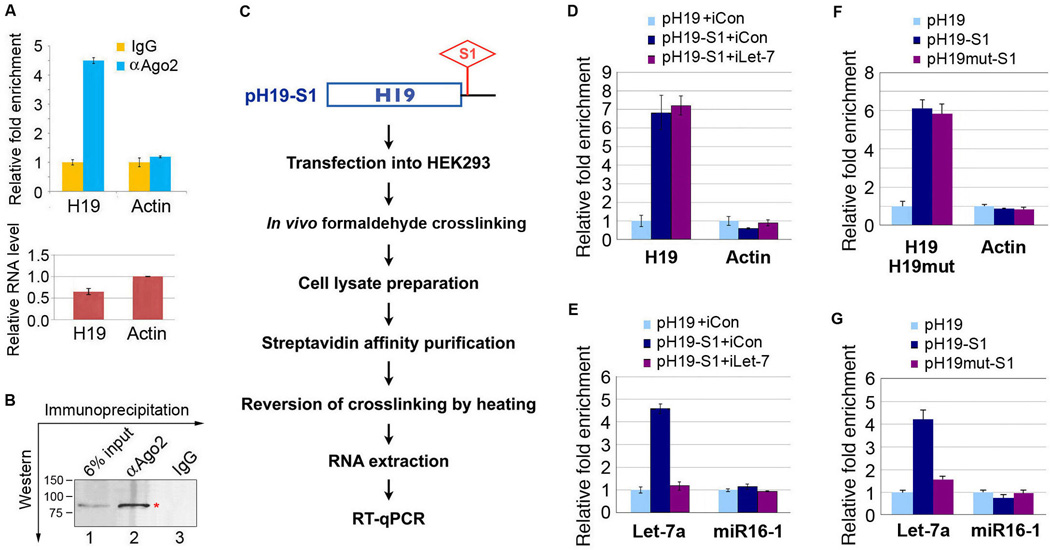
(A) Co-IP with mouse monoclonal anti-Ago2 (αAgo2) or preimmune IgG from extracts of retinoic acid (RA)-induced PA-1 cells. RNA levels in immunoprecipitates were determined by reverse transcription and quantitative real-time PCR (RT-qPCR). Top, levels of H19 and beta-actin RNA are presented as fold enrichment in αAgo2 relative to IgG immunoprecipitates. Bottom, relative RNA levels of H19 and beta-actin in RA-induced PA-1. Numbers are mean ± SD (n=3). See also Table S1. (B) Immunoprecipitation using αAgo2 (lane 2) or IgG (lane 3), followed by Western blot analysis using a rabbit monoclonal anti-Ago2. 6% input was loaded in lane 1. The Ago2 band is marked with a red asterisk. Molecular markers in kD are on the left. (C) Schematic outline of purification of H19-associated RNPs and RNA component identification. (D–G) The indicated plasmids (pH19, pH19-S1, or pH19mut) were each transfected into HEK293, with or without cotransfection of iCon or iLet-7. Cells were subjected to in vivo crosslinking, followed by affinity purification of H19-associated RNPs. RNAs extracted from RNPs were subjected to RT-qPCR. Relative abundance of indicated RNAs associated with tagged vs. untagged H19 (or H19mut) are plotted as relative fold enrichment after normalization against beta-tubulin mRNA levels (D, F) or U6B snRNA (RNU6B) levels (E, G). Numbers are mean ± SD (n=3).
To further demonstrate the physical interaction between H19 and let-7, in vivo crosslinking combined with affinity purification experiments were carried out using an S1 aptamer-tagged H19 (pH19-S1) (Figure 3C). Thus, pH19-S1 (tagged) or pH19 (untagged) were transfected into HEK293 cells, with or without cotransfection of let-7 inhibitor (iLet-7, or a control inhibitor iCon), followed by crosslinking and affinity purification of H19-associated ribonucleoprotein complexes (RNPs). RNAs were extracted from the RNPs and subjected to reverse transcription and real-time PCR (RT-qPCR) analysis. Effective affinity purification of tagged H19 RNPs was demonstrated by a ~7-fold preferential enrichment of the tagged vs. untagged H19 in the purified RNPs (Figure 3D, left column, purple and dark blue bars vs. light blue bar), while such an enrichment was not seen with beta-actin mRNA (right column). Let-7a was also preferentially enriched by ~5-fold in the tagged vs. untagged H19 fraction (Figure 3E, left column, dark blue bar vs. light blue bar). Importantly, the enrichment was abolished in the presence of iLet-7 (Figure 3E, left column, purple bar vs. dark blue bar). No enrichment of miR16-1 in tagged H19 was observed (right column). Crosslinking experiments using a tagged H19 mutant (pH19mut-S1) (Figure 3F, G) showed significantly reduced recovery of let-7 (Figure 3G, left column, purple bar vs. dark blue bar). Together, these results strongly suggest that H19 physically interacts with let-7 in vivo and that functional let-7 sites are required for promoting this association.
H19 affects expression of endogenous let-7 targets
To determine whether H19 affects the expression of endogenous let-7 targets, pH19 was transfected into HEK293 cells and the levels of expression of two known let-7 targets Dicer (Forman et al., 2008; Tokumaru et al., 2008) and Hmga2 (Lee and Dutta, 2007) were analyzed. The transfected H19 was expressed at a level ~2.5-fold that of beta-tubulin mRNA (Supplemental Figure S1B, 2nd column from left, red bar vs. blue bar). A 5.5- and 2.3-fold increase in the protein level for DICER and HMGA2, respectively, in response to H19 expression was observed 48 h post-transfection (Figure 4A), while the levels of the respective mRNAs were not changed (Figure 4B). miRNA-mediated repression generally involves translational repression followed by RNA degradation. However, examples of translational repression without RNA destabilization have also been documented (Filipowicz et al., 2008; Izaurralde, 2012). This apparent discrepancy is likely due to different cell types and/or experimental conditions used (see below), consistent with context-dependent miRNA effects (Ebert and Sharp, 2012; Mukherji et al., 2011). Together, these results suggest that ectopically expressed H19 inhibits endogenous let-7 function, leading to derepression of Dicer and Hmga2. In H19-expressing cells the levels of the seven let-7 subtypes predicted to bind H19 were not significantly changed (Figure 4C).
