Abstract
Free full text
Next-generation sequencing and large genome assemblies
Abstract
The next-generation sequencing (NGS) revolution has drastically reduced time and cost requirements for sequencing of large genomes, and also qualitatively changed the problem of assembly. This article reviews the state of the art in de novo genome assembly, paying particular attention to mammalian-sized genomes. The strengths and weaknesses of the main sequencing platforms are highlighted, leading to a discussion of assembly and the new challenges associated with NGS data. Current approaches to assembly are outlined and the various software packages available are introduced and compared. The question of whether quality assemblies can be produced using short-read NGS data alone, or whether it must be combined with more expensive sequencing techniques, is considered. Prospects for future assemblers and tests of assembly performance are also discussed.
Genome assembly continues to be one of the central problems of bioinformatics. This is owing, in large part, to the continuing development of the sequencing technology that provides ‘reads’ of short sequences of DNA, from which the genome is inferred. Larger sets of data, and changes in the properties of reads such as length and errors, bring with them new challenges for assembly. For the earliest sequencing efforts using the whole-genome shotgun (WGS) approach, in which reads are generated from random locations across the entire genome, assembly could be dealt with by arranging print-outs of the reads by hand. Through the next three decades, Sanger capillary sequencing gained substantially in throughput, and WGS became practical for increasingly large and complex genomes, from tens of kilobases in the early 1980s to gigabases by 2001 [1]. In line with this, assembly went on to use not only increasingly powerful computational means, but also increasingly time and memory-efficient assemblers.
A further revolution in sequencing began around 2005, when second-generation sequencing (SGS) technologies began to produce massive throughput at far lower costs than Sanger sequencing, enabling a mammalian genome to be sequenced in a matter of days [2]. De novo assemblies of the Panda [3] and Turkey [4] genomes have now been made using SGS data alone, and several human resequencing projects have been completed [5-7]. The disadvantages of the new technologies lie primarily in the short lengths of reads and, in some cases, higher error rate. So-called third-generation sequencing technology is now available, and promises similar throughput, lower costs and longer read lengths, as well as novel read types. These innovations promise to change the game once more.
Assembly is not at all a trivial task. Repeated sequences of DNA make it difficult to infer the relative positions in the genome corresponding to reads, and they occur far more often in real genomes than they would in a sequence of independently randomly generated bases. Overcoming this problem, as well as correcting for errors in reads and taking heterozygosity into account, all while staying within the bounds of practical computability, make assembly a complex and difficult challenge, which is often qualitatively altered by advances in technology. In particular, many assemblers designed to handle Sanger reads were found to be impractical when dealing with next-generation sequencing (NGS) data. In response to this, several new assemblers have been developed, employing qualitatively new approaches, and the field continues to develop rapidly. It is thus of interest at this time to ask whether the resulting de novo assemblies are of good enough quality to replace assemblies based on more expensive techniques, at least for certain purposes. It is also of use to compare the strengths and weaknesses of existing methods.
In the field of pharmacogenomics, data from DNA sequencing are used to find genetic variations associated with drug efficacy and toxicity. The area has been pushed forward by the rapid development of NGS technologies. Reference-guided alignment methods can detect SNPs and short indel variants. However, the SNPs identified to date have been found to account for only 30–50% of the observed variations in drug response [8]. The copy number variation (CNV), another major form of human genetic variation, and its significance in pharmacogenomics, needs to be fully investigated. It is known that CNVs involve some known metabolizing enzymes, such as CYP2D6, GSTM1 and potential drug targets such as CCL3L1, and can influence the phenotype through alteration in gene dosage, structure and expression [9]. However, the identification of CNVs using NGS data poses significant challenges, particularly for large insertion sequences. Despite this, the first analysis of structural variation detection by whole-genome de novo assembly was recently reported [10]. The findings demonstrate that whole-genome de novo assembly is a feasible approach to deriving more comprehensive maps of genetic variation. More recently, a graph-based assembly method, which uses a human reference as well as homology among individual samples, was developed to detect different forms of variation from a population [11]. There are several earlier reviews on assembly using NGS data [12-14]. Reviews concerning particular applications of these methods are also available, including finding genetic variations in plants [15] and the study of cancer [16,17].
