Abstract
Free full text

Prostaglandin D2 regulates human type 2 innate lymphoid cell chemotaxis
To the Editor:
Type 2 innate lymphoid cells (ILC2) were identified in 2010 and include natural helper cells, nuocytes, and innate type 2 helper cells.1–3 ILC2 do not express known T-cell, B-cell, or natural killer–cell lineage markers (lineage-negative) and produce large amounts of IL-5 and IL-13 in response to cytokines IL-33, IL-25, or both.1–3 Importantly, ILC2 have been shown to contribute to airway hyperresponsiveness and type 2 lung inflammatory responses in mice infected with influenza virus, and after challenge with multiple allergens including Alternaria, papain, house dust mite, and ovalbumin, suggesting a potential role for ILC2 in the pathogenesis of allergic inflammation and asthma (reviewed elsewhere4). Studies of ILC2 in humans have demonstrated their presence in peripheral blood, the gastrointestinal tract, lung, bronchoalveolar lavage (BAL), and nasal polyps.5–7 Our group7 and others8 have reported that ILC2 in human peripheral blood highly express the master TH2 cytokine transcription factor GATA-3, suggesting that ILC2 are primed for rapid and robust TH2 cytokine production in humans. ILC2 are cells derived from the bone marrow that circulate in the blood and localize to tissues relevant to allergic inflammation including the lung, BAL, and nasal polyps. Although recent studies have provided an insight into ILC2 development and cytokine production, mechanisms that regulate ILC2 recruitment to tissues have not previously been reported.
As human ILC2 express the chemokine receptor CRTH2 (the chemoattractant receptor homologous molecule expressed on TH2 lymphocytes) that binds to prostaglandin D2 (PGD2), we hypothesized that PGD2 may promote one of the steps of human peripheral blood recruitment to tissues, namely, chemotaxis in mucosal sites in which PGD2 is highly expressed. Because PGD2 is released by several cell types important to allergic inflammation including mast cells, macrophages, and eosinophils, activation of these cell types at mucosal sites in the upper or lower airway could promote PGD2-mediated chemotaxis in tissues.
To determine whether PGD2 induced chemotaxis of human peripheral blood ILC2, we recruited 4 atopic subjects (immediate hypersensitivity skin test positive to dust mite, cat, or grass pollen), aged 34.3 ± 18.4 years (3 males and 1 female), and 6 nonatopic healthy volunteers aged 33.3 ± 6.6 years (3 males and 3 females), who each donated blood in a protocol approved by the University of California San Diego Human Subjects Protection Committee. PBMCs were isolated by using density gradient centrifugation in Vacutainer Cell Preparation Tubes with sodium citrate (BD, Franklin Lakes, NJ). PBMCs (1 × 106) in 200 µL RPMI media were applied in duplicate to the upper chamber of a 5-µm pore-size 24-well plate (BD). The lower chambers contained varying concentrations of PGD2 (0, 5, or 25 nM) (Cayman Chemical, Ann Arbor, Mich) each dissolved in 5% dimethyl sulfoxide in PBS. After incubation of PGD2 in the lower chamber for 90 minutes at 37°C, the 5-µm pore-size filter and any remaining cells on the filter were discarded. The plate was then placed on ice for 30 minutes, and cells in the lower chamber were collected from the plate by washing followed by staining for fluorescence-activated cell sorting analysis to detect the number of ILC2 that had chemotaxed into the lower chamber.
To detect ILC2, cells in the lower chamber were stained with a fluorescein isothiocyanate lineage cocktail (CD3, CD14, CD16, CD19, CD20, CD56; BD), TCRgd (BD), CD4, CD11b, CD235a, and FceRI (eBioscience, San Diego, Calif), Alexa-647 conjugated anti-CD127 (eBioscience), and biotin-conjugated CRTH2 (Miltenyi, Auburn, Calif) followed by streptavidin phycoerythrin as previously described in this laboratory to detect human peripheral blood ILC2.7 This staining protocol defines ILC2 as lineage-negative lymphocytes (do not express T-cell, B-cell, natural killer–cell, mast-cell, and basophil markers) that do express CRTH2 as well as the IL-7 receptor CD127 (Fig 1). Flow cytometry was performed by using the BD Accuri FACS machine. ILC2 represent approximately 1% to 20% of lineage-negative lymphocytes and 0.01% to 0.04% of total lymphocytes in the PBMC populations applied to the upper chamber.