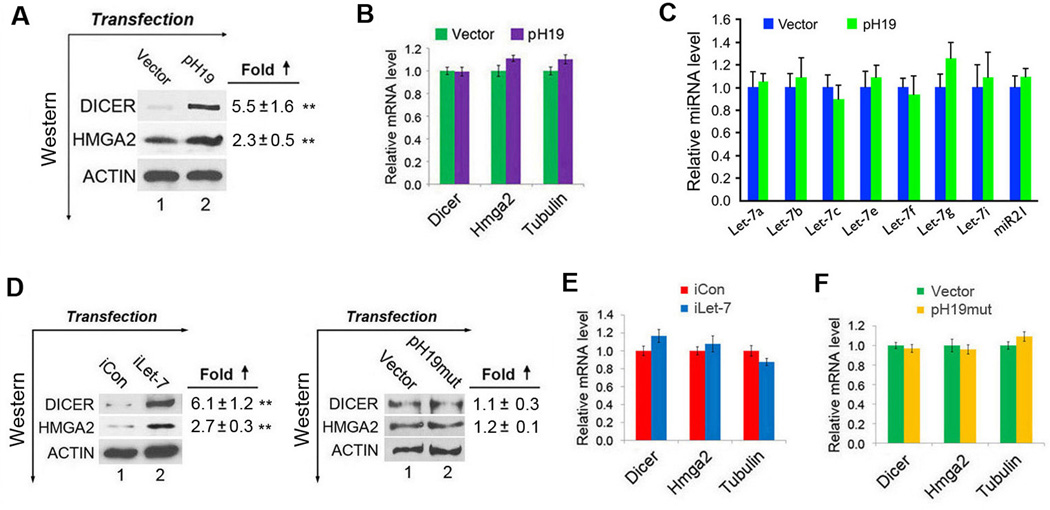
(A–C) Empty vector or pH19 were transfected into HEK293 cells. Protein and RNA were extracted 48 h later and levels determined by Western blot (A) and RT-qPCR (B, C). In A, the fold increases in the protein levels relative to vector-transfected controls after normalization against beta-actin loading controls are marked on the right. Numbers are mean ± SD (n=3). **, p < 0.01. One-sample t tests were performed to compare each data point with the controls. See also Figure S1C. In B, mRNA levels were normalized against those of beta-actin. Numbers are mean ± SD (n=3). Group t tests were performed to compare means of each mRNA between vector- and pH19-transfected cells. See also Figure S1D. In C, miRNA levels were normalized against those of U6B and presented as mean ± SD (n=3). Group t tests were performed to compare means of each miRNA between vector- and pH19-transfected cells. See also Figure S1E. (D) iCon, iLet-7, vector, or pH19mut were transfected into HEK293 cells. Protein and RNA were extracted 48 h later and levels determined by Western blot (D) and RT-qPCR (E, F). The fold increases in the protein levels relative to controls (iCon- or vector-transfected) after normalization against the beta-actin loading control are marked on the right. Numbers are mean ± SD (n=3).
To confirm that the observed effects were indeed due to impaired let-7 function, transfection experiments using iLet-7 (or iCon as a negative control) were performed. As expected, increased protein levels of both DICER and HMGA2 were observed in iLet-7 treated vs. iCon-treated HEK293 cells (Figure 4D, left panel, compare lane 2 to lane 1 in the top two blots). Further, when pH19mut in which all four let-7 binding sites were inactivated was used, derepression of Dicer and Hmga2 was not observed (Figure 4D, right panel, compare lane 2 to lane 1 in the top two blots), despite the fact that the mutant H19 was expressed at a level comparable to that of wild-type H19 (Supplemental Figure S1B, compare the left second to the last column). Again, the mRNA levels of Dicer and Hmga2 were not affected (Figure 4E, F), consistent with previous reports of translational repression without RNA destabilization (Filipowicz et al., 2008; Izaurralde, 2012) and context-dependent miRNA effects (Ebert and Sharp, 2012; Mukherji et al., 2011). These data suggest that the predicted let-7 binding sites in H19 are required for derepression of endogenous targets. Finally, derepression of DICER and HMGA2 in response to ectopic H19 expression was also observed when primary cultures of human umbilical vein endothelial cells (HUVEC) were tested (Supplemental Figure S1C–E). Taken together, these results strongly suggest that derepression of endogenous let-7 targets Dicer and Hmga2 was due to a direct “sponge” effect of H19.
Next, we asked whether down-regulating endogenous H19 would promote let-7 function. We first carried out reporter gene assays in PA-1 cells. PA-1 cells were induced to differentiate by retinoic acid (RA), followed by transfection of siRNAs specific for human H19 (siH19) or control siRNA (siCon). During differentiation, endogenous H19 typically rises to a level similar to that of beta-tubulin mRNA (Supplemental Figure S1B, 2nd column from right, red bar vs. blue bar). Approximately 48 h after the induction, the cells were transfected with reporter psiCHECK2-let-7 4× or psiCHECK-miR20 (as a negative control), together with let-7 miRNA, and luciferase activities were measured 18 h later. The levels of both endogenous let-7 and miR20 were not affected by H19 down-regulation (Supplemental Figure S1F). As shown in Figure 5A, a ~95% knockdown of H19 48 h following transfection with siH19 was observed (left column, red bar vs. blue bar), while beta-actin mRNA was not affected (right column). Importantly, H19 knockdown was accompanied by a ~50% decrease in the Rluc activity of psiCHECK2-let-7 4×, as compared to that of siCon-transfected cells (Figure 5B, column #1, red bar vs. blue bar). In contrast, no Rluc activity change was observed with psiCHECK2-miR20 (column #2, red bar vs. blue bar), suggesting that H19 knockdown enables exogenously introduced let-7 to more effectively repress psiCHECK2-let-7 4×. Next, we asked whether H19 knockdown would activate endogenous let-7, using DICER as a read-out. Thus, H19 was downregulated in PA-1 cells using siH19 (or siH19b targeted to a different region of H19). Decreased DICER expression was observed when siH19 (Figure 5C, D) (or siH19b, Supplemental Figure S3B, C) was used. Together, these results strongly point to the notion that H19 knockdown desequesters let-7, leading to heightened let-7 function.
