Abstract
Free full text

Culturing of Human Nasal Epithelial Cells at the Air Liquid Interface
Abstract
In vitro models using human primary epithelial cells are essential in understanding key functions of the respiratory epithelium in the context of microbial infections or inhaled agents. Direct comparisons of cells obtained from diseased populations allow us to characterize different phenotypes and dissect the underlying mechanisms mediating changes in epithelial cell function. Culturing epithelial cells from the human tracheobronchial region has been well documented, but is limited by the availability of human lung tissue or invasiveness associated with obtaining the bronchial brushes biopsies. Nasal epithelial cells are obtained through much less invasive superficial nasal scrape biopsies and subjects can be biopsied multiple times with no significant side effects. Additionally, the nose is the entry point to the respiratory system and therefore one of the first sites to be exposed to any kind of air-borne stressor, such as microbial agents, pollutants, or allergens.
Briefly, nasal epithelial cells obtained from human volunteers are expanded on coated tissue culture plates, and then transferred onto cell culture inserts. Upon reaching confluency, cells continue to be cultured at the air-liquid interface (ALI), for several weeks, which creates more physiologically relevant conditions. The ALI culture condition uses defined media leading to a differentiated epithelium that exhibits morphological and functional characteristics similar to the human nasal epithelium, with both ciliated and mucus producing cells. Tissue culture inserts with differentiated nasal epithelial cells can be manipulated in a variety of ways depending on the research questions (treatment with pharmacological agents, transduction with lentiviral vectors, exposure to gases, or infection with microbial agents) and analyzed for numerous different endpoints ranging from cellular and molecular pathways, functional changes, morphology, etc.
In vitro models of differentiated human nasal epithelial cells will enable investigators to address novel and important research questions by using organotypic experimental models that largely mimic the nasal epithelium in vivo.

Introduction
Goal of the technique
Epithelial cells (ECs) in the airways are among the first sites to be directly exposed to inhaled environmental stressors, such as pathogens, allergens or air pollutants. ECs are more than a structural barrier: They act as a switchboard to initiate and regulate the respiratory immune response1-3 via the release of soluble mediators4-6 and the expression of ligands/receptors on their surfac 7-9. While immortalized cell lines can be used as models to study respiratory ECs in vitro, they fail to differentiate into heterogeneous cell populations composed of polarized ciliated and mucus producing cells, similar to what is observed in vivo. To recapitulate important characteristics of the respiratory epithelium observed in vivo, primary airway ECs can be used. Therefore, our goal was to optimize our method utilizing nasal ECs (NECs) obtained from human volunteers. The NECs are expanded and cultured in vitro, yielding a cell culture model of differentiated NECs which mimics many of the features of the nasal epithelium seen in vivo.
Rationale
The use of in vitro models with human primary ECs mimicking in vivo situation - as described in this study - gives unique possibilities to study disease specific differences of ECs, physiological parameters associated with the airway epithelium, or the interactions between ECs and other cell types, such as dendritic cell 10, natural killer cells11 or macrophages. Several existing methods utilize bronchial ECs, which can be obtained invasively via brush biopsies during bronchoscopies, or from otherwise discarded lung tissue12-17. In addition, commercial sources to obtain fully differentiated human airway epithelial cell cultures exist (EpiAirway model from MatTek, Ashland, MA , Clonetics from Lonza, Basel, Switzerland, MucilAir from Epithelix Sars, Geneva Switzerland), where investigators can choose from different donors. However, because of the limited pool of commercially available epithelial cells, limited or prescribed set of parameters, the cost associated with commercially obtaining primary human airway ECs, and the limited access to discarded lung tissue or freshly obtained human bronchial biopsy tissue, prohibit many investigators from conducting studies in fully differentiated human airway ECs. Therefore, we set out to develop a technique that will 1) utilize non-invasively obtained human NECs, 2) provide a protocol yielding cultures of differentiated human airway epithelium, and 3) be reproducible in most lab settings with an existing tissue culture infrastructure.