NGS technologies & platforms
DNA sequencing is a fast-moving area with technologies and platforms being updated at a blistering pace. The hallmark of NGS has been a massive increase in throughput and decrease in price as compared with previous technologies: SGS sequencing can now be 10,000-times cheaper per base than typical Sanger capillary sequencing. As far as assembly is concerned, the available platforms are distinguished by possible read length, biases in coverage and error profile. Below, we outline the characteristic features of the most commonly used NGS platforms. While exact specifications are likely to change rapidly, Glenn gives details of state of the art as of May 2011 [18]; an update is planned for May 2012. Some of those figures are given in Table 1 along with updated information from other sources cited below including, where noted, the instrument manufacturers.
Table 1
| Platform | Read length | Throughput/ run | Approximate time/run | Machine cost (US$) | Reagent cost (US$) | Reagent cost/Gb (US$) | Primary error | Base error rates |
|---|---|---|---|---|---|---|---|---|
| HiSeq™ 2000 | 100 bp | 600 Gb | 11 days | 690,000 | 23,470 | 40 | Substitution | ~1–2% over 100 bp |
| SOLiD™ 4 | 75 bp | 100 Gb | 12 days | 475,000 | 8128 | <110 | A–T bias | 0.06% |
| SOLiD™ 4hq | 75 bp | 300 Gb | 14 days | 595,000 | 10,503 | 70 | A–T bias | 0.01% |
| SOLiD™ PI | 75 bp | 77 Gb | 8 days | 349,000 | 6101 | 80 | A–T bias | 0.01% |
| 454GSFLX Titanium XL+ | 700 mean bp, ≤1000 bp | 700 Mb | 23 h | 500,000 | 6200 | 7000 | Indel | 0.5% |
| lonTorrent™ PGM™ 316 | 200 bp | 100 Mb | ~2 h | 50,000 | 750 | <7500 | Indel | 1.2% over 150 bp |
| lonTorrent™ PGM™ 318 | 200 bp | 1 Gb | ~2 h | 50,000 | 925 | <925 | Indel | 1.2% over 150 bp |
| MiSeq® | 150 bp | >1 Gb | 27 h | 125,000 | 750 | 740 | Substitution | ~1–2% over 100 bp |
| 454 GS Junior | 400 mean bp | 35 Mb | 12 h | 108,000 | 1100 | 22,000 | Indel | 1.00% |
| PacBioRS (early 2012) | 2700 mean bp, ≤5000 bp | 90 Mb per cell | <1 day (?) | 695,000 | 110–1700 | 11,000- 340,000 | CG deletion | 13.00% |
Currently available next-generation sequencing technologies. All specifications are liable to rapid change. Cost information is taken from Glenn [18], but can vary widely by country and individual deal. The read lengths given are taken from manufacturers websites [101-105], and are the maximum over all currently available protocols with the system in question. The same holds for throughput, and times are the shortest for the given throughput. Base error rate, from the same sources, should be taken as a rough indicator only as the details of errors vary. It is an average for the read, or for a fixed number of bases from the start of the read (which is stated in this case).
The first next-generation DNA sequencing machine, the GS20, was introduced to the market by 454 Life Sciences (Basel, Switzerland) in 2005. The technology is based on a large-scale parallel pyrosequencing system, which relies on fixing nebulized and adapter-ligated DNA fragments to small DNA-capture beads in a water-in-oil emulsion. The DNA fixed to these beads is then amplified by PCR. The very latest 454 GS FLX Titanium XL+ claims an average read length of 700 bp with some reads up to 1000 bp in length [101]. With an advantage in sequencing length, it enables a variety of applications including de novo whole-genome sequencing, re-sequencing of whole genomes and target DNA regions, metagenomics and RNA analysis. Characteristic errors include exact number in homopolymer lengths, an error that is a type of indel specific to the 454 sequencing method [19].
