Abstract
Free full text

Antigen-specific B cell receptor sensitizes B cells to infection by influenza virus
Abstract
Influenza A virus-specific B lymphocytes and the antibodies they produce protect against infection 1. However, the outcome of interactions between a flu hemagglutinin (HA)-specific B cell via its receptor (BCR) and virus is unclear. Through somatic cell nuclear transfer (SCNT) we generated mice that harbor B cells with a BCR specific for the HA of A/WSN/33 (FluBI mice). Their B cells secrete an IgG2b that neutralizes infectious virus. While B cells from FluBI and control mice bind equivalent amounts of virus through interactions of HA with surface-disposed sialic acids, the A/WSN/33 virus infects only the HA-specific B cells. Mere binding of virus is not sufficient for infection of B cells: this requires interactions of the BCR with HA, causing both disruption of antibody secretion and FluBI B cell death within 18 hours. In mice infected with A/WSN/33, lung-resident FluBI B cells are infected by the virus, thus delaying the onset of protective antibody release into the lungs, while FluBI cells in the draining lymph node are not infected and proliferate. We propose that influenza targets and kills influenza-specific B cells in the lung, thus allowing the virus to gain purchase prior to the initiation of an effective adaptive response.
Memory B lymphocytes contribute to the protective immune response to flu infection by producing immunoglobulins that bind and neutralize the virus 1. The lung of an exposed individual contains influenza-specific memory B cells that bind virus, differentiate into plasma cells and secrete either IgG or IgA locally, reducing the spread of virus 2,3. However, the fate of virus-specific B cells that encounter live influenza virus remains unknown.
The low frequency of antigen-specific B cells has hampered analysis of the interactions between live virus, flu antigens and the primary B cells specific for them 2. To detect influenza virus-specific B cells, we used sortase-mediated labeling to install Alexa647 fluorophore onto the HA protein 4-6. Virus was disrupted with detergent, HA-Alexa647 was purified by immunoprecipitation and dialyzed to form fluorescent flu micelles (ED Fig. 1a-d). These flu micelles did not stain splenocytes from uninfected mice, but did stain a small number of CD19+ cells in spleens of mice infected with influenza and boosted multiple times with A/WSN/33 in incomplete Freund's adjuvant (ED Fig. 1e).
Virus-specific CD19+ B cells, isolated by fluorescence-activated cell sorting (Fig. 1a), were used as a source of nuclei for SCNT 7-9. We transferred the nuclei of these B cells into enucleated oocytes and derived ES cells 7-9 that harbor the VDJ/VJ (heavy chain/light chain) rearrangements of the original donor B cell to produce chimeric mice. We screened offspring of the founder chimeras by ELISA for the presence of anti-flu antibodies and obtained one animal that showed high titers of IgG2b flu-specific antibodies (ED Fig. 2) and IgG2b+IgM- B cells in the absence of infection (Fig. 1b). We backcrossed this mouse to C57BL/6 to secure germline transmission of the VDJ/VJ pair and hereafter refer to the line as FluBI. We know of no other mouse model that harbors B cells of known pathogen specificity or whose primary B cells produce IgG2b.
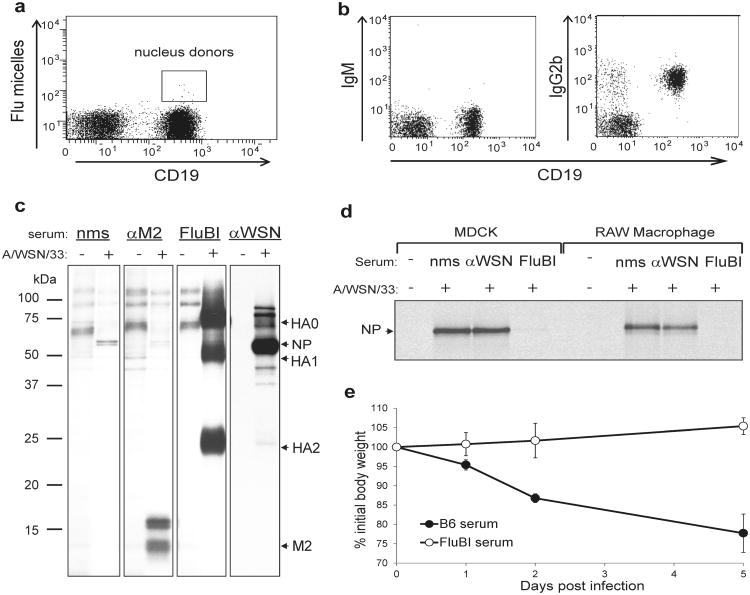
a) A B6x129F1 mouse was infected intranasally with A/WSN/33 and immunized i.p. at days 7, 14, and 21 post-infection with disrupted A/WSN/33 in incomplete Freund's adjuvant. Splenocytes were harvested at day 28 post-infection, and stained with anti-CD19 and Alexa647-labeled HA flu glycoprotein micelles. Cells indicated were sorted and used as donor nuclei for SCNT.
b) Peripheral blood from FluBI mice with germline transmission of the rearranged VDJ and VJ genes was stained with the indicated antibodies and analyzed by cytofluorimetry.
c) MDCK cells were infected with A/WSN/33 and labeled with 35S-cysteine/methionine for 4 hrs prior to lysis. Lysates were immunoprecipitated with monoclonal anti-M2 (αM2), FluBI serum (FluBI), or serum from uninfected (nms) or A/WSN/33 infected mice (αWSN). Immunoprecipitates were analyzed by SDS-PAGE and autoradiography. All panels were from the same gel; αWSN panels shown are from a shorter exposure time.
d) A/WSN/33 virus was incubated with the indicated serums prior to infection of MDCK cells or RAW macrophages. At 2hpi, cells were labeled with 35S-cysteine/methionine for 2 hrs, lysed and immunoprecipitated with αWSN serum.
e) Balb/c mice received 100μL of serum IV from wild type or FluBI mice, prior to intranasal challenge with A/WSN/33 (2×105 pfu/mouse). n = 5; error bars are SD.