Advantages over Existing Techniques/Models
Obtaining fresh NECs, as compared to bronchial or small airway ECs, is much less invasive, individuals can be biopsied multiple times with no significant side effects, and superficial nasal scrape biopsies can be conducted in diseased populations which would otherwise not qualify for research-related bronchoscopies. Noninvasive superficial scrape biopsies to obtain NECs allow the investigator to set donor specification a priori, including age, gender, disease status, etc. in accordance with the research objective to be accomplished. Thus, when designing research studies investigators are not limited by clinically available tissues/epithelial specimen, purchase expensive differentiated cell culture models from commercial vendors, or design invasive bronchoscopy studies. In addition, the ease to obtain NECs increases the ability to conduct experiments in cells from different donors, which enhances statistical validation of observed findings. Lastly, the nose is one of the first sites to be exposed to any kind of air-borne stressors. Thus, generating organotypic in vitro culture systems mimicking the nasal epithelium are useful for studying inhalation of viral particles, pollutants, allergen, or gases, which can be easily achieved by using this cell culture system.
Protocol
1. Prepare Plastic Ware: Coat Plate
Add 500 μl PureCol (1:100 diluted with sterile water) to each well of a 12-well plate.
Incubate at 37 °C in an incubator for 30 min, remove the PureCol and rinse with HBSS right before the cells are added to the plate (see step 3.1 below).
2. Obtaining and Pretreating Biopsy of Human NECs
Nasal scrape biopsy
Obtain superficial ECs lining the nasal turbinates under direct vision through a 9 mm reusable polypropylene nasal speculum (Model 22009) on an operating otoscope with speculum (Model 21700). This device provides optimal visualization of the nasal turbinates and flexibility of motion.
Perform the biopsies with the subject seated upright in a straight-backed chair with head tilted back slightly or lying in a supine position on an examination table.
Insert the otoscope speculum into the nostril and the inferior turbinate visualized with illumination.
Insert a sterile thermoplastic curette through the speculum with the tip extended distally to the back of the turbinate.
Using gentle pressure on the inferior surface of the turbinate, the curette is drawn across the mucosal surface 5x and retracted. A successful retrieval of mucosal cells held by capillary action will be evident in the cup of the curette.
Place sample in a 15 ml conical tube containing 8 ml of RPMI 1640, transport on ice.
Use forceps to retrieve the probe, dislodge tissue from rhino probe with a P-1000 pipette, discard probe
Centrifuge to pellet (4 °C, 400 x g, 5 min), aspirate supernatant (attention: careful not to aspirate the pellet).
Resuspend the pellet in 1 ml BEGM with 10 μl of 100x DNase 1.
Incubate at RT for 20 min.
Centrifuge (4 °C, 400 x g, 5 min), do NOT aspirate the supernatant!
3. Seed the Cells on Plastic (Day 0)
Remove the PureCol from the plate and rinse with HBSS (see also step 1.2).
Add a minimum volume of BEGM++ media (80-100 μl, until a meniscus of media appears in the center of the well) into 1-4 wells (depending on biopsy size) of the coated plate.
Use a P-1000 pipette to carefully remove the pellet of the biopsy tissue from the tube, without aspirating too much media.
Transfer the pellet into the middle of the wells, keep the biopsy together as a cluster of cells, and do not break up to make a single cell suspension. A good biopsy yields up to 4 wells that can be seeded. Put the plate into tissue culture incubator (37 °C, 5% CO2, >90% relative humidity).
Check after two hours to assure that there is enough media (without disturbing the meniscus).
Day1, 24 hr post-seeding: collect all non-adherent cells of all wells (tilt plate and collect media with cells in a P-1000, collect all wells in a 1.5 ml centrifuge tube).
Centrifuge (4 °C, 400 x g, 5 min).
While centrifuging cell suspension: add about 250 μl of BEGM++ and 250 μl of BEGM+ media mixture to the cells seeded on day 0.