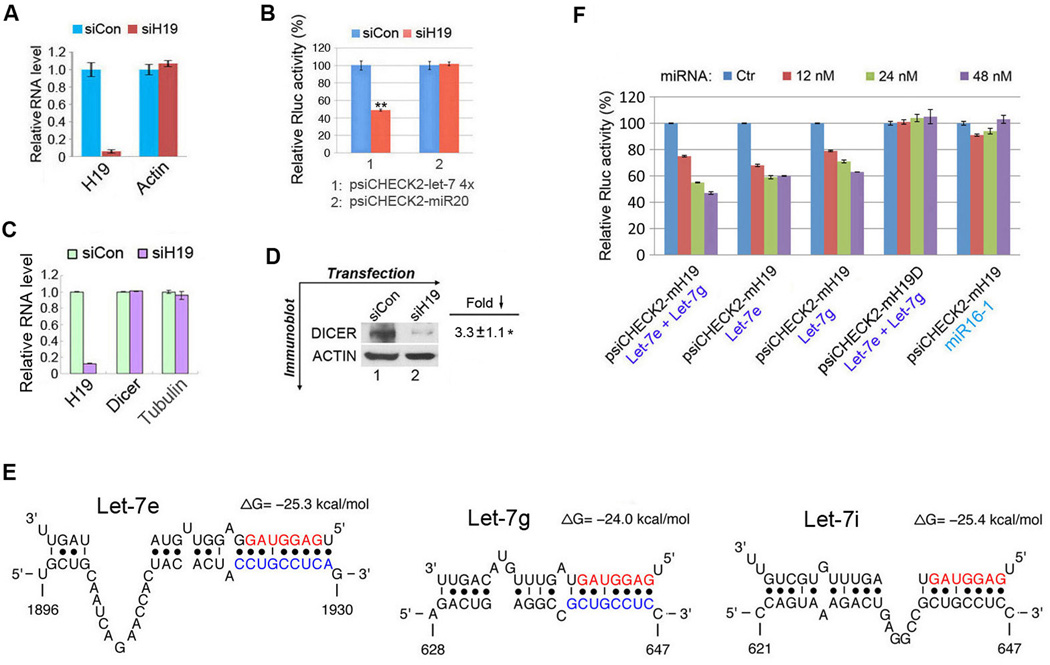
(A, B) PA-1 cells were RA-induced to express endogenous H19, followed by siRNA transfection. Cells were subjected to a second transfection 48 h later with psiCHECK2-let-7 4× (column 1) or psiCHECK2-miR20 (column 2), together with 48 nM of let-7. RNA levels (A) and luciferase activities (B) were determined 18 h later. Numbers are mean ± SD (n=3). **, p < 0.01. See also Figure S1F. (C, D) PA-1 cells were induced to express endogenous H19, followed by siRNA transfection. RNA (C) and protein (D) levels were measured 72 h later. Numbers are mean ± SD (n=3). *, p < 0.05. See also Figure S3B, C. (E) Bioinformatics predicted let-7 binding sites at three distinct positions in mouse H19. The nucleotides deleted in psiCHECK2-mH19D are marked in blue. See also Figure S4A–D. (F) Luciferase assay results. Numbers are mean ± SD (n=3).
Let-7 regulation by H19 is conserved in human and mouse
To address whether the modulation of let-7 by H19 is evolutionarily conserved, bioinformatic analyses were performed and putative let-7 binding sites in H19 were identified in other mammals including mouse (Figure 5E), rat, monkey, chimpanzee, and bovine (Supplemental Figure S4A–D). A segment from mouse H19 containing binding sites for both let-7g and let-7e was inserted into the reporter to create psiCHECK2-mH19 and tested. Let-7e and let-7g, transfected either singly or in combination, inhibited Rluc activity in a dose-dependent fashion (Figure 5F, columns 1–3 from left). When a mutant reporter (psiCHECK2-mH19D) missing the sequences complementary to the seed region of both let-7e and let-7g was tested, a dose-dependent inhibition was not observed (column 4). No dose-dependent inhibition was observed when miR16-1 was tested on psiCHECK2-mH19 (column 5). Together, these results suggest that the predicted let-7-binding sites in the mouse H19 are likely functional.
H19 depletion causes accelerated muscle differentiation
To demonstrate H19 regulation of let-7 is physiologically relevant, we turned to the mouse myogenic C2C12 cell line. Consistent with previous reports (Davis et al., 1987; Milligan et al., 2000), H19 expression was strongly induced during myoblast differentiation, beginning at day 1, peaking at day 3, and declining by day 4 of differentiation (Figure 6A, blue curve). A concomitant induction in let-7 was also observed (red curve). Since high let-7 levels are generally associated with increased differentiated states (Roush and Slack, 2008), we hypothesized that H19 might serve to inhibit let-7 activity, thereby preventing precocious differentiation. If this were the case, we would expect to see let-7 targets become downregulated as a result of H19 depletion. Thus, we transfected siRNA specific for mouse H19 (simH19) or control siRNA (siCon) into day 1 differentiating C2C12 myoblasts. RNAs were harvested 40 h later and subjected to microarray experiments. We analyzed microarray data (Supplemental Table S2) using ArraySylamer, a web-server implementation of the Sylamer algorithm that uses the hypergeometric P-values to measure significance of motif enrichment in the 3’-UTRs of ranked gene sets. Results are consistent with sponge effects by H19 on let-7 (Supplemental Figure S5A, B). Our qPCR analysis on several known let-7 targets (Forman et al., 2008; Frost and Olson, 2011; Lee and Dutta, 2007; Tokumaru et al., 2008; Zhu et al., 2011) showed decreased levels of these targets in either simH19- or let-7-transfected cells (Figure 6B, compare green or purple bars to blue bars in the second through fifth columns from left). The specificity of simH19 was confirmed by experiments using a second siRNA targeted to another region of mouse H19 (Supplemental Figure S6). It is noteworthy that in differentiating mouse myoblasts the mRNA levels of let-7 targets Dicer and Hmga2 decreased in response to increased let-7 function, a phenomenon that was not observed in proliferating HEK293 (Figure 4) or HUVEC (Supplemental Figure S1C, D) cells, consistent with context-dependent miRNA effects (Ebert and Sharp, 2012; Mukherji et al., 2011). We also noticed that let-7 overexpression led to decreased H19 expression (Figure 6B, left 1st column, compare purple bar to blue bar), suggesting possible miRNP-mediated degradation of H19.