The sequences of the rearranged heavy and light chain genes (ED Fig. 3) show 7 and 4 somatic mutations in the VH and Vκ segments, respectively. We established the specificity of the FluBI IgG2b antibody by immunoprecipitation from lysates of 35S-cysteine/methionine labeled, A/WSN/33 -infected MDCK cells (Fig. 1c). The antibody retrieves HA0 and its cleavage products 10 HA1 and HA2. FluBI IgG2b antibody purified from hybridomas generated from FluBI;RAG2-/- splenocytes gave similar results (Supplemental Methods). The serum from FluBI mice neutralizes A/WSN/33 in vitro (Fig. 1d) and in vivo (Fig. 1e). Cytofluorimetry of B cell populations in lymph node, spleen and bone marrow from FluBI mice showed a complete absence of B-1a B cells, while other B cell subsets were near-normal in distribution and number (ED Fig. 4). As shown for OBI mice 7, the presence of a functionally rearranged γ2b heavy chain locus does not compromise B cell development, despite the deletion of the μ, δ,γ3 and γ1 constant regions in FluBI mice.
To determine the fate of HA-specific B cells upon encounter with virus, we obtained B cells from the FluBI mouse and from OBI mice, whose B cells produce an IgG1 specific for ovalbumin 7. Prior to infection, we activated cells overnight with anti-CD40 to improve biosynthetic labeling, used to assess viral antigen synthesis. At 2 hpi FluBI B cells infected with A/WSN/33 synthesize vastly more HA, NP and M2 (inset) than OBI B cells (Fig. 2a). In FluBI B cells, the levels of NP were comparable to those obtained from A/WSN/33 -infected MDCK cells, an indication that replication of A/WSN/33 in FluBI B cells is robust (Fig. 2a, ED Fig. 5). Neither FluBI nor OBI B cells were infected by the closely related strain A/Puerto Rico/8/1934 (H1N1) or with A/Udorn/307/1972 (H3N2) (Fig. 2b, ED Figs. 6, 7).
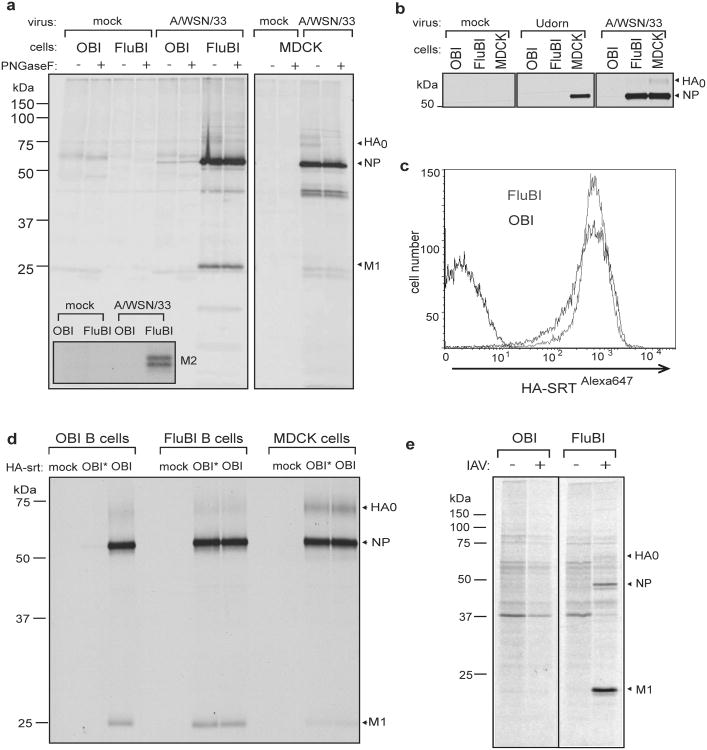
a) CD40-activated OBI and FluBI B cells and MDCK cells were incubated with A/WSN/33 at an MOI 1.0 for 30min on ice, washed, and transferred to 37° C in RPMI (0.2% BSA). At 2hpi, cells were labeled with 35S-cysteine/methionine for 2 hrs, immunoprecipiated with αWSN serum or anti-M2 antibody (inset), digested with PNGaseF, and analyzed by SDS-PAGE and autoradiography.
b) MDCK cells and CD40-activated OBI or FluBI B cells were incubated with A/WSN/33 or A/Udorn/307/1972 (H3N2) virus and analyzed as in a).
c) CD40-activated OBI or FluBI B cells were incubated with HA-SRTAlexa647 virus for 30 min on ice and analyzed by cytofluorimetry.
d) CD40-activated OBI or FluBI B cells or MDCK cells were incubated on ice for 30 min with HA-SRT virus modified with a 17-mer peptide containing the OBI epitope (OBI) or a mutant version that no longer binds to OBI (OBI*). Infection was analyzed as in a).
e) CD40-activated OBI or FluBI B cells were infected with A/WSN/33 at a MOI of 1.0 and labeled with 35S-cysteine/methionine at 4 hpi for 4 hrs. Released virus particles were recovered by adsorption to chicken erythrocytes and analyzed by SDS-PAGE and autoradiography.