Repeat steps 3.1-3.5 for the non-adherent cells.
Day 2-5: change media of the cells daily; add 500 μl media to each well, reduce the amount of BEGM++ media stepwise (day 2 after seeding: 125 μl BEGM++ plus 375 μl BEGM+, day3 after seeding and the following days: 73 μl BEGM++ plus 427 μl BEGM+).
4. Expanding the Cells in the Flasks
Monitor cells daily; once they have reached 80-90% confluency (usually 5-7 days after the biopsy, if they take longer they won't grow successfully) lift cells as following: carefully aspirate media; add 500 μl of 0.25% trypsin per well, keep them at 37 °C until the cells are detached (it usually takes 2-3 min).
Prepare the needed volume of Soy Bean Trypsin Inhibitor (SBTI) (750 μl per 1 ml Trypsin) in a 15 ml tube.
Dislodge cells by repeatedly pipetting up and down the trypsin solution; transfer detached cells in the 15 ml conical tube with SBTI.
Rinse all wells with in total 1 ml HBSS, add HBSS to the tube with the cells.
Centrifuge to pellet (4 °C, 400 x g, 5 min), aspirate the media, resuspend in 1 ml BEGM+ media and vortex the tube.
Expand cells in a T25 or T75 flask depending on pellet size (this is p1; approximately two confluent wells go into one T75 flask, one confluent well is enough for one T25, usually 2 confluent wells yield one T75).
Add BEGM+ to the flasks (5 ml each for a T75, 3 ml each for a T25).
Add the cell suspension to the flasks. Transfer the flasks into a tissue culture incubator.
24 hr post-flask-seeding: add additional BEGM+ media to the flask (3 ml to a T25, 10 ml to a T75).
maintain the cells in the flasks: Remove media (aspirate from the corner) and add BEGM+ media (5 ml/T25, 20 ml/T75). The media needs to be changed every 2-3 days. Maintain the cells in the flask until they are about 80% confluent (confluency may not be even throughout the entire flask; normally this takes between 7-10 days; if they are not confluent after 10 days they won't grow successfully).
5. Seed the Cells on Tissue Culture Inserts
Remove the media from the flask and add Trypsin (3 ml for T25, 4 ml for T75 flask).
Incubate at 37 °C until the cells are detached (about 2-3 min).
Prepare SBTI (750 μl per 1 ml Trypsin > 2.25 ml for T25, 3 ml for T75) in a 15 ml tube.
Dislodge cells by taping the flask and pipetting up and down and transfer to conical tube with the SBTI.
Rinse the flask with HBSS (1 ml for T25, 2 ml for T75), add the HBSS to the 15 ml tube.
Centrifuge to pellet (4 °C, 400 x g, 5 min), remove supernatant carefully.
Prepare the plates: decide how many plates are going to be seeded, label the plates with sample code and number, passage number (i.e. p2) and date. Add 700 μl BEGM+ media to the basal chamber.
Resuspend the cell pellet in 1 ml BEGM ALI media by vortexing the tube.
Count the cells with a hemocytometer (a T25 usually produces about 4 million cells; a T75 can have up to 20 million cells depending on the islet; use 1-2 million of cells for one 12-well plate) and dilute the cell suspension with BEGM ALI media to the total volume needed for the amount of plates that should be seeded (1.2 x 106 cells in 2.5 ml media per plate).(Note: If part of the cells want to be frozen, label the tube and remove cells here: we usually freeze 2 x 106 cells in 2 ml of freezing media)
Add 200 μl cell suspension to the apical side of each uncoated Corning 12well cell insert (>this will be about 80,000-150,000 cells/insert; 12 mm transwells with 0.4 μm pore polyester (Polyethylene Terephthalate (PET)) membrane insert); transfer into tissue culture incubator.
After 24 hr, change the apical media (200 μl expansion medium).
Maintaining the cell cultures: Change the media (BEGM ALI media; 700 μl basolateral and 200 μl apical) every other day. Inserts have to be maintained submerged until cells are totally confluent (no holes in monolayer are visible). Depending on cell number this usually takes between 2-6 days, if it takes longer the cells will most likely not be viable.