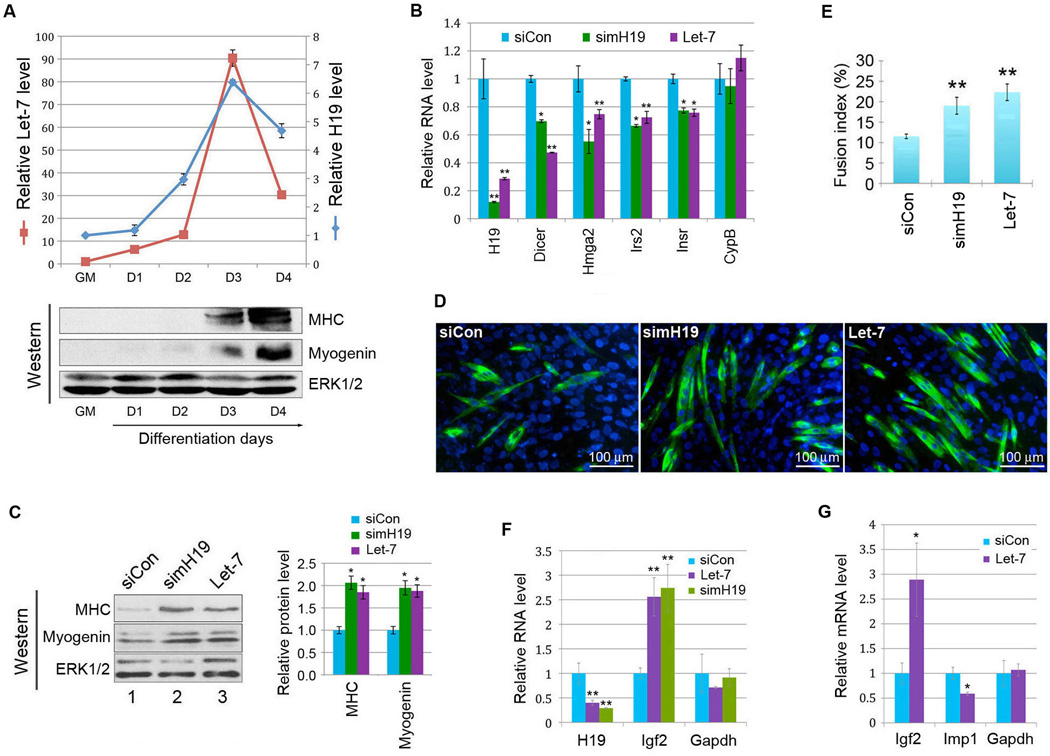
(A) C2C12 myoblasts were grown in growth medium (GM) or allowed to differentiate for 4 days. RNA and protein were harvested at the indicated time points and analyzed. Top, the expression profiles of H19 RNA and let-7a miRNA were analyzed by RT-qPCR and results are presented with levels at GM set as 1. Numbers are mean ± SD (n=3). Bottom, the expression profiles of muscle differentiation markers myosin heavy chain (MHC) and myogenin are shown by Western blot analysis. ERK1/2 were used as loading controls. (B) C2C12 myoblasts at day 1 differentiation were tranfected with siCon, simH19, or let-7. RNAs were harvested 40 h later and analyzed by RT-qPCR. Relative RNA levels are presented after normalization against beta-tubulin mRNA. Numbers are mean ± SD (n=3). *, p < 0.05; **, p < 0.01. See also Figures S5, S6, and Table S2. (C) Day 1 differentiating C2C12 myoblasts were transfected with siCon, simH19, or let-7. Proteins were harvested 3 days later and analyzed. Representative Western blot gels are shown on the left. Bar graphs on the right are quantitation of three separate Western blot experiments. Numbers are mean ± SD (n=3). *, p < 0.05. (D) MHC staining representative of two separate experiments. (E) Quantitation of myotube formation from D. Numbers are mean ± SD (n=5). **, p < 0.01. (F) RT-qPCR results of differentiating C2C12 myoblasts transfected with siCon, let-7, or simH19 as described in B. Numbers are mean ± SD (n=3). **, p < 0.01. (G) RT-qPCR results of proliferating C2C12 myoblasts transfected with siCon or let-7. Numbers are mean ± SD (n=3). *, p < 0.05.
Next, we asked whether H19 depletion would produce a phenotype that mimics let-7 overexpression. Thus, differentiating C2C12 myoblasts were transfected with siCon, simH19, or let-7, followed by the assessment of myogenic differentiation. Both H19 depletion and let-7 overexpression led to significantly increased expression of muscle differentiation markers myosin heavy chain and myogenin at day 4 of differentiation (Figure 6C, left panel, compare lanes 2 and 3 to lane 1 in the top and middle blots; and right panel, compare green and purple bars to blue bars), suggesting enhanced myogenesis. Consistent with this finding, we observed increased multinucleated myotube formation in simH19- and let-7-transfected myoblasts (Figure 6D, compare right two panels to the left). This was reflected quantitatively as an increase in the fusion index as compared with siCon-transfected myoblasts (Figure 6E). Together, these results underscore the physiological significance of the H19-let-7 interaction while uncovering a previously unexpected regulatory network involving the H19/let-7 axis in myogenic differentiation.