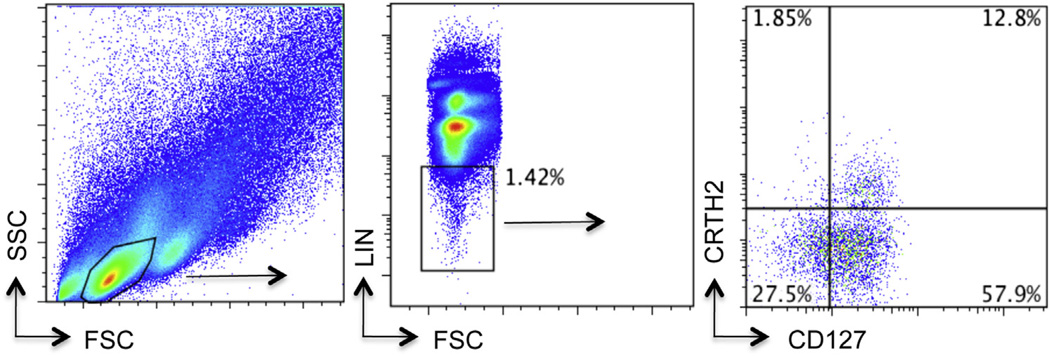
ILC2 FACS: Lymphocytes were identified from whole PBMCs (left panel), and lineage-negative cells gated (middle panel). Lineage-negative lymphocytes were further assessed for the expression of CD127 and CRTH2 (right panel). ILC2 were identified as lineage-negative CD127+ CRTH2+ lymphocytes and represent 12.8% of the cells in this subject (range 1%–20% in all subjects). FACS, Fluorescence-activated cell sorting; FSC, forward scatter; LIN, lineage; SSC, side scatter.
In allergic subjects we detected a dose-dependent increase in ILC2 migration in the presence of 5 to 25 nMof PGD2 (Fig 2, A). Higher concentrations of PGD2 did not increase ILC2 chemotaxis (data not shown). PGD2 also induced chemotaxis of ILC2 in normal healthy controls, but to a lesser degree than in allergic donors (Fig 2, B). ILC2 in allergic individuals may therefore be primed for chemotaxis. Preincubation of PBMCs containing ILC2 with ramatroban, a pharmacologic CRTH2 antagonist,9,10 blocked ILC2 chemotaxis in response to PGD2 (see Fig E1 in this article’s Online Repository at www.jacionline.org). In experiments in which PGD2 was added to both the upper and lower wells, the predominant effect of 25 nM of PGD2 was on ILC2 chemotaxis (P < .01) (see Fig E2 in this article’s Online Repository at www.jacionline.org). Previous studies have demonstrated that the effect of PGD2 on eosinophils and basophils is predominantly chemotactic11 but also chemokinetic especially at low 1 to 10 nM concentrations of PGD2.12 Because we studied impure preparations of ILC2, we were not able to determine whether PGD2 acts directly on ILC2 or indirectly through other cells present in the PBMCs studied.
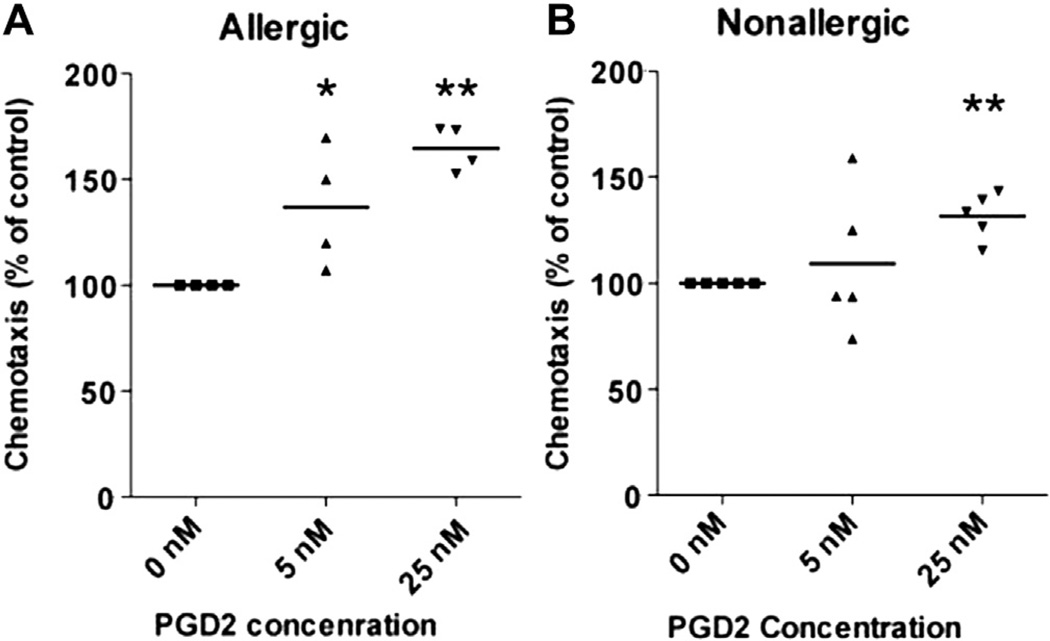
ILC2 chemotaxis to PGD2: PBMCs (Fig 2, A: n = 4 allergic subjects; Fig 2, B: n = 6 nonallergic controls) were incubated in duplicate in the upper chamber of a 5-µm pore-size plate. Varying concentrations of PGD2 were placed in the lower chamber. Each PGD2 concentration (5 nM PGD2, 25 nM PGD2), as well as control 0 nM PGD2, was dissolved in 5% DMSO in PBS. The number of ILC2 in the lower chamber was assessed by using FACS at 90 minutes. DMSO, Dimethyl sulfoxide; FACS, fluorescence-activated cell sorting. *P < .05 and **P < .01.