The increased levels of antigen detected in FluBI B cells might result from improved binding of virus via BCR-HA interactions. To measure virus binding, we incubated anti CD40-activated B cells with Alexa647 labeled HA-SRT virus (HA-SRTAlexa647) 4 and measured bound virus by cytofluorimetry. Virus bound equally well to FluBI and OBI B cells (Fig. 2c). To determine whether increased susceptibility of FluBI B cells to infection is indeed BCR-dependent, we generated HA-SRT virus, transacylated 4 at the C-terminus of HA1 with a synthetic 17-residue ovalbumin peptide that comprises the epitope recognized by the ovalbumin-specific OBI B cells (HA-SRTwtOBI). This virus should now also bind to the BCR expressed on the surface of OBI B cells. For comparison we labeled HA-SRT virus with a mutant version of the OBI 17-mer peptide (HA-SRTmtOBI) no longer recognized by the OBI IgG1. We then exposed FluBI and OBI B cells to either HA-SRTwtOBI or HA-SRTmtOBI virus (Fig. 2d). As expected, the two HA-SRT viruses infected FluBI B cells equally, regardless of the identity of the peptide epitope installed. In contrast, only HA-SRTwtOBI infected OBI B cells. The level of infection in OBI B cells exposed to HA-SRTwtOBI was similar to that seen in FluBI B cells. The presence of a BCR that recognizes HA, native or modified to impart BCR reactivity, thus causes susceptibility to influenza infection. Mere adsorption of virus to the cell surface through interactions with sialic acids may therefore not suffice to gain entry 11-13, and interaction with an internalizing receptor is important 14-17. Antigen-occupied BCRs are indeed efficiently internalized 18, especially when engaged by a multivalent ligand, and could thus improve virus entry and infection.
Do infected B cells produce virus or virus-like particles (VLPs)? We infected OBI and FluBI B cells with A/WSN/33 and biosynthetically labeled them 4-6 hpi. We incubated the culture supernatants with chicken erythrocytes to recover released virus and VLPs via interactions of HA with erythrocyte-borne sialic acids, and analyzed adsorbed materials by SDS-PAGE and autoradiography to detect the presence of viral structural proteins (Fig. 2e). NP and M1 proteins were present in material released by FluBI but not OBI B cells, indicating that infection of FluBI B cells was productive.
We also looked for secreted antibody from A/WSN/33-exposed B cells (Fig. 3a). OBI cells showed no change in their ability to secrete antibody after exposure to A/WSN/33, presumably because they are poorly infected, if at all. Not only did FluBI B cells exposed to A/WSN/33 show decreased levels of secreted IgG2b, they also died within 18 hours post-infection (Fig. 3b) under conditions where OBI B cells survived.
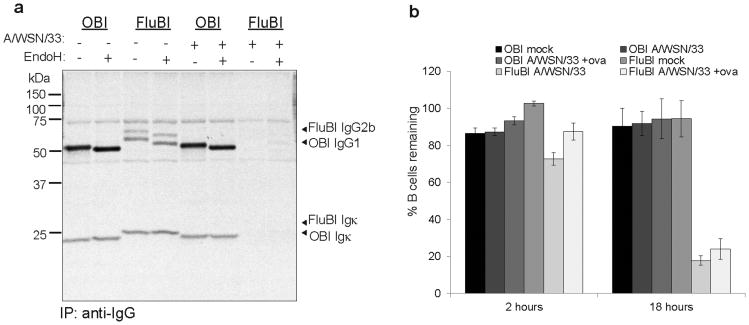
a) CD40-activated OBI or FluBI B cells were incubated with A/WSN/33 for 30min on ice, washed and cultured in RPMI (0.2% BSA) for 2 hrs. Cells were labeled with 35S-cysteine/methionine for 2 hrs, and immunoglobulins were recovered from supernatants using Protein-G agarose.
b) CD40-activated OBI or FluBI B cells were incubated with A/WSN/33 for 30 min on ice, washed and mixed at a 1:1 ratio with mock-infected MHCII-GFP CD40-activated B cells. Ovalbumin (100μg/ml) was included where indicated. Cells were cultured in complete RPMI for 2 or 18 hrs, stained with anti-CD19 and 7-AAD, and analyzed by cytofluorimetry. Live cells were calculated as (#GFP-CD19+ cells/#GFP+CD19+ cells)/ (#GFP-CD19+ cells/#GFP+CD19+ cells at time 0) × 100. Error bars are SD of triplicate cultures.
To explore the consequences of in vivo exposure of FluBI cells to virus and track FluBI B cells, we transferred Celltrace violet-marked OBI (IgG1+) and FluBI (IgG1-) B cells, obtained from transnuclear animals crossed onto the MHC II GFP background, into naïve recipients. The number of B cells transferred was chosen such that it still allowed detection of transferred cells in lungs and mediastinal lymph nodes from infected mice. Mice were inoculated intranasally with live or irradiated A/WSN/33, mixed with ovalbumin, to ensure engagement of the respective donor BCRs. We then measured cell number, proliferation and expression of viral antigens on donor MHC II GFP+ B cells (Fig. 4). The ovalbumin-specific OBI B cells showed robust proliferation in both spleen and mediastinal lymph nodes while FluBI B cells proliferated to a greater extent in the mediastinal lymph nodes in response to live virus, likely due to the increased amount of antigen present locally following viral replication in the lungs. By day 6 post-infection, proliferating FluBI B cells in the lymph nodes had differentiated into CD138+ plasmablasts (ED Fig. 8). Mice that received irradiated A/WSN/33 showed less proliferation of FluBI cells in mediastinal lymph nodes due to the absence of antigen synthesis, but more proliferation of FluBI cells in the lungs at 3 dpi than animals that received live virus (Fig. 4a; Day 3 lungs live: 2.7 +/- 0.43% n=6 vs. Day 3 lungs irradiated: 6.5 +/- 1.3% n=5; p=0.015).