6. Establish Air Liquid Interface Once the Cells are Completely Confluent (When the Cells on the Transwells are Completely Confluent)
Once the cells are completely confluent replace both apical and basolateral media with BEGM ALI media supplemented with 500nM retinoic acid for 48 hr.
48 hr after adding the retinoic acid supplemented BEGM ALI media: Prepare PneumaCult-ALI Maintenance Medium (following manufacturer's instructions): Calculate the volume of medium needed (for one plate usually 8.5 ml for basal compartments); add per 1 ml PneumaCult Complete Base Medium:
10 μl PneumaCult 100x Maintenance Supplement
2 μl Heparin (of a 2 mg/ml stock solution)
5 μl Hydrocortisone (of a 96 μl/ml stock solution).
Remove all media and add 700 μl PneumaCult-ALI maintenance media to the basolateral side only (no medium on the apical side).
Maintain the cell cultures for 4 weeks at the ALI:
Change the media every other day (Monday, Wednesday and Friday schedule works well for us).
After one week at ALI, once a week (Wednesdays) the apical surface is rinsed: add 200 μl HBSS to the apical side, keep them for 10 min in the incubator, carefully aspirate the HBSS (use a 9 in glass pipette attached to the vacuum trap in the Biological safety cabinet, use 200 μl tips on the tip of the pipette to suction off the HBSS).
Note: Change tips between plates, as well as changing the tips when going from apical surface versus basolateral. After about 2 weeks at the ALI, cells start to differentiate and cilia appear. Cell cultures reach full differentiation after about 4 weeks at the ALI.
Representative Results
NECs cultured following the here described protocol re-differentiate into a heterogeneous layer of ECs composed of ciliated and non-ciliated cells, representing the in vivo situation (Figure 1). Some of the differentiated NECs are mucus producing (cells staining in blue using AB/PAS, Figure 1B). Even though the here presented protocol is optimized, differentiation can vary with regards to the distribution of the overall cell populations (i.e. different ratios of ciliated cells: mucus-producing cells: non-ciliated cells) or even yield a more squamous epithelium. These variations can depend on the donor population and therefore represent recapitulation of an underlying disease phenotype, as we have previously demonstrated using NECs from smokers 18. Figure 2 shows examples of suboptimal re-differentiated NECs using a different protocol and media formulations. The cells are squamous and neither cuboidal nor ciliated and do not mimic a pseudostratified respiratory epithelium (Figure 2).
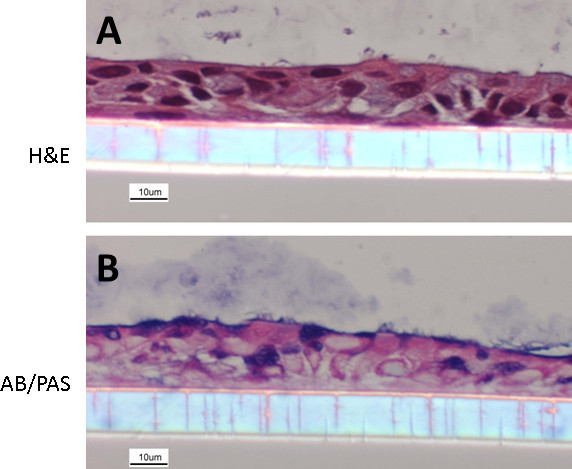
Figure 1. Re-differentiated human NEC culture. NECs were fixed with 4% PFA and embedded in paraffin. Cultures were stained with hematoxylin and eosin (A) and Alcian blue/periodic acid-Schiff (B) (following methods described in19). The images show NECs with a clear organization, a cuboidal structure, mucus-producing goblet cells as well as ciliated ECs.

Figure 2. Hematoxylin and eosin stained paraffin-embedded section of suboptimal cultures. NEC cultures with suboptimal differentiation. Cells appear squamous, no cuboidal structure, no ciliated cells.