In search of potential downstream effectors of H19/let-7-mediated regulation during muscle differentiation, we focused our attention on insulin-like growth factors (IGF1 and IGF2), major regulators of pre- and postnatal muscle development and growth (Braun and Gautel, 2011; Chang, 2007). IGF1 and IGF2 act through the same receptor, IGF1R, to promote myogenesis (Braun and Gautel, 2011; Chang, 2007). As a potent autocrine/paracrine regulator whose expression is rapidly induced early during differentiation, IGF2 stimulates muscle differentiation both in vivo (Borensztein et al., 2013; Kalista et al., 2012; Merrick et al., 2007) and in vitro (Rosen et al., 1993; Stewart et al., 1996; Wilson et al., 2003; Yoshiko et al., 2002). Importantly, let-7 has been shown to be an indirect upstream regulator of Igf2 expression (Jung et al., 2010; Lu et al., 2007; Lu et al., 2011). Indeed, a positive in vivo correlation of expression between let-7 and Igf2 has been documented in both epithelial ovarian cancer and breast cancer (Lu et al., 2007; Lu et al., 2011). In addition, experiments using HeLa cells transfected with let-7 have demonstrated an up-regulation of Igf2 expression as a result of let-7 overexpression (Lu et al., 2011). This up-regulation is thought to be mediated by the Igf2 mRNA-binding protein IMP1, which binds specifically to the 5′-UTR of Igf2 mRNA and inhibits its expression (Jung et al., 2010; Nielsen et al., 1999). Imp1 mRNA is a validated target of let-7 inhibition (Boyerinas et al., 2008; Ioannidis et al., 2005). Based on these previous studies we hypothesized that Igf2 might be a downstream effector of let-7-mediated regulation during myogenesis. In line with this view, we observed increased Igf2 expression in differentiating myoblasts transfected with let-7 or simH19 (Figure 6F, middle column, compare purple or green bar with blue bar). To rule out myotube differentiation-dependent phenomena (as both H19 knockdown and let-7 overexpression accelerate myoblast differentiation {Figure 6C–E} which can lead to increased Igf2 expression), we performed let-7 transfection experiments in proliferating myoblasts that express low levels of H19. An increase in Igf2 expression in response to let-7 overexpression was evident as well (Figure 6G, left column, compare purple bar with blue bar), suggesting that Igf2 is a downstream effector of let-7, consistent with previous reports (Ioannidis et al., 2005; Jung et al., 2010; Lu et al., 2007; Lu et al., 2011). Importantly, let-7 overexpression led to decreased expression of Imp1 (Figure 6G, middle column, compare purple bar with blue bar), which might contribute, at least in part, to increased Igf2 expression (Figure 6G, left column). In differentiating myoblasts which express high levels of endogenous let-7 (Figure 6A, top panel), we did not detect Imp1 expression, consistent with Imp1 mRNA being a direct target of let-7 inhibition (Boyerinas et al., 2008; Ioannidis et al., 2005). Taken together, these results suggest that Igf2 is a downstream effector of H19/let-7-mediated regulation in myoblasts.
Given the wide-range of putative targets of let-7 and the complex nature of signaling factors and pathways involved in pre- and postnatal muscle development (Braun and Gautel, 2011; Chang, 2007), we believe that other genes also function as downstream effectors of the H19/let-7-mediated regulation in muscle. Indeed, Hmga2 knockout mice display skeletal muscle hypotrophy due to impaired myoblast proliferation, whereas Dicer knockout mice show decreased skeletal muscle mass with abnormal myofiber morphology, partly as a result of increased apoptosis in muscle cells (Li et al., 2012; O'Rourke et al., 2007). In line with a role in promoting myoblast proliferation or survival, both Hmga2 and Dicer are highly expressed in proliferating myoblasts and their expression declines upon differentiation (Li et al., 2012; O'Rourke et al., 2007; Sago et al., 2004). We find that both Hmga2 and Dicer mRNA levels decrease in C2C12 myoblasts transfected with simH19 or let-7 (Figure 6B, 2nd and 3rd columns from left, compare green and purple bars with blue bars), consistent with Hmga2 and Dicer being additional downstream effectors of H19/let-7-mediated regulation. Future studies dissecting detailed signal transduction pathways mediated by these effectors will be necessary to firmly establish this conclusion.
DISCUSSION
We find that the conserved, imprinted H19 lncRNA, which is highly expressed in the developing embryo and adult muscle, binds to let-7 and inhibits its function, acting as a molecular sponge. The physiological significance of this interaction is underscored using a myogenic differentiation model system where H19 depletion accelerates muscle differentiation, a phenotype recapitulated by let-7 overexpression. It is long known that cellular miRNA abundance could be titrated for regulatory effect using artificial transcripts (miRNA sponges) that contain tandem repeats of miRNA-binding sites (Ebert and Sharp, 2010). Recently, a new class of endogenously expressed noncoding RNAs, called circular RNAs, has been demonstrated to act as potent miRNA sponges (Hansen et al., 2013; Memczak et al., 2013). In addition, mRNAs, transcribed pseudogenes, and other lncRNAs may function as miRNA sponges (Cesana et al., 2011; Poliseno et al., 2010; Tay et al., 2011). We find that H19, a developmentally regulated and abundantly expressed lncRNA, is a natural sponge for let-7.
Although the let-7-binding sites on H19 we identify appear to be “weak” and not significantly conserved, as predicted by the currently available algorithms, they do in fact show appreciable levels of sponge effects using both reporter and endogenous genes as readouts, and under different conditions as well as in different cell types derived from different species. In addition, our crosslinking affinity purification experiments demonstrating an in vivo physical interaction between H19 and let-7 provide further support for H19’s sponge activity. Importantly, our results are consistent with data from genetic testing (Didiano and Hobert, 2006), mammalian studies (Lal et al., 2009; Tay et al., 2008) and high throughput RIP-Chip and CLIP studies (Chi et al., 2009; Hafner et al., 2010; Landthaler et al., 2008; Loeb et al., 2012). A comprehensive CLIP data analysis strongly suggests a substantial involvement of noncanonical sites in gene regulation (Liu et al., 2013). For example, lin-41, the first known target for let-7, only has two noncanonical sites (Vella et al., 2004). Among genetically identified let-7 targets in worms, 40% lack conserved binding sites (Hammell, 2010). Work from David Bartel’s group suggests that a large fraction of nonconserved seed sites can be functional and some might represent important species-specific repression (Bartel, 2009). Thus, our findings reported here add to both the growing list of functional noncanonical sites and the list of nonconserved seed sites.
In addition to contributing to expansion of the repertoire of endogenous miRNA sponges, the significance of our findings is several-fold. First, high levels of H19 expression have been detected in fetal skeletal and cardiac muscle, in addition to fetal liver, yet the physiological significance of this expression is unknown (Gabory et al., 2010). Our results suggest that H19 likely plays a role in controlling timing of muscle differentiation in vivo by modulating the action of let-7. Recently, a muscle-specific lncRNA, called Linc-MD1, was reported to regulate myoblast differentiation by acting as a “sponge” for miR-133 and miR-135 (Cesana et al., 2011). Whether there is any crosstalk between the pathways regulated by these two lncRNAs remains to be investigated.