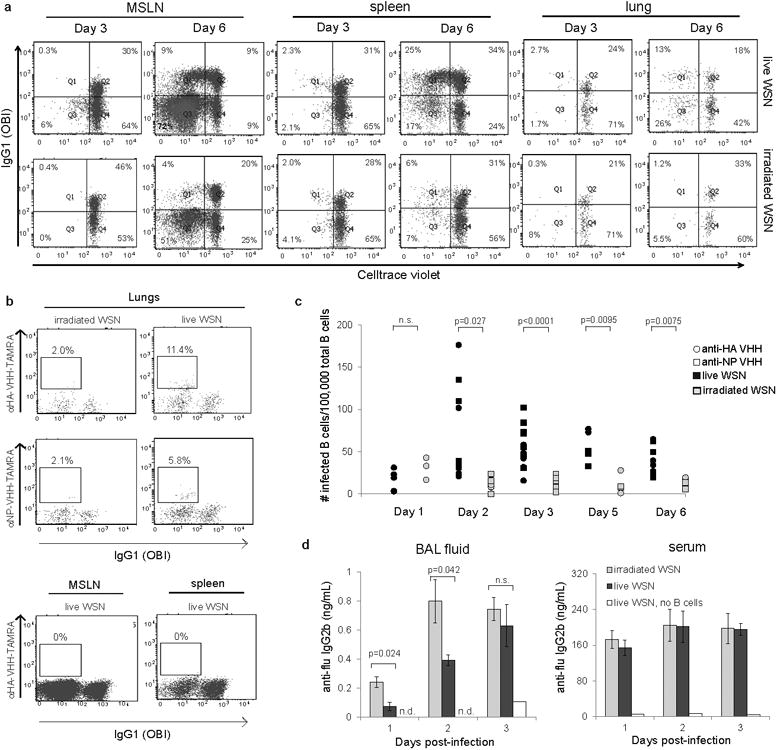
Naïve B cells from OBI;MHCII-GFP and FluBI;MHCII-GFP mice were stained with Celltrace violet, mixed in a 1:1 ratio, and transferred IV into C57BL/6 mice (107 total cells per recipient). Recipient mice were infected intranasally with live or irradiated A/WSN/33 (2×105 pfu/mouse) mixed with ovalbumin (100 μg/mouse) and sacrificed at 3 or 6 days post-infection. Cells were harvested from spleen, mediastinal lymph nodes (MSLN) and lungs.
a) Plots are gated on GFP+ transferred cells. Dilution of violet dye indicates proliferation. Anti-IgG1distinguishes OBI from FluBI B cells. Plots are representative of 4 mice per group.
b) Cells from a) were permeabilized, fixed and stained with single domain antibodies recognizing HA (αHA-VHH) or NP (αNP-VHH). Plots are representative of 5 mice per group.
c) Quantification of viral antigen-positive B cells as shown in b. p values were determined using a one-sided t-test with Bonferroni correction.
d) C57BL/6 mice were administered 107 FluBI B cells IV and infected intranasally with live or UV-irradiated A/WSN/33. Mice were sacrificed at 1, 2 or 3 days post-infection. Bronchoalveolar lavage (BAL) fluid and serum were analyzed by ELISA for A/WSN/33 reactivity using HRP-coupled anti-IgG2b. n.d., not detected. p values were determined using a two-sided t-test with Bonferroni correction. Day 1, n=5; Day 2, n=7 irradiated, n=8 live; Day 3, n=4. Error bars are SEM.
To detect intracellular influenza viral proteins as an additional measure of infection, we generated HA- and NP-specific heavy-chain only antibody fragments (VHHs) from an influenza-immunized alpaca (ED Fig. 9). These small, single domain VHHs are C-terminally modified with an LPETG motif for direct coupling with TAMRA using sortase 19 and detect flu antigens in FluBI B cells infected in vitro (ED Fig. 10). In mice infected with live A/WSN/33 we observed HA- and NP-positive, infected FluBI cells in the lung (Fig. 4b,c). Although virus is reportedly delivered to the MSLN by dendritic cells 20,21, no flu-antigen positive B cells were found in the MSLN (Fig. 4b). Not unexpectedly, mice exposed to irradiated A/WSN/33 lacked flu antigen-positive B cells in either location. Co-transferred OBI cells were present in the lungs of infected mice, but were HA- and NP-negative.
To determine whether infection of lung-resident flu-specific B cells affects antibody production in vivo, we measured in bronchoalveolar lavage (BAL) fluid the presence of flu-specific IgG2b, secreted by the transferred FluBI B cells, and found that irradiated virus elicited a stronger and more rapid initial response than live virus, consistent with the ability of live, but not irradiated virus to kill FluBI B cells. Serum levels of flu-specific IgG2b were equivalent in mice receiving live versus irradiated virus, suggesting that loss of flu-specific IgG2b is restricted to the lung bronchoalveolar space.
FluBI cells, specific for HA and secreting a neutralizing antibody, themselves succumb to infection mediated by the surface-disposed BCR. The rapid death of A/WSN/33 -specific FluBI cells provides respite for the virus at the lung epithelium, a site to which antigen-specific B cells are recruited in the course of infection and remain as sentinels thereafter 2,22,23. Infection and killing of a fraction of the rare antigen-specific B cells impairs the kinetics of the memory response, and confers an advantage to the virus with its replication cycle measured in hours. The ability of a pathogen to exploit this mode of entry and eliminate the initial wave of the very B cells capable of counteracting the infection is an efficient means of ensuring a window for replication and horizontal transmission. It is unlikely to be limited to influenza virus.