Discussion
Respiratory ECs provide not only a physical barrier to protect the human body from exogenous stimuli20, but also act as a switchboard to initiate and regulate respiratory immune defense responses1-3 via secretion of soluble mediators or direct receptor-ligand cell-cell interactions. In order to further study the role of ECs in the immune response, to examine potential differences among ECs from diseased populations, or to study the effects of exogenous stressors on ECs, it is important to recapitulate the in vivo situation. The human airways contain more than 40 different cell types20 including different epithelial cells, such as ciliated cells, goblet cells and basal cells20. Our protocol describes how a superficial nasal scrape biopsy can be processed to yield NECs that can be expanded and cultured to undergo re-differentiation, yielding NEC cultures, which largely mimic the nasal epithelium in vivo.
The use of this in vitro model of organotypic primary human differentiated NECs provides unique possibilities for various applications. It allows studying disease specific differences of NECs (e.g. cystic fibrosis, asthma, smoking), underlying mechanisms of a disease, as well as controlled exposures to air pollutants (e.g. ozone, diesel exhaust, cigarette smoke)18,21-23. In addition, differentiated NECs grown on tissue culture inserts lend themselves to co-culture with other cells types (e.g. dendritic cells, natural killer cells, or macrophages)10,11, allowing to examine cell-cell interactions in a controlled setting.
We describe the culturing and re-differentiating of NECs, and not bronchial epithelial cells, which have been used in many previously published studies24-27. We see various reasons why the use of NECs can be advantageous as compared to the use of bronchial ECs. First, obtaining superficial nasal scrape biopsies is less expensive than performing a bronchoscopy to obtain brush biopsies. Second, for a variety of pre-existing diseases (e.g. sever asthmatics) it is unrealistic to carry out a research-related bronchoscopy to obtained lower airway epithelial cells. However, the less invasive procedure of scraping the inferior surface of the inferior nasal turbinate with a Rhino Probe can be performed in these subjects. Third, nasal scrape biopsies can be performed on the same individuals multiple times (usually about 4 weeks in between biopsies) without any significant side effects28-30 . Fourth, NECs are among the first cells to be exposed to inhaled environmental stressors, such as air pollutants, pathogens or allergens and therefore represent valuable in vitro models to study the effects of these stressors on airway epithelial cells.
Various cell culture systems of re-differentiated human ECs are commercially available (e.g. EpiAirway model from MatTek, Ashland, MA , Clonetics from Lonza, Basel, Switzerland, MucilAir from Epithelix Sars, Geneva Switzerland). Those commercially available cell cultures remain stable for a long time period and are well characterized. However, they are expensive and the studies are usually limited to a low number of samples from different individuals. Additionally, it is difficult to control the donor population, which depends on the availability presented by the supplier. Freshly obtaining NECs with superficial scrape biopsies allows the investigator to control subject characteristics (such as age, gender, race, weight, body mass index, disease status, medication use, etc.) and to re-call volunteers back for a potential follow-up.