Second, given the abundant and broad expression pattern of H19 during embryogenesis, and given that the expression of both H19 and let-7 is developmentally regulated in a cell- and tissue-specific manner (Bussing et al., 2008; Gabory et al., 2010), we postulate that the H19/let-7 regulation may also contribute to other developmental processes. For example, H19 deletion mice (H19Δ3) display an increase in both prenatal and postnatal growth compared to wt littermates, which is accompanied by an overexpression of Igf2 and several other genes of the imprinted gene network (IGN) involved in embryonic development and growth (Gabory et al., 2009). Remarkably, when a H19 transgene expressed from a site outside of the H19-Igf2 locus was introduced back to the H19 deletion animals, the overgrowth phenotype, together with the IGN gene overexpression, was restored to the wt level (Gabory et al., 2009). While a cis-acting effect of the transgenic H19 expression has been ruled out (Gabory et al., 2009), a transcriptional and/or posttranscriptional regulation of Igf2 and other IGN genes by H19 lncRNA in contributing to the in vivo growth regulation remains possible. This is suggested by the notion that there exists a positive correlation of expression between let-7 and Igf2 both in vivo (Lu et al., 2007; Lu et al., 2011) and in vitro (Figure 6F, G, and (Lu et al., 2011)), and that downregulation of H19 increases Igf2 expression (Figure 6F) and phenocopies let-7 overexpression (Figure 6D, E). It is therefore tempting to speculate that H19Δ3 mice may have heightened let-7 activity, which in turn leads to increased Igf2 expression, thereby contributing to the overgrowth phenotype.
Third, a high level of H19 expression persists in adult skeletal muscle (Gabory et al., 2010; Onyango and Feinberg, 2011). Because let-7 has been implicated in regulating glucose metabolism in muscle (Frost and Olson, 2011; Zhu et al., 2011), we speculate that H19 may modulate let-7 action in this organ, thereby contributing to glucose metabolism regulation. Fourth, H19 contains putative binding sites for additional miRNAs (Supplemental Table S1), suggesting that H19 may also regulate other miRNAs, perhaps in a tissue/cell and developmental stage-dependent manner. It seems clear that H19 is likely to have role(s) in gene regulation beyond those of titrating let-7 and serving as a reservoir for miR675.
EXPERIMENTAL PROCEDURES
Bioinformatic analysis
MiRNA-binding sites on H19 were predicted using a web-based program RNAhybrid (Rehmsmeier et al., 2004). To examine the evolutionary conservation of the binding sites in human H19 lncRNA, we used conversation scores for each nucleotide of H19 lncRNA. For the NCBI human H19 lncRNA sequence (NR_002196, Build 37.3), the nucleotide-specific conservation score files were downloaded from UCSC genome browser (http://genome.ucsc.edu/). These files were generated by the PhastCons program (Siepel et al., 2005) through multiple-sequence alignments of 33 mammalian genomes to the human genome (hg19). For Sylamer analysis, we used the ArraySylamer program (http://bioinformatics.oxfordjournals.org/content/early/2010/09/24/bioinformatics.btq545.full.pdf). See also Supplemental Experimental Procedures.
Luciferase assays, immunoprecipitation, Western blot, and RT-qPCR
These were carried out as previously described (Qiu et al., 2010). See also Supplemental Experimental Procedures.
In vivo formaldehyde crosslinking and affinity purification
These were carried out as preciously described (Vasudevan et al., 2007) with modifications. See also Supplemental Experimental Procedures.
C2C12 myotube derivation, immunostaining, and fusion index
See Supplemental Experimental Procedures for detailed description.
Microarray experiments
Total RNAs were extracted from siRNA transfected C2C12 myotubes. Hybridization and signal acquisition of the GeneChip Mouse Exon 1.0 ST exon array (Affymetrix) containing a total of 29000 gene transcripts were performed. Each array experiment was performed in triplicates. The signal intensities were normalized by the RMA method using the Expression Console 1.1.2 (Affymetrix).
Statistical Analysis
All data are presented as mean ± SD. Data were analyzed using two-tailed Student’s t test. P values at 0.05 or less were considered significant.
ACKNOWLEDGEMENTS
We thank Wei Ma for helping with cloning and Western blot analysis, Seth Guller for human term placental cDNA, Gerald Shulman for mouse muscle samples, Yukihide Tomari for psiCHECK2-let-7 4×, Kunhua Chen (Exonbio) for site-directed mutagenesis of full-length human H19, and the Computational Molecular Biology and Statistics Core at the Wadsworth Center for supporting computing resources. This work was supported by the following grants: CT Innovations Inc. grant 09SCAYALE14 and Albert McKern Scholar Award 1063338 to Y.H., Bennack-Polan Foundation R11756 to A.N.K., National Science Foundation grant DBI-0650991 to Y.D., and National Institutes of Health grants GM099811 to Y.D. and GM099801 to A.M.B..
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures, two tables and Supplemental Experimental Procedures and can be found with this article online.
GenBank Accession Number
The accession number for the microarray data reported in this paper is GSE49539.