The release of Illumina’s (CA, USA) Genome Analyzer in 2007 marked a true revolution for genome sequencing, in which short reads became significant to genomic applications. The technology is based on reversible dye terminators. DNA molecules are first attached to primers on a slide and amplified so that local clonal colonies are formed (bridge amplification). Four types of reversible terminator (RT)-bases are added, and nonincorporated nucleotides are washed away. Unlike pyrosequencing, the DNA can only be extended one nucleotide at a time, similar to Sanger sequencing. Read length and throughput have undergone rapid changes in the last few years, from 35 bp length reads with 1 Gb throughput using the Genome Analyzer to protocols now available yeilding 100 bp reads with 600 Gb using HiSeq 2000. These protocols generate read pairs (see below) [102]. Base substitutions are the most common error type for this platform [20]. Owing to its high accuracy (base error rate of raw sequencing data <1%) and relatively low costs, these platforms have been widely used for applications in resequencing [21,22], de novo assembly and RNA-seq analysis [23,24] among others.
Life Technologies’ (CA, USA) SOLiD™ technology employs sequencing by ligation. Here, a pool of all possible oligonucleotides of a fixed length are labeled according to the sequenced position. Oligonucleotides are annealed and ligated; the preferential ligation by DNA ligase for matching sequences results in a signal informative of the nucleotide at that position. SOLiD generally has more reads than its competitors, but with a shorter read length [103]. Most importantly, the use of color spaces rather than sequence bases in the earlier versions of platforms hampered its applications in de novo assemblies.
So-called ‘third-generation’ technologies directly sequence individual DNA molecules rather than relying on amplification prior to sequencing. The recently released PacBio RS system can produce 35–45 megabases of data per SMRT® cell with an average read length of 1500 bp. The latest C2 chemistry can produce reads with an average read length of 2700 bp [104]. The method used is sequencing by synthesis, which has a high base error rate of ~13–15% in raw data. However, the high base error rate can be traded in for read length, basically by reading the same sequence more than once and/or by means of computational processes. As well as reads of the usual type, ‘strobe reads’ can be produced, which cover larger ranges in the genome but contain several unsequenced gaps whose size is approximately known.
The Ion Torrent™ Personal Genome Machine™ (PGM™) is another third-generation platform that uses standard sequencing chemistry, but with a novel, semiconductor based detection system [25]. The method of sequencing is based on the detection of hydrogen ions that are released during the polymerisation of DNA. This technology already claims read lengths of approximately 200 bp with high accuracy, and the latest PGM 318 chip can produce 1.0 Gb of data in a 2-h run [105].
With low machine costs, short sequencing time and reasonable amount of throughput, desktop sequencers such as Ion Torrent PGM, and its second-generation technology competitors, Illumina’s MiSeq® and 454 GS Junior, offer exciting prospects for diagnostic sequencing in future medical care. With the 318 chip, IonTorrent competes with MiSeq on throughput and cost, and MiSeq’s read length is a little shorter at 150 bp. The 454 GS Junior achieves longer reads, with a mean length of 400 bp, at the penalty of lower throughput and, as a result, higher cost per Gb.