Online Supplemental Materials and Methods
Reagents
Anti A/WSN/33 serum was generated by infecting BALB/C mice with A/WSN/33 (2×105 pfu/mouse). Anti-M2 antibody (14C2) was purchased from Santa Cruz Biotechnology. OBI peptides were provided by the MIT biopolymers facility. The amino acid sequence that comprises the OBI epitope is as follows: GGGFDKLPGFGDSIEAQGGK. The mutant sequence that fails to bind to the OBI antibody is as follows (substitutions denoted in bold): GGGFDKLPGAGASIEAQGGK. Sortase was expressed in Escherichia coli and purified as described 24. The GGGK-Alexa647 peptide used to label HAsrt was provided by Martin Witte. Anti IgG-FITC antibody (A1101) was purchased from Molecular Probes. Chicken Erythrocytes (cRBCs) were purchased from Lampire Biological Laboratories. Express35S protein labeling mix was purchased from Perkin Elmer. Methionine and cysteine-free RPMI, Optimem, HEPES buffer and non-essential amino acids (NEAA) were purchased from Life Technologies. Endoglycosidase H (EndoH) and PNGasF were purchased from New England Biolabs. Protein-G agarose and EDTA-free complete protease inhibitor cocktail was purchased from Roche Diagnostics.
Virus propagation and infection
A/Puerto Rico/8/1934 (H1N1) and A/Udorn/307/1972 (H3N2) viral stocks were a kind gift from Dr. Xiaowei Zhuang. MDCK cells and RAW cells were originally obtained from the ATCC, and are tested for mycoplasma every 3-6 months. A/WSN/33 and HA-SRT virus 4 were propagated in MDCK cells grown in Optimem and supplemented with 1μg/mL TPCK-treated trypsin. Titers of A/WSN/33 and HAsrt virus stocks were determined by plaque assay on MDCK cells. For the plaque assay, MDCK cells were cultured in 24-well dishes until sub-confluent. Cells were washed 2× in PBS supplemented with Ca2+ and Mg2+ (PBS+) and infected with 10 fold serial dilutions of virus in PBS+ supplemented with 0.25% BSA for 1 hour at room temperature. Cells were washed 1× in PBS+ then overlayed with plaque media (1xMEM, 0.25% BSA, 0.8% agar, 0.5ug/ml trypsin-TPCK) and placed at 37° C. After 24 to 48 hours, the agar overlay was removed and the cells were fixed with 3% paraformaldehyde and permeabilized using PBS 0.5% NP-40. Influenza plaques were stained using monoclonal antibody against NP conjugated to FITC then visualized and quantified by fluorescent microscopy. Labeled HA-SRT virus was quantified using hemagglutination assay against a standard containing a known quantity of A/WSN/33. For all in vitro experimental infections virus was diluted in PBS+/BSA(0.25%) and supplemented with 1μg/mL TPCK-treated trypsin. In the case of MDCK cells, cells were trypsinized and infections carried out in suspension. Virus and cells were incubated on ice for 30min. Cells were washed with PBS+, and resuspended in DMEM with 0.2% BSA, 100mM HEPES and NEAA (or RPMI with 0.2% BSA, 100mM HEPES and NEAA in experiments where B cells were used).
Sortase labeling of HA-SRT virus
HA-SRT virus 4 was incubated with Sortase A (150μM) and the indicated nucleophile (500μM) in sortase labeling buffer (100mM Tris pH7.4, 150mM NaCl, 10mM Ca2+) supplemented with 0.2% BSA at 37° C for 1 hour. The labeled virus was then concentrated over a 20% sucrose cushion and resuspended in sortase labeling buffer supplemented with 0.2%BSA.
Neutralization assay
1×105 PFU A/WSN/33 was incubated with 20μL of the indicated test serum for 30 minutes on ice. The virus/serum mixture was then overlayed onto 1×106 MDCK or RAW macrophages and incubated for 30 minutes at room temperature. Cells were washed and incubated at 37° C in DMEM (0.2%BSA). At 2hpi, both cell types were biosynthetically labeled for two hours and subsequently lysed. Immunopreciptates were prepared with anti-WSN serum and analyzed by SDS-PAGE and autoradiography.
Animal care
All mice were housed at the Whitehead Institute for Biomedical Research and were maintained according to protocols approved by the MIT Committee on Animal Care. A/WSN/33 infected animals were housed in an approved quarantine room at Whitehead Institute. C57BL/6 and Balb/c mice were purchased from Jackson Labs. RAG2-/- mice (RAGTN12) were purchased from Taconic. MHCII-GFP mice 25 and OBI mice 7 have been described.
In vivo influenza infections
8-10 week old male mice were anesthetized by a single dose of avertin (1.25% tribromoethanol, GIBCO), 250 mg/kg body weight, delivered by intraperitoneal injection. A/WSN/33 (2×10^5 pfu/mouse unless otherwise stated) or A/Puerto Rico/8/1934 (2×105 pfu) was administered intranasally. Infected Balb/c mice were weighed daily, and mice losing more than 20% of starting weight were euthanized. Infected C57BL/6 mice are intrinsically more resistant to influenza infection 26, and did not show more than 5% weight loss; C57BL/6 mice used as hosts for adoptive transfer (IV) of FluBI or OBI B cells were infected with A/WSN/33 within 24 hours of donor cell transfer. Mice that received adoptive transfer of FluBI and/or OBI B cells were allowed to intermix freely in a large cage before separating mice into groups to receive irradiated or live flu virus. Infections were performed by a non-blinded investigator. Successfully administration of intranasal virus was verified by recruitment of FluBI cells to the MSLN. <5% of mice showed failure of intranasal delivery and were excluded from analysis, as per pre-established criteria.