REFERENCES
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. [Europe PMC free article] [Abstract] [Google Scholar]
- Borensztein M, Monnier P, Court F, Louault Y, Ripoche MA, Tiret L, Yao Z, Tapscott SJ, Forne T, Montarras D, et al. Myod and H19-Igf2 locus interactions are required for diaphragm formation in the mouse. Development. 2013;140:1231–1239. [Europe PMC free article] [Abstract] [Google Scholar]
- Boyerinas B, Park SM, Shomron N, Hedegaard MM, Vinther J, Andersen JS, Feig C, Xu J, Burge CB, Peter ME. Identification of let-7-regulated oncofetal genes. Cancer Res. 2008;68:2587–2591. [Abstract] [Google Scholar]
- Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990;10:28–36. [Europe PMC free article] [Abstract] [Google Scholar]
- Braun T, Gautel M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat Rev Mol Cell Biol. 2011;12:349–361. [Abstract] [Google Scholar]
- Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008;14:400–409. [Abstract] [Google Scholar]
- Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. [Europe PMC free article] [Abstract] [Google Scholar]
- Chang KC. Key signalling factors and pathways in the molecular determination of skeletal muscle phenotype. Animal. 2007;1:681–698. [Abstract] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. [Europe PMC free article] [Abstract] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. [Abstract] [Google Scholar]
- Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat Struct Mol Biol. 2006;13:849–851. [Abstract] [Google Scholar]
- Ebert MS, Sharp PA. Emerging roles for natural microRNA sponges. Curr Biol. 2010;20:R858–R861. [Europe PMC free article] [Abstract] [Google Scholar]
- Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. [Europe PMC free article] [Abstract] [Google Scholar]
- Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol. 2012;19:586–593. [Abstract] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. [Abstract] [Google Scholar]
- Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci U S A. 2008;105:14879–14884. [Europe PMC free article] [Abstract] [Google Scholar]
- Frost RJ, Olson EN. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci U S A. 2011;108:21075–21080. [Europe PMC free article] [Abstract] [Google Scholar]
- Gabory A, Jammes H, Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. Bioessays. 2010;32:473–480. [Abstract] [Google Scholar]
- Gabory A, Ripoche MA, Le Digarcher A, Watrin F, Ziyyat A, Forne T, Jammes H, Ainscough JF, Surani MA, Journot L, et al. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development. 2009;136:3413–3421. [Abstract] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. [Europe PMC free article] [Abstract] [Google Scholar]
- Hammell M. Computational methods to identify miRNA targets. Semin Cell Dev Biol. 2010;21:738–744. [Europe PMC free article] [Abstract] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013 [Abstract] [Google Scholar]
- Ioannidis P, Mahaira LG, Perez SA, Gritzapis AD, Sotiropoulou PA, Kavalakis GJ, Antsaklis AI, Baxevanis CN, Papamichail M. CRD-BP/IMP1 expression characterizes cord blood CD34+ stem cells and affects c-myc and IGF-II expression in MCF-7 cancer cells. J Biol Chem. 2005;280:20086–20093. [Abstract] [Google Scholar]
- Iwasaki S, Kawamata T, Tomari Y. Drosophila argonaute1 and argonaute2 employ distinct mechanisms for translational repression. Mol Cell. 2009;34:58–67. [Abstract] [Google Scholar]
- Izaurralde E. Elucidating the temporal order of silencing. EMBO Rep. 2012 [Europe PMC free article] [Abstract] [Google Scholar]
- Jung YH, Gupta MK, Shin JY, Uhm SJ, Lee HT. MicroRNA signature in testes-derived male germ-line stem cells. Mol Hum Reprod. 2010;16:804–810. [Abstract] [Google Scholar]
- Kalista S, Schakman O, Gilson H, Lause P, Demeulder B, Bertrand L, Pende M, Thissen JP. The type 1 insulin-like growth factor receptor (IGF-IR) pathway is mandatory for the follistatin-induced skeletal muscle hypertrophy. Endocrinology. 2012;153:241–253. [Abstract] [Google Scholar]
- Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012;14:659–665. [Europe PMC free article] [Abstract] [Google Scholar]
- Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O'Day E, Chowdhury D, Dykxhoorn DM, Tsai P, Hofmann O, et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, other cell-cycle genes via binding to "seedless" 3'UTR microRNA recognition elements. Mol Cell. 2009;35:610–625. [Europe PMC free article] [Abstract] [Google Scholar]
- Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 2008;14:2580–2596. [Europe PMC free article] [Abstract] [Google Scholar]
- Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–1030. [Europe PMC free article] [Abstract] [Google Scholar]
- Li Z, Gilbert JA, Zhang Y, Zhang M, Qiu Q, Ramanujan K, Shavlakadze T, Eash JK, Scaramozza A, Goddeeris MM, et al. An HMGA2-IGF2BP2 axis regulates myoblast proliferation and myogenesis. Dev Cell. 2012;23:1176–1188. [Europe PMC free article] [Abstract] [Google Scholar]
- Liu C, Mallick B, Long D, Rennie WA, Wolenc A, Carmack CS, Ding Y. CLIP-based prediction of mammalian microRNA binding sites. Nucleic Acids Res. 2013 [Europe PMC free article] [Abstract] [Google Scholar]
- Loeb GB, Khan AA, Canner D, Hiatt JB, Shendure J, Darnell RB, Leslie CS, Rudensky AY. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol Cell. 2012;48:760–770. [Europe PMC free article] [Abstract] [Google Scholar]
- Lu L, Katsaros D, de la Longrais IA, Sochirca O, Yu H. Hypermethylation of let-7a-3 in epithelial ovarian cancer is associated with low insulin-like growth factor-II expression and favorable prognosis. Cancer Res. 2007;67:10117–10122. [Abstract] [Google Scholar]
- Lu L, Katsaros D, Zhu Y, Hoffman A, Luca S, Marion CE, Mu L, Risch H, Yu H. Let-7a regulation of insulin-like growth factors in breast cancer. Breast Cancer Res Treat. 2011;126:687–694. [Abstract] [Google Scholar]
- Matouk IJ, DeGroot N, Mezan S, Ayesh S, Abu-lail R, Hochberg A, Galun E. The H19 non-coding RNA is essential for human tumor growth. PLoS One. 2007;2:e845. [Europe PMC free article] [Abstract] [Google Scholar]
- McKarney LA, Overall ML, Dziadek M. Expression of H19 and Igf2 genes in uniparental mouse ES cells during in vitro and in vivo differentiation. Differentiation. 1996;60:75–86. [Abstract] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013 [Abstract] [Google Scholar]
- Merrick D, Ting T, Stadler LK, Smith J. A role for Insulin-like growth factor 2 in specification of the fast skeletal muscle fibre. BMC Dev Biol. 2007;7:65. [Europe PMC free article] [Abstract] [Google Scholar]