Most NGS platforms require that template DNA is short, typically 200–1000 bp (short insert size) and that each template contains forward and reverse primer-binding sites. Libraries can be constructed so that the sequencing machine reads the DNA starting from both ends of the template fragment, producing two reads that overlap or are separated by a short gap of approximately known length. This process is called paired-end sequencing for short insert sizes. ‘Mate-pair’ libraries, prepared using more complex techniques, provide for larger separations between pairs of reads. The insert size in mate pair libraries varies from 2 to 40 Kb [26,27]. Using Bacterial Artificial Chromosome (BAC) techniques, inserts of 150 Kb can be produced, but at higher cost. The mate-pair type of data is essential for establishing long-range continuity in de novo assemblies, especially with short reads where other long-range information is lacking. Errors, however, are common for long insert sizes. A large proportion of read pairs can be ‘chimeric’ (from random, unrelated places in the genome). Duplicates of read pairs are often found, reducing true coverage. Thirdly, for some protocols employing DNA fragment circularization, it can happen that two reads are made unexpectedly close to each other and with the wrong orientation (‘cross-biotin’ pairs). The variance of the insert size also affects the usefulness of pair information, as does any departure of the distribution of insert size from the normal distribution, which can be pronounced with many common protocols.
Overview of assembly methods
Assembly would be an easy task, if it could be determined whether (and by how much) given reads correspond to overlapping positions on the genome. Reads are said to ‘overlap’ if there is a match between the sequence at the beginning of one read and the end of the other that is long enough to be reliably distinguished from a random event. This is the case if they are from overlapping locations on the genome, but the converse is not true: the reads may have arisen from two different copies of the same sequence. This complicates assembly. Consider a genome that contains the concatenation of the three sequences A, X and B, and elsewhere contains the concatenation of the sequences C, X and D. If the sequence X is longer than the longest read, overlap information alone cannot be used to rule out possibility that the genome contains the sequence A, X, D, which may not in fact be part of the genome. For this reason, assembled sequences must end at the boundaries of such repeats.
Figure 1 shows how this problem affects the best attainable quality of assemblies for four genomes: human (Homo sapiens), mouse (Mus musculus), fruit fly (Drosophila melanogaster) and the malaria parasite (Plasmodium falciparum). It shows a statistic summarising the lengths of sequences from the genome that could be successfully reconstructed, if the only factor obstructing assembly was ambiguity caused by repeats (as opposed to sequencing errors, errors caused by heuristic computing methods and so on). For each position in the reference genome, we first calculated the uniqueness of the following sequence of length y in both strands (forward and reverse complement) and then marked those unique positions over the whole genome. Continuous intervals of marked positions were treated as assembled sequences; in other words, these contiguous sequences end at the boundaries of repeats in the genome that are longer than y bases. The lengths of the resulting pieces are summarized using the N50 statistic. The N50 of a set of sequences is the maximum length for which a subset of longer or equal sequences can be found whose combined length is over half of the total length of all sequences. The result is an upper bound on the N50 of assemblies produced from WGS data with reads of length y.
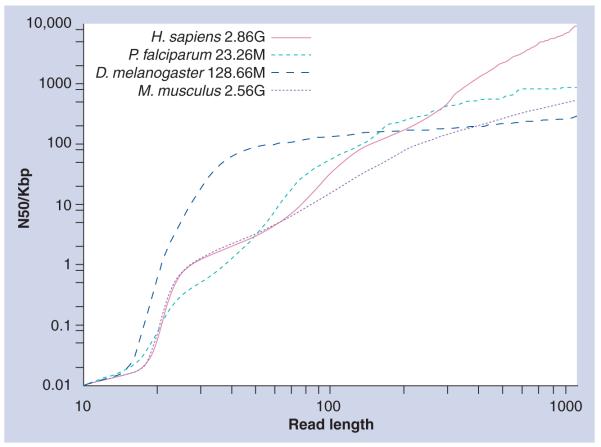
For a set of sequences, the N50 is the number of bases in the longest sequence such that 50% of the total bases are contained in this sequence or longer sequences. Here, the N50 is given for the set of contiguous sequences of bases in each genome that are covered by a unique segment of sequence at a given length y. Owing to the ambiguities in ordering caused by nonunique sequences, this provides an upper bound on the N50 that is possible for whole-genome sequencing assembly when using reads below this length, and gives an indication of the advantages to be gained from longer read length in some cases.