In general, all published protocols for re-differentiating human airway (either bronchial or nasal) ECs at the ALI include similar procedures: obtaining ECs, expanding the cells in tissue culture flasks, and transferring them to porous cell culture inserts for the differentiation at the ALI. The protocols vary in the type of media, the growth factor supplements, the number of passaging, the coating of the inserts, and the activation of the NECs before establishing the ALI. While in one protocol, after obtaining and digesting, the cells are seeded directly on uncoated inserts and only kept a few days at the ALI before use13; other protocols include culturing for several weeks to re-differentiate at the ALI12,14,15. There is no consistency about coating the inserts with collagen or other extracellular matrix components: while some protocols require coating12,15 others use uncoated inserts13,14. However, culture conditions are of great importance, especially the incubator settings. The relative humidity, which needs to be >90%, and the CO2, which should be adjusted at 5% are very important factors to avoid drying out of the ALI cell culture and help buffer the media. In order to share our experience, we want to mention here that there is no 100% success rate. Some nasal scrape biopsy samples do not adhere to the tissue culture plate, while some NEC samples will not adhere to the inserts or will never reach confluency. Over the years, our overall success rate using the protocol described here is approximately 80%. Our experience shows that culturing of NECs from defined diseased populations, such as smokers, is more challenging than NECs from healthy non-smokers
Our protocol for culturing of primary human NECs incorporates several key steps which we optimized over the years: 1. The NECs are only expanded twice before they are seeded on the tissue culture inserts. 2. The inserts are used uncoated. 3. Once seeded on tissue culture plates, the amount of NuSerum is stepwise reduced to avoid that the cells lift off from the tissue culture plates. 4. The NECs are kept at least 25-28 days at the ALI before using them in experiments to assure complete re-differentiation. In summary, the protocol described here includes an optimized protocol to culture and re-differentiate human NECs at the ALI.
Acknowledgments
The authors would like to thank Lisa Dailey for the helpful inputs.
This work was supported by grants from the National Institutes of Health (ES013611 and HL095163), the Flight Attendant Medical Research Institute (FAMRI), and the Environmental Protection Agency (CR83346301) and declares no conflict of interest. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Although the research described in this article has been funded in part by the U.S. Environmental Protection Agency through cooperative agreement CR83346301 with the Center for Environmental Medicine, Asthma and Lung Biology at the University of North Carolina-Chapel Hill, it has not been subjected to the agency's required peer and policy review and therefore does not necessarily reflect the views of the agency, and no official endorsement should be inferred. Mention of trade names or commercial products does not constitute endorsement or recommendation for use. Loretta Müller is supported by the Swiss National Science Foundation with a personal grant.
References
- Swamy M, Jamora C, Havran W, Hayday A. Epithelial decision makers: in search of the 'epimmunome. Nat. Immunol. 2010;11:656–665. [Europe PMC free article] [Abstract] [Google Scholar]
- Vareille M, Kieninger E, Edwards MR, Regamey N. The airway epithelium: soldier in the fight against respiratory viruses. Clin. Microbiol Rev. 2011;24:210–229. [Europe PMC free article] [Abstract] [Google Scholar]
- Muller L, Jaspers I. Epithelial cells, the "switchboard" of respiratory immune defense responses: Effects of air pollutants. Swiss Med. Wkly. 2012. [Europe PMC free article] [Abstract]