- Milligan L, Antoine E, Bisbal C, Weber M, Brunel C, Forne T, Cathala G. H19 gene expression is up-regulated exclusively by stabilization of the RNA during muscle cell differentiation. Oncogene. 2000;19:5810–5816. [Abstract] [Google Scholar]
- Mukherji S, Ebert MS, Zheng GX, Tsang JS, Sharp PA, van Oudenaarden A. MicroRNAs can generate thresholds in target gene expression. Nat Genet. 2011;43:854–859. [Europe PMC free article] [Abstract] [Google Scholar]
- Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol. 1999;19:1262–1270. [Europe PMC free article] [Abstract] [Google Scholar]
- O'Rourke JR, Georges SA, Seay HR, Tapscott SJ, McManus MT, Goldhamer DJ, Swanson MS, Harfe BD. Essential role for Dicer during skeletal muscle development. Dev Biol. 2007;311:359–368. [Europe PMC free article] [Abstract] [Google Scholar]
- Onyango P, Feinberg AP. A nucleolar protein, H19 opposite tumor suppressor (HOTS), is a tumor growth inhibitor encoded by a human imprinted H19 antisense transcript. Proc Natl Acad Sci U S A. 2011;108:16759–16764. [Europe PMC free article] [Abstract] [Google Scholar]
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. [Europe PMC free article] [Abstract] [Google Scholar]
- Rosen KM, Wentworth BM, Rosenthal N, Villa-Komaroff L. Specific, temporally regulated expression of the insulin-like growth factor II gene during muscle cell differentiation. Endocrinology. 1993;133:474–481. [Abstract] [Google Scholar]
- Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–516. [Abstract] [Google Scholar]
- Sago N, Omi K, Tamura Y, Kunugi H, Toyo-oka T, Tokunaga K, Hohjoh H. RNAi induction and activation in mammalian muscle cells where Dicer and eIF2C translation initiation factors are barely expressed. Biochem Biophys Res Commun. 2004;319:50–57. [Abstract] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. [Europe PMC free article] [Abstract] [Google Scholar]
- Schmitter D, Filkowski J, Sewer A, Pillai RS, Oakeley EJ, Zavolan M, Svoboda P, Filipowicz W. Effects of Dicer and Argonaute down-regulation on mRNA levels in human HEK293 cells. Nucleic Acids Res. 2006;34:4801–4815. [Europe PMC free article] [Abstract] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. [Europe PMC free article] [Abstract] [Google Scholar]
- Stewart CE, James PL, Fant ME, Rotwein P. Overexpression of insulin-like growth factor-II induces accelerated myoblast differentiation. J Cell Physiol. 1996;169:23–32. [Abstract] [Google Scholar]
- Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–357. [Europe PMC free article] [Abstract] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. [Abstract] [Google Scholar]
- Tokumaru S, Suzuki M, Yamada H, Nagino M, Takahashi T. let-7 regulates Dicer expression and constitutes a negative feedback loop. Carcinogenesis. 2008;29:2073–2077. [Abstract] [Google Scholar]
- Varrault A, Gueydan C, Delalbre A, Bellmann A, Houssami S, Aknin C, Severac D, Chotard L, Kahli M, Le Digarcher A, et al. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev Cell. 2006;11:711–722. [Abstract] [Google Scholar]
- Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3'UTR. Genes Dev. 2004;18:132–137. [Europe PMC free article] [Abstract] [Google Scholar]
- Wilson EM, Hsieh MM, Rotwein P. Autocrine growth factor signaling by insulin-like growth factor-II mediates MyoD-stimulated myocyte maturation. J Biol Chem. 2003;278:41109–41113. [Abstract] [Google Scholar]
- Yoshiko Y, Hirao K, Maeda N. Differentiation in C(2)C(12) myoblasts depends on the expression of endogenous IGFs and not serum depletion. Am J Physiol Cell Physiol. 2002;283:C1278–C1286. [Abstract] [Google Scholar]
- Yoshimizu T, Miroglio A, Ripoche MA, Gabory A, Vernucci M, Riccio A, Colnot S, Godard C, Terris B, Jammes H, et al. The H19 locus acts in vivo as a tumor suppressor. Proc Natl Acad Sci U S A. 2008;105:12417–12422. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhu H, Shyh-Chang N, Segre AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG, et al. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147:81–94. [Europe PMC free article] [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.molcel.2013.08.027
Read article for free, from open access legal sources, via Unpaywall:
http://www.cell.com/article/S1097276513006278/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Functional Bidirectionality of ERV-Derived Long Non-Coding RNAs in Humans.
Int J Mol Sci, 25(19):10481, 29 Sep 2024
Cited by: 0 articles | PMID: 39408810 | PMCID: PMC11476766
Review Free full text in Europe PMC
Non-coding RNAs as potential targets in metformin therapy for cancer.
Cancer Cell Int, 24(1):333, 01 Oct 2024
Cited by: 0 articles | PMID: 39354464 | PMCID: PMC11445969
Review Free full text in Europe PMC
LnCRNAs in the Regulation of Endometrial Receptivity for Embryo Implantation.
JBRA Assist Reprod, 28(3):503-510, 26 Aug 2024
Cited by: 0 articles | PMID: 38875127 | PMCID: PMC11349255
Review Free full text in Europe PMC
Risk of Fat Mass- and Obesity-Associated Gene-Dependent Obesogenic Programming by Formula Feeding Compared to Breastfeeding.
Nutrients, 16(15):2451, 28 Jul 2024
Cited by: 1 article | PMID: 39125332 | PMCID: PMC11314333
Review Free full text in Europe PMC
Role of long noncoding RNAs in diabetes-associated peripheral arterial disease.
Cardiovasc Diabetol, 23(1):274, 24 Jul 2024
Cited by: 0 articles | PMID: 39049097 | PMCID: PMC11271017
Review Free full text in Europe PMC
Go to all (685) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
RefSeq - NCBI Reference Sequence Database
- (1 citation) RefSeq - NR_002196
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The developmental transcriptome sequencing of bovine skeletal muscle reveals a long noncoding RNA, lncMD, promotes muscle differentiation by sponging miR-125b.
Biochim Biophys Acta, 1863(11):2835-2845, 31 Aug 2016
Cited by: 59 articles | PMID: 27589905
LncRNA H19/miR-let-7 axis participates in the regulation of ox-LDL-induced endothelial cell injury via targeting periostin.
Int Immunopharmacol, 72:496-503, 01 May 2019
Cited by: 30 articles | PMID: 31054453
The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration.
Genes Dev, 28(5):491-501, 14 Feb 2014
Cited by: 313 articles | PMID: 24532688 | PMCID: PMC3950346
The Interplay of LncRNA-H19 and Its Binding Partners in Physiological Process and Gastric Carcinogenesis.
Int J Mol Sci, 18(2):E450, 20 Feb 2017
Cited by: 50 articles | PMID: 28230721 | PMCID: PMC5343984
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NHLBI NIH HHS (1)
Grant ID: R01 HL115148
NICHD NIH HHS (3)
Grant ID: L50 HD081739
Grant ID: R01 HD101475
Grant ID: K12 HD000849
NIDDK NIH HHS (1)
Grant ID: P01 DK057751
NIGMS NIH HHS (4)
Grant ID: GM099801
Grant ID: R01 GM099801
Grant ID: R01 GM099811
Grant ID: GM099811