Here we see the gains in ideal assembly quality that can be made with longer read length: for the human genome the possible N50 ranges from approximately 3–32 Kb as we increase read length of single-end reads from 50 to 100. Similarly, using reads of length 1000, an N50 of 8978 Kb is ideally achievable. It is also evident that repeat structure varies considerably even in mammals. Realistically, assembly quality is further limited by read errors and suboptimal assembly algorithms.
Early assemblers for viral genomes used a simple ‘greedy’ algorithm; however, larger and more repetitive genomes called for a more cautious approach, in which two sequences are not merged if either one of the sequences can be extended in conflicting ways. This stage results in a number of separate sequences or ‘unitigs’ that terminate at the boundaries of repeats, or more generally a set of contiguous sequences from the genome called ‘contigs’ [28]. Read errors can also be corrected during assembly by comparing overlapping sequences and choosing the most likely version of any difference by various means. A ‘scaffolding’ stage follows, in which read-pair data is used to find the approximate distances between nonrepetitive contigs in the genome, producing a sequence with ‘gaps’ of undetermined bases. Repeats are retained and can be inserted back into the sequence when the order of the contigs bordering each copy is no longer ambiguous, and then overlaps between remaining reads are used in attempts to close any remaining gaps. Repeat contigs can be identified, up to some error, by the relatively high density of reads mapping to them, and in principle by the multiple ways of extending the contig by overlap and/or multiple conflicting contigs connected to them by read pairs. The process can be expressed using an ‘overlap graph’ in which the reads are nodes and the (directed) edges represent overlaps (Figure 2a & 2b). The genome corresponds to one of the paths through the graph. The division into stages of assembly (including error correction), scaffolding and gap closure has remained in place up to the present.
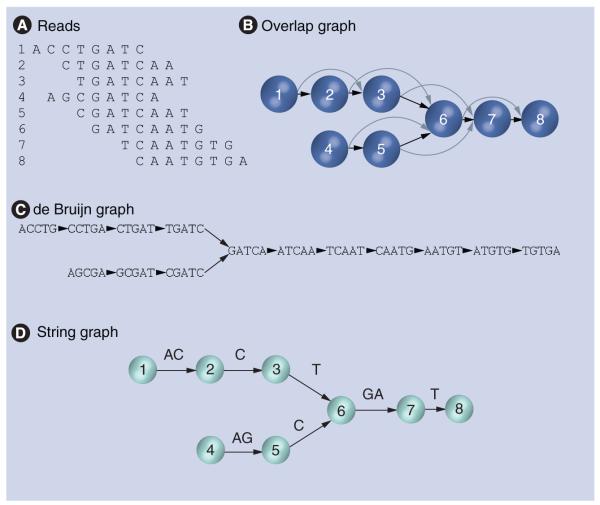
(A) Eight aligned reads are shown. (B) The corresponding overlap graph, in which nodes correspond to reads and edges to overlaps, in this case overlaps of five or more bases (transitive edges, meaning overlaps that are covered by a set of shorter overlaps, are shown as curved arrows). (C) The de Bruijn graph, in which nodes are k-mers and edges indicate that some read contains both k-mers consecutively. Note that reads such as two add nothing to the de Bruijn graph. The basic idea of the string graph is illustrated under (D). Here, the graph topology is the same as in (A) with transitive edges removed; however, but nodes correspond to the beginnings of reads, and edges are labeled by the string between these two points in the case that those reads overlap, rather than the whole read being associated to each node. All sequences supported by reads and overlaps can be recovered from this labeling (along with terminal reads such as read 8 in the example given in this figure): following the graph backwards adds the sequence necessary to complete the previous read. Both the de Bruijn and string graphs can be further simplified by merging linear subgraphs. Treatment of reverse complement sequences has been neglected here for clarity.