- Sanders CJ, Doherty PC, Thomas PG. Respiratory epithelial cells in innate immunity to influenza virus infection. Cell Tissue Res. 2011;343:13–21. [Abstract] [Google Scholar]
- Robertson MJ. Role of chemokines in the biology of natural killer cells. J. Leukoc. Biol. 2002;71:173–183. [Abstract] [Google Scholar]
- Adachi M, Matsukura S, Tokunaga H, Kokubu F. Expression of cytokines on human bronchial epithelial cells induced by influenza virus. A. Int. Arch. Allergy Immunol. 1997;113:307–311. [Abstract] [Google Scholar]
- Borchers MT, Harris NL, Wesselkamper SC, Vitucci M, Cosman D. NKG2D ligands are expressed on stressed human airway epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 2006;291:L222–L231. [Abstract] [Google Scholar]
- Groh V, et al. Costimulation of CD8alphabeta T cells by NKG2D via engagement by MIC induced on virus-infected cells. Nat. Immunol. 2001;2:255–260. [Abstract] [Google Scholar]
- Obeidy P, Sharland AF. NKG2D and its ligands. Int J. Biochem. Cell Biol. 2009;41:2364–2367. [Abstract] [Google Scholar]
- Horvath KM, Brighton LE, Zhang W, Carson JL, Jaspers I. Epithelial Cells From Smokers Modify Dendritic Cell Responses in the Context of Influenza Infection 1. Am. J. Resp. Cell. 2010. [Europe PMC free article] [Abstract]
- Muller L, Brighton LE, Jaspers I. Ozone exposed epithelial cells modify co-cultured natural killer cells. AJP Lung. 2013. [Europe PMC free article] [Abstract]
- Turi JL. Oxidative stress activates anion exchange protein 2 and AP-1 in airway epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 2002;283:791–798. [Abstract] [Google Scholar]
- Zimmermann GS, Neurohr C, Villena-Hermoza H, Hatz R, Behr J. Anti-inflammatory effects of antibacterials on human Bronchial epithelial cells. Respir. Res. 2009;10:89. [Europe PMC free article] [Abstract] [Google Scholar]
- Lopez-Souza N, Avila PC, Widdicombe JH. Polarized cultures of human airway epithelium from nasal scrapings and bronchial brushings. In Vitro Cell Dev. Biol. Anim. 2003;39(2003):266–269. [Abstract] [Google Scholar]
- Gray TE, Guzman K, Davis CW, Abdullah LH, Nettesheim P. Mucociliary differentiation of serially passaged normal human tracheobronchial epithelial cells. Am. J. Respir. Cell Mo.l Biol. 1996;14:104–112. [Abstract] [Google Scholar]
- Adler KB, Schwarz JE, Whitcutt MJ, Wu R. A New Chamber System for Maintaining Differentiated Guinea-Pig Respiratory Epithelial-Cells between Air and Liquid-Phases. Biotechniques. 1987;5:462. [Google Scholar]
- Whitcutt MJ, Adler KB, Wu R. A biphasic chamber system for maintaining polarity of differentiation of cultured respiratory tract epithelial cells. In Vitro Cell. Dev. Biol. 1988;24:420–428. [Abstract] [Google Scholar]
- Jaspers I. Reduced expression of IRF7 in nasal epithelial cells from smokers after infection with influenza. Am. J. Respir. Cell Mol. Biol. 2010;43:368–375. [Europe PMC free article] [Abstract] [Google Scholar]
- Muller L, Brighton LE, Jaspers I. Ozone exposed epithelial cells modify cocultured natural killer cells. Am. J. Physiol. Lung Cell Mol. Physiol. 2013;304:332–341. [Europe PMC free article] [Abstract] [Google Scholar]
- Ochs M, Weibel ER. In: Fishman's Pulmoary Diseases and Disorders. Fishmanetal AP, et al., editors. Vol. 4. New York, NY: Mc Graw Hill; 2008. pp. 23–69. [Google Scholar]
- Kesic MJ, Meyer M, Bauer R, Jaspers I. Exposure to Ozone Modulates Human Airway Protease/Antiprotease Balance Contributing to Increased Influenza A Infection. PLoS ONE. 2012;7:e35108. [Europe PMC free article] [Abstract] [Google Scholar]
- Jaspers I. Diesel exhaust enhances influenza virus infections in respiratory epithelial cells. Toxicol. Sci. 2005;85:990–1002. [Abstract] [Google Scholar]
- Jaspers I, Flescher E, Chen LC. Ozone-induced IL-8 expression and transcription factor binding in respiratory epithelial cells. Am. J. Physiol. 1997;272:L504–L511. [Abstract] [Google Scholar]
- Blume C, et al. Barrier responses of human bronchial epithelial cells to grass pollen exposure. Eur. Respir. J. 2012. [Abstract]