TN mouse generation
TN mice were generated as previously described 7-9,27,28. Briefly, flu-specific B cells were sorted by FACS and used as a source of donor nuclei for SCNT according to the protocol established by the Wakayama group 29. The mitotic spindle was removed from mouse oocytes and replaced with donor nuclei. The nucleus-transplanted oocytes were then activated in medium containing strontium and trichostatin A, and allowed to develop in culture to the blastocyst stage. Because the live birth rate of SCNT blastocysts transferred into pseudopregnant females is quite low, SCNT blastocysts were used instead to derive embryonic stem (ES) cell lines. These ES cell lines were then injected into wild type B6xDBA F1 blastocysts and implanted into pseudopregnant females. The resulting chimeric pups were mated to C57BL/6 females to establish the FluBI line. All animals used were backcrossed 4-5 generations onto C57BL/6 or C57BL/6;RAG2-/- backgrounds.
Flu micelles
HA-SRT virus was incubated with Sortase A (150μM) and Alexa647 nucleophile (500μM) in sortase labeling buffer (100mM Tris pH7.4, 150mM NaCl, 10mM Ca2+) supplemented with 0.2% BSA at 37° C for 1 hour. HAsrtAlexa647 virus was disrupted Triton ×100 and incubated with 400μg anti-Alexa647 overnight. HA-Alexa647 was then recovered using protein G-sepharose. Bound proteins were eluted with 0.1M Glycine pH2.8. Detergent was dialyzed to form protein micelles 30.
Flow cytometry
Cells were harvested from spleen, pooled mesenteric and cervical lymph nodes, mediastinal lymph nodes, lung, peritoneal cavity and bone marrow. Lung tissue was digested in RPMI with 1% (w/v) collagenase D (Sigma) for 30-60 minutes at 37°C prior to mechanical dissociation with a 40μm cell strainer. Cell preparations were subjected to hypotonic lysis to remove erythrocytes, stained and analyzed using a FACS Fortessa (BD). Celltrace violet was purchased from Invitrogen. All antibodies were from BD Pharmingen. For intracellular staining, cells were fixed and permeabilized using Cytofix/Cytoperm (BD) according to the manufacturer's instructions.
Flu-specific ELISAs
High-binding 96-well microtiter plates (Costar) were coated overnight at 4° with A/WSN/33 (2×104 pfu/mL) in PBS. Plates were washed 3 times with wash buffer (PBS, 0.05% Tween-20), blocked with 10% fetal bovine serum for 1 hour at room temperature, washed 3 times, and incubated with samples. Bronchoalveolar lavage (BAL) fluid samples were collected by inserting a 24 gauge catheter into an incision in the trachea, filling the lungs with 1 mL PBS and recovering 0.7-0.8 mL of lavage fluid. BAL fluid samples were used neat. Serum samples were used at 1:10 dilution. FluBI antibody purified from hybridoma supernatants was used as standard. Plates were incubated with samples at room temperature for 2 hours, washed 5 times, and incubated with HRP-coupled anti-IgG2b secondary reagent for 1 hour. Plates were washed 7 times, and detected using 3,3′,5,5′-Tetramethylbenzidine (TMB) Liquid Substrate (Sigma).
Cell culture
B cells were purified from pooled spleen and lymph nodes by negative selection using anti-CD43 magnetic beads (Miltenyi Biotec). B cells were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated FBS, 2 mM l-glutamine, 100 U/ml penicillin G sodium, 100 μg/ml streptomycin sulfate, 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, and 0.1 mM 2-ME. For differentiation of B cells into plasmablasts, anti-CD40 (BD clone HM40-3, 1μg/mL) was added to the culture medium. MDCK cells and RAW macrophages were cultured in DMEM supplemented with 10%FCS, 100mM HEPES and NEAA. For experiments where B cells and MDCK cells were compared side by side, MDCK cells were cultured during the experiment using RPMI media.
Biosynthetic labeling and immunoprecipitation
Cells were starved for 15 minutes in DMEM (Cys-, Met-) or RPMI (Cys-, Met-), supplemented with 10% FCS and then labeled with 35S-methionine/cysteine (276 μCi of Express35S protein labeling mix per 1×106 cells. Cells were washed and lysed in NP40 lysis buffer (25mM Tris pH7.4, 150mM NaCl, 5mM MgCl2, 0.5% NP-40). For immunoprecipitation, lysates were incubated with 20μL of Protein-G agarose and the antibody stated or immune serum. Monoclonal anti M2 antibody was used at 2.5 μg/mL. For all other immunoprecipitations, we used 1 μl of serum per ml of lysate. Incubations with antisera were performed for 3 hours at 4° C. The protein G sepharose beads were washed three times in lysis buffer resuspended in SDS sample buffer. Where glycosidase treatment was required, digestions were performed following manufacturer's instructions (New England Biolabs), prior to addition of SDS sample buffer. To recover released virions form culture supernatants, the amount of supernatant used was normalized to the amount of radioactivity incorporated into cells. The adjusted volumes of supernatant were incubated with chicken red blood cells at a 1:50 dilution for three hours at 4° C. The erythrocytes were washed by centrifugation (3×) with PBS and lysed in NP40-containing lysis buffer. Immunoprecipitates and materials adsorbed onto chicken red blood cells were analyzed by SDS-PAGE and autoradiography.
Production of FluBI hybridoma
Spleen cells from FluBI;RAG2-/- mice were stimulated with 40μg/mL LPS and 20ng/mL IL4 for 5 days, and were fused with NSObcl2 cells (a gift from Betty Diamond, The Feinstein Institute for Medical Research) and selected in medium supplemented with 20% heat inactivated FCS and Hypoxanthine aminopterin thymidine (HAT) and grown in 10% CO2 for 3 weeks, before transfer of positive clones to hypoxantine thymidine (HT) supplemented medium containing 10% heat inactivated FCS . Resulting hybridomas were screened for A/WSN/33 reactivity by ELISA using HRP-coupled anti-IgG2b or anti-Igκ (Southern Biotech) secondary antibodies for detection.