New problems arose in the new era of high-throughput sequencing. Assembly is confounded by locations in which there are not enough overlaps to extend the sequence with confidence, and shorter read lengths imply a larger expected number of these coverage gaps when the average coverage is held constant. For Sanger reads, models show that, ideally, it is sufficient for each base in a mammalian sized-genome to be covered by at least three reads on average (written as 3× coverage) [29]; however, for the new short reads, this figure rises to around 30×. Correction of the larger error rate also requires a higher coverage. In practice assemblies of large genomes used coverage of between 7× and 10× in the previous sequencing era, and have begun to use 50× coverage, 100× coverage or even higher with NGS technology. Furthermore, no amount of coverage will make repeats disappear, and with shorter reads, less repeats can be resolved without turning to read-pair data, introducing a new problem for generating long contigs. While it quickly became possible to produce such datasets cheaply in the laboratory, even for large genomes, assembling them proved impossible for most of the previously existing tools.
This problem led to the wide adoption of de Bruijn graph methods [30]. In this approach, instead of storing information about reads and overlaps explicitly, the nodes of the graph are sequences of a fixed length k or ‘k-mers’. All such k-mers that appear in some read are included, and an edge is placed between all pairs of k-mers that appear consecutively in some read. Again, the genome corresponds to a path in this graph. This structure is sketched in Figure 2C. A read whose (k+1)-mers are all contained in other reads adds nothing to the graph, and so memory requirements scale well with coverage. The same is true for processing time: constructing the de Bruijn graph only requires recording the k-mers in the read, rather than explicitly constructing scored overlaps for each pair of reads. Unambiguous contigs are now represented by nonbranching paths, while the ambiguities at the boundaries of repeats are explicitly represented in the graph as branch nodes. Most popular assemblers merge nonbranching paths of k-mers into one node, thereby saving further space. Scaffolding and gap closure can proceed after these unambiguous contigs are found.
Read errors pose a problem for this improved scaling behaviour. A single-base error in the middle of a read changes k of its k-mers to ones which are likely to be uncommon in the other reads. Many assemblers make use of this very property to find such errors [31], although genuine k-mers can be lost without further conditions. Using the topology of the graph to find errors improves this and is now widely implemented [32,33]. For example, errors at the end of reads (which are common in, e.g., Illumina reads) correspond to short chains of k-mers that only connect to the rest of the graph at one end ‘tips’, while errors in the middle of reads give two paths starting and ending at nearby locations with similar sequence. A complication here is that, for diploid or polyploid genomes, sequences that should be mapped to the same position in the genome may have genuine discrepancies. Most pipelines do not attempt to explicitly construct the alternative alelles for regions of sequence variation when building contigs, instead producing one representative haplotype. Because of this, when there is sequence variation assemblers must either construct two contigs or treat the differences as error and merge the contigs. The former can lead to misassembly errors, and so generally efforts are made to avoid this occurrence.
One drawback of the de Bruijn graph approach is the loss of information from reads. Repeats longer than a k-mer cannot be resolved using only the de Bruijn graph described above, even if reads bridge the repeat (in assembly terminology, the approach is not ‘read consistent’). This is a problem that some de Bruijn assemblers remedy by adding information on reads’ paths through the graph, at the cost of more computational resources.
The choice of k involves a number of trade-offs. The longer the k-mers are, the fewer edges are needed, decreasing computational requirements. But with greater length, more bits are required to store individual k-mers. On balance, it is generally true that the use of large k-mers requires more memory for the same assembler. The main advantage of larger k is the retention of more information about short repeats; however, only read overlaps of more than k-1 bases are reflected in the graph, and so, as well as greater memory requirements, larger k means that more coverage is needed to find enough overlaps. This is not a problem when read lengths are as small as 25 bp; however, typical values of k used in studies have not stayed approximately the same as read lengths as they have approached 100 bp. The alternative is to use smaller k, which either means losing read information on short repeats (which is the main reason for preferring longer read lengths in the first place) or retaining even more information from reads.