Thus, PGD2 induces chemotaxis of human peripheral blood ILC2 that express the PGD2 receptor CRTH2. PGD2 plays a role in allergic disease and has previously been shown to induce chemotaxis of TH2 cells, eosinophils, and basophils via CRTH2.11 Our novel findings suggest that human ILC2 chemotaxis is also induced by PGD2. Human ILC2 have been detected in blood, lung, BAL, and nasal polyps, and potently produce TH2 cytokines.5,6,8 The mechanism by which human (or mouse) ILC2 migrate from the bone marrow through the circulation into the tissues has not been reported. Our studies suggest that PGD2 may contribute to one step of the tissue recruitment of ILC2, namely, the chemotaxis of ILC2 to tissue sites in which PGD2 is released such as the upper and lower airway in asthma and allergic rhinitis. CRTH2 antagonists currently undergoing clinical trials in asthma and allergic rhinitis for their inhibitory effects on TH2 cell recruitment may also inhibit ILC2 recruitment into tissues.
Acknowledgments
This study was funded by the National Institutes of Health (NIH) grant numbers AI 38425, AI 70535, AI 107779, and AI 72115 to D.B., NIH KO8 AI 080938 and American Lung Association/American Academy of Allergy, Asthma & Immunology Allergic Respiratory Diseases Award to T.D., and NIH grant ULRR031980 and UL1TR000100 of Clinical and Translational Science Award funding to University of California San Diego Clinical and Translational Research Institute.
T. Doherty has received research support from the National Institutes of Health (NIH) and the American Academy of Allergy, Asthma & Immunology/ALA and has grants/grants pending with the NIH. R. Baum has received research support from the NIH. D. Broide has received research support from the NIH, consultancy fees from Kyowa Kirin California, and has grants/grants pending with the NIH, Immune Tolerance Network, and the Department of Defense.
Footnotes
Disclosure of potential conflict of interest: J. E. Chang claims no relevant conflicts of interest.
REFERENCES
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.jaci.2013.09.020
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3943597?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1016/j.jaci.2013.09.020
Article citations
Tuft cells in the intestine, immunity and beyond.
Nat Rev Gastroenterol Hepatol, 26 Sep 2024
Cited by: 0 articles | PMID: 39327439
Review
Deciphering the Interplay between the Epithelial Barrier, Immune Cells, and Metabolic Mediators in Allergic Disease.
Int J Mol Sci, 25(13):6913, 24 Jun 2024
Cited by: 1 article | PMID: 39000023
Review
Exploring the immunopathology of type 2 inflammatory airway diseases.
Front Immunol, 15:1285598, 12 Apr 2024
Cited by: 0 articles | PMID: 38680486 | PMCID: PMC11045947
Review Free full text in Europe PMC
Mast Cells in Aspirin-Exacerbated Respiratory Disease.
Curr Allergy Asthma Rep, 24(2):73-80, 13 Jan 2024
Cited by: 0 articles | PMID: 38217825
Review
Phospholipid scramblase-1 regulates innate type 2 inflammation in mouse lungs via CRTH2-dependent mechanisms.
J Clin Invest, 133(15):e169583, 01 Aug 2023
Cited by: 0 articles | PMID: 37289545
Go to all (82) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The prostaglandin D₂ receptor CRTH2 regulates accumulation of group 2 innate lymphoid cells in the inflamed lung.
Mucosal Immunol, 8(6):1313-1323, 08 Apr 2015
Cited by: 148 articles | PMID: 25850654 | PMCID: PMC4598246
Cysteinyl leukotriene E4 activates human group 2 innate lymphoid cells and enhances the effect of prostaglandin D2 and epithelial cytokines.
J Allergy Clin Immunol, 140(4):1090-1100.e11, 20 Jan 2017
Cited by: 82 articles | PMID: 28115217 | PMCID: PMC5624780
Prostaglandin D2 inhibits fibroblast migration.
Eur Respir J, 19(4):684-689, 01 Apr 2002
Cited by: 24 articles | PMID: 11998998
The impact of the prostaglandin D2 receptor 2 and its downstream effects on the pathophysiology of asthma.
Allergy, 75(4):761-768, 20 Aug 2019
Cited by: 15 articles | PMID: 31355946
Review
Funding
Funders who supported this work.
NCATS NIH HHS (2)
Grant ID: UL1 TR000100
Grant ID: UL1TR000100
NCRR NIH HHS (1)
Grant ID: UL1 RR031980
NIAID NIH HHS (10)
Grant ID: AI 107779
Grant ID: R37 AI038425
Grant ID: KO8 AI 080938
Grant ID: K08 AI080938
Grant ID: R01 AI072115
Grant ID: R01 AI038425
Grant ID: R01 AI107779
Grant ID: AI 72115
Grant ID: AI 70535
Grant ID: U19 AI070535