Statistics
Center values are mean. p<0.05 defined as significant. Standard two-sided t-test was used throughout unless otherwise noted. Sample size was based on variability from pilot studies.
Supplementary Material
1
Extended Data Figure 1: Flu micelles stain HA-specific B cells:a) Schematic for preparation of glycoprotein micelles from HA-SRTAlexa647 virus.
b) Immunoprecipitation of HA-Alexa647 with anti- Alexa647 monoclonal antibody. Triton ×100-disrupted virions were incubated with 400μg anti-Alexa647 overnight and HA-Alexa647 was then recovered using protein G-sepharose. Bound proteins were eluted with 0.1M Glycine pH2.8. W=wash; E=elution.
c) Typhoon image of the fractions obtained from a linear sucrose gradient after 20h centrifugation (107,900×g).
d) Fraction 8 from the sucrose gradient was concentrated and sucrose-depleted by centrifugation over a 30kD filter (Amicon UltraCel). The preparation was stained with phosphotungstate and examined by transmission electron microscopy (150,000× magnification).
e) Splenocytes from mice infected with A/WSN/33 or control mice were stained with anti-CD19 and HA-Alexa647 micelles and analyzed by cytofluorimetry. Plots are representative of 6 mice per group.
10
Extended Data Figure 10: Flu-specific VHHs can stain infected FluBI B cells:B cells from OBI or FluBI mice were cultured for 24 hrs in RPMI containing anti-CD40 (1μg/mL) prior to exposure to A/WSN/33. OBI B cells, FluBI B cells and MDCK cells were incubated with A/WSN/33 at an MOI 1.0 for 30min on ice, washed once with PBS, and transferred to 37° C in RPMI (0.2% BSA). At 5 hours post infection, cells were washed, permeabilized, fixed and stained using TAMRA-conjugated flu-specific VHHs (1μg/50μL). Infected MDCK cells were analyzed in parallel as a positive control. Cells were analyzed by cytofluorimetry using a BD Fortessa.
2
Extended Data Figure 2: FluBI antibody is of the IgG2b subclass:ELISA plates were coated with A/WSN/33-infected MDCK cell lysate and exposed to 1:100 diluted serum from a single C57BL/6 (wt), FluBI, FluBI;RAG2-/-, or wild type mouse infected with A/WSN/33. Plates were washed and probed with isotype-specific secondary antibodies. Uninfected wild type mice have flu-reactive antibodies of the IgM subclass. Flu-specific IgE was not detected in any sample. Error bars are SD of samples analyzed in triplicate.
3
Extended Data Figure 3: Sequence of the VDJ and VJ segments of the FluBI antibody:Genomic DNA was prepared from tails of FluBI mice. The heavy and light chain rearrangements were first identified by amplifying and sequencing of the segments with degenerate primers: for heavy chain: forward 5′-ARGCCTGGGRCTTCAGTGAAG-3′ and reverse 5′-AGGCTCTGAGATCCCTAGACAG-3′; for light chain: forward 5′-GGCTGCAGSTTCAGTGGCAGTGGRTCWGGRAC-3′ and reverse 5′-ATGCGACGTCAACTGATAATGAGCCCTCTCC-3′. Then the full sequences of the rearranged heavy and light chain segments were obtained using specific primers: forward 5′- ttactgagcacacaggacctc-3′ and reverse 5′-AGGCTCTGAGATCCCTAGACAG-3′; for light chain: forward 5′-cagcccatattctcccatgt-3′ and reverse 5′-ATGCGACGTCAACTGATAATGAGCCCTCTCC-3′. Amplified products were agarose gel-purified and sequenced. Sequences were aligned to the NCBI mouse V,D, and J genes using IgBlast. Sequences were deposited in GenBank (accession numbers KF419287 and KF419288).
4
Extended Data Figure 4: FluBI mice lack B-1a B cells, but show near-normal development of follicular B cells:Cells were isolated from spleen, lymph node (LN, pooled mesenteric and cervical), peritoneal cavity and bone marrow of FluBI, FluBI RAG2-/-, or C57BL/6 mice. Erythrocytes were lysed and cells were stained with the indicated antibodies and 7AAD viability dye. LN plots were gated on total live cells. All other populations were gated on CD19+ live cells. Numbers indicated the percent of cells in the indicated gates. B-1a B cells (CD5+) are absent and B-1b B cells (CD5-,CD11b+) are reduced in the peritoneal cavity of FluBI and FluBI RAG2-/- mice. Plots are representative of 5 mice per group.
5
Extended Data Figure 5: FluBI B cells are infected by A/WSN/33:CD40-activated OBI or FluBI B cells were incubated A/WSN/33 virus at an MOI 1.0 for 30 min on ice. Cells were then washed and incubated at 37° C in RPMI (0.2% BSA). At 2hpi, cells were fixed, permeabilized and stained with anti-IgG and TAMRA-conjugated anti-NP (VHH54, derived from alpaca; see ED Fig. 9). a) Cells were visualized by confocal microscopy. b) Cells from a were scored as VHH54 positive or negative. Error bars represent SD of positive cells counted/ field (3 fields counted; ~200 total cells were counted per group).
6
Extended Data Figure 6: Antibody secreted by FluBI B cells does not cross-react with other strains of influenza virus:ELISA plates were coated with A/WSN/33 (H1N1), A/Udorn/307/1972 (H3N2), or A/Puerto Rico/8/1934 (H1N1) overnight at 4°. Plates were then washed, blocked with 10% fetal bovine serum, and exposed to FluBI hybridoma supernatant or WSN infected serum at the indicated dilutions. Bound antibody was detected using HRP-coupled anti-IgG2b secondary reagent.