- Bauer RN. Influenza enhances caspase-1 in bronchial epithelial cells from asthmatic volunteers and is associated with pathogenesis. J. Allergy Clin. Immunol. 2012;130:958–967. [Europe PMC free article] [Abstract] [Google Scholar]
- Ostrowski LE, Stewart D, Hazucha M. Interferon gamma stimulates accumulation of gas phase nitric oxide in differentiated cultures of normal and cystic fibrosis airway epithelial cells. Lung. 2012;190:563–571. [Europe PMC free article] [Abstract] [Google Scholar]
- Jardim MJ, Fry RC, Jaspers I, Dailey L, Diaz-Sanchez D. Disruption of microRNA expression in human airway cells by diesel exhaust particles is linked to tumorigenesis-associated pathways. Environ. Health Perspect. 2009;117:1745–1751. [Europe PMC free article] [Abstract] [Google Scholar]
- Horvath KM, Brighton LE, Herbst M, Noah TL, Jaspers I. Live Attenuated Influenza Virus (LAIV) induces different mucosal T cell function in nonsmokers and smokers. Clin. Immunol. 2012;142:232–236. [Europe PMC free article] [Abstract] [Google Scholar]
- Horvath KM. Nasal lavage natural killer cell function is suppressed in smokers after live attenuated influenza virus. Respir. Res. 2011;12:102. [Europe PMC free article] [Abstract] [Google Scholar]
- Noah TL. Tobacco smoke exposure and altered nasal responses to live attenuated influenza virus. Environ. Health Perspect. 2011;119:78–83. [Europe PMC free article] [Abstract] [Google Scholar]
Articles from Journal of Visualized Experiments : JoVE are provided here courtesy of MyJoVE Corporation
Full text links
Read article at publisher's site: https://doi.org/10.3791/50646
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3938966?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.3791/50646
Article citations
Understanding the cellular dynamics, engineering perspectives and translation prospects in bioprinting epithelial tissues.
Bioact Mater, 43:195-224, 24 Sep 2024
Cited by: 0 articles | PMID: 39386221 | PMCID: PMC11462153
Review Free full text in Europe PMC
Bioengineering Human Upper Respiratory Mucosa: A Systematic Review of the State of the Art of Cell Culture Techniques.
Bioengineering (Basel), 11(8):826, 13 Aug 2024
Cited by: 0 articles | PMID: 39199784 | PMCID: PMC11352167
Review Free full text in Europe PMC
Building a human lung from pluripotent stem cells to model respiratory viral infections.
Respir Res, 25(1):277, 15 Jul 2024
Cited by: 0 articles | PMID: 39010108 | PMCID: PMC11251358
Review Free full text in Europe PMC
Characteristics of upper respiratory tract rhinovirus in children with allergic rhinitis and its role in disease severity.
Microbiol Spectr, 12(7):e0385323, 23 May 2024
Cited by: 0 articles | PMID: 38780281 | PMCID: PMC11218526
Emissions from plastic incineration induce inflammation, oxidative stress, and impaired bioenergetics in primary human respiratory epithelial cells.
Toxicol Sci, 199(2):301-315, 01 May 2024
Cited by: 1 article | PMID: 38539046
Go to all (76) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Ciliated Epithelial Cell Differentiation at Air-Liquid Interface Using Commercially Available Culture Media.
Methods Mol Biol, 2109:275-291, 01 Jan 2020
Cited by: 22 articles | PMID: 31707647 | PMCID: PMC7116769
Air-Liquid-Interface Differentiated Human Nose Epithelium: A Robust Primary Tissue Culture Model of SARS-CoV-2 Infection.
Int J Mol Sci, 23(2):835, 13 Jan 2022
Cited by: 13 articles | PMID: 35055020 | PMCID: PMC8776210
Characterization of pediatric cystic fibrosis airway epithelial cell cultures at the air-liquid interface obtained by non-invasive nasal cytology brush sampling.
Respir Res, 18(1):215, 28 Dec 2017
Cited by: 17 articles | PMID: 29282053 | PMCID: PMC5745630
From Submerged Cultures to 3D Cell Culture Models: Evolution of Nasal Epithelial Cells in Asthma Research and Virus Infection.
Viruses, 13(3):387, 28 Feb 2021
Cited by: 7 articles | PMID: 33670992 | PMCID: PMC7997270
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NHLBI NIH HHS (2)
Grant ID: HL095163
Grant ID: R01 HL095163
NIA NIH HHS (1)
Grant ID: R03 AG045088
NIEHS NIH HHS (2)
Grant ID: ES013611
Grant ID: R01 ES013611