7
Extended Data Figure 7: FluBI B cells are not infected with A/Puerto Rico/8/1934 virus in vivo:C57BL/6 mice were administered 5×106 MHC II GFP+ FluBI B cells 2 hours prior to intranasal infection with 2×105 pfu/mouse of either A/WSN/33 (WSN) or A/Puerto Rico/8/1934 (PR8). Mice were sacrificed 3 days post-infection, and lung resident cells were stained with anti-CD19 and TAMRA-conjugated VHH68 (anti-HA) or TAMRA-conjugated VHH52/54 (anti-NP). a) Representative plots gated on CD19+ cells. b) Quantification of flu-antigen positive cells as shown in a. n=3. Error bars are SD. p=0.06 using two-sided t test.
8
Extended Data Figure 8: Proliferating FluBI cells in the mediastinal lymph node are plasmablasts:a) Mediastinal lymph node cells from Day 6 post live infection mice described in Figure 4 were analyzed by confocal microscopy. GFP+ cells displayed a morphology consistent with plasmablasts.
b) MSLN cells from Day 6 post live infection mice described in Figure 4 were analyzed by cytofluorimetry. Proliferating (violet low) cells were B220low and CD138+.
9
Extended Data Figure 9: Alpaca-derived VHHs recognize HA and NP from A/WSN/33:a) An alpaca was immunized with ethanol-fixed influenza virus. Phage display libraries were constructed from selectively amplified VHH specific cDNA using peripheral blood lymphocytes as starting material, and panned twice against sortase labeled influenza HA-SRTbiotin virus bound to streptavidin coupled beads. VHH sequences obtained from specific binders were expressed with a sortase recognition motif to allow direct conjugation of biotin or fluorophores.
b) VHH54 and VHH68 conjugated directly to agarose beads were used to precipitate lysates of A/WSN/33 infected, 35S-cysteine/methionine labeled MDCK cells.
Acknowledgments
SKD and CG were funded by the Cancer Research Institute. JJC was funded by the Human Frontiers Science Program. FA, RJ and HLP are funded by grants from the NIH. FA is an HHMI investigator. SKD and HLP are funded by the AACR-Pancreatic Cancer Action Network. We are grateful to Patti Wisniewski for cell sorting, to John Jackson for mouse husbandry, to George Bell for statistical analysis, to Nicki Watson for electron microscopy, and to Martin Witte for sortase nucleophiles.
Footnotes
Author contributions: SKD and JA contributed equally. SCNT was performed by SKD. SKD, JA, RAK, MWP, MB, AFA performed experiments. AMA generated hybridomas. JRI and JJC generated flu-specific VHHs. CG sequenced the BCR loci. FWA and RJ provided advice and reagents. SKD, JA, and HLP designed experiments, analyzed data and wrote the paper.
Author information: Reprints and permissions information is available at www.nature.com/reprints.
The authors have no competing financial interests.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nature12637
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3863936
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Antiviral memory B cells exhibit enhanced innate immune response facilitated by epigenetic memory.
Sci Adv, 10(13):eadk0858, 29 Mar 2024
Cited by: 0 articles | PMID: 38552009 | PMCID: PMC10980274
Pandemic influenza A (H1N1) virus causes abortive infection of primary human T cells.
Emerg Microbes Infect, 11(1):1191-1204, 01 Dec 2022
Cited by: 10 articles | PMID: 35317717 | PMCID: PMC9045768
Single-cell genome-wide association reveals that a nonsynonymous variant in ERAP1 confers increased susceptibility to influenza virus.
Cell Genom, 2(11):100207, 01 Nov 2022
Cited by: 3 articles | PMID: 36465279 | PMCID: PMC9718543
Elucidating the characteristics of Mx1 and resistance to influenza A virus subtype H1N1 in the newly developed KWM/Hym mice.
Lab Anim Res, 38(1):28, 08 Sep 2022
Cited by: 1 article | PMID: 36076303 | PMCID: PMC9454180
T Cell Epitope Discovery in the Context of Distinct and Unique Indigenous HLA Profiles.
Front Immunol, 13:812393, 06 May 2022
Cited by: 4 articles | PMID: 35603215 | PMCID: PMC9121770
Review Free full text in Europe PMC
Go to all (51) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (2)
- (1 citation) ENA - KF419288
- (1 citation) ENA - KF419287
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
An influenza haemagglutinin-specific IgG enhances class I MHC-restricted CTL killing in vitro.
Immunology, 73(1):12-18, 01 May 1991
Cited by: 7 articles | PMID: 1646161
Virus-specific antigen presentation by different subsets of cells from lung and mediastinal lymph node tissues of influenza virus-infected mice.
J Virol, 69(10):6359-6366, 01 Oct 1995
Cited by: 76 articles | PMID: 7666537 | PMCID: PMC189535
Lethal coinfection of influenza virus and Streptococcus pneumoniae lowers antibody response to influenza virus in lung and reduces numbers of germinal center B cells, T follicular helper cells, and plasma cells in mediastinal lymph Node.
J Virol, 89(4):2013-2023, 26 Nov 2014
Cited by: 16 articles | PMID: 25428873 | PMCID: PMC4338877
Functional balance between haemagglutinin and neuraminidase in influenza virus infections.
Rev Med Virol, 12(3):159-166, 01 May 2002
Cited by: 392 articles | PMID: 11987141
Review
Funding
Funders who supported this work.
Howard Hughes Medical Institute
NIAID NIH HHS (2)
Grant ID: R01 AI087879
Grant ID: R01 AI033456
NICHD NIH HHS (2)
Grant ID: R37 HD045022
Grant ID: R01 HD045022
NIGMS NIH HHS (2)
Grant ID: DP1 GM106409
Grant ID: R01 GM100518