The string graph, represented in Figure 2D, is another way to compress read and overlap data [34]. Here, the overlaps of all pairs of reads must be calculated. Unlike the overlap graph, however, the edges in the graph carry the sequence information and the nodes represent the beginning or ends of overlaps. First, reads that are contained in other reads can be discarded as they add nothing to the set of possible genomic sequences (neglecting error correction). These sequences can be represented as concatenations of the ‘overhangs’ of overlaps, where an overhang is the part of one overlapping read not covered by the other. The string graph has nodes corresponding to the start and end point of each read (i.e., the boundaries of the overhangs) and edges corresponding to the overhangs running between them labeled by the corresponding sequence. Non-minimal overhangs that contain several smaller ones add no extra implied sequences and can be discarded, saving memory in comparison with the overlap graph approach. Algorithms for this stage scale linearly in time with the number of edges. After this stage, nonbranching paths in the string graph can be merged into one edge corresponding to a unitig in the overlap graph approach. A common simplifying assumption here (and in the de Bruijn approach) is that the genome is the shortest nonbranching path through the graph that contains all edges (or nodes in the de Bruijn graph). This has the advantage of picking out a unique order of copies of contigs when that path is unique, although it is not obvious how close to reality this assumption will typically be. Scaffolding and gap closure follow the assembly stage as above.
Like the de Bruijn graph, boundaries of repeats are branch nodes in the string graph. However, the string graph does not lose information from reads on short repeats. The disadvantages include the need to calculate all overlaps on a pairwise basis rather than comparing k-mers in each read to the set of previously found k-mers, although efficient new algorithms massively reduce time and memory requirements for this.
The other stages of assembly also change with NGS data. As with assembly, simple greedy algorithms for scaffolding can fail because of repeats, and more sophisticated approaches make use of the graph of connections between contigs in one way or another. Beyond this general point, the situation is different from read assembly. The main problem is a result of the error-prone nature of NGS mate pair libraries: distinguishing the genuine relationship between contigs implied by good mate pairs from spurious connections caused by errors.
Third-generation sequencing promises to cheaply generate data with higher read lengths; however, with a larger volume of data needing to be dealt with quickly, the possibility of cheaply generating more coverage to suppress errors, and recent algorithmic innovations, this will not simply mean a return to earlier methods. With significantly longer reads, string-graph methods would become more attractive compared with de Bruijn-graph methods.
Next-generation assemblers
When the implications of NGS technology became apparent, several assemblers were designed to deal with the new problems. The Euler assembler [30] was the first to employ de Bruijn graphs for WGS assembly, and proved capable of assembling bacterial genomes. Velvet [32] and ALLPATHS [35] improved assembly in terms of speed, contig and scaffold length and avoidance of misassembly. Both implement graph topology-based error correction and, instead of storing the paths of reads, these assemblers employ short read-pair data to resolve short repeats, finding long contigs that are joined by several reads pairs and then extending them along available paths towards each other when this can be done uniquely, using different algorithms. This allowed assemblies of bacterial-sized genomes and BACs from short-read data.
ABySS followed the innovations with de Bruijn methods, but also introduced a distributed representation of the graph, allowing message passing interface parallelization [36]. Greater exploitation of computational resources enabled ABySS to assemble a human genome from short read data for the first time. SOAPdenovo is another assembler using a similar overall strategy that is also able to assemble large genomes [37].
The CABOG [38] and variant MSR-CA pipelines are updates of the Celera overlap-based assembler designed for a combination of read types, which showed some success with short-read data for genomes in the 100 Mb range. CABOG has now been used to assemble the Tasmanian devil genome using a combination of Illumina and 454 reads [39]. The CABOG pipeline will also attempt to construct multiple alelles for regions of sequence variation after contig assembly [40]. MSR-CA uses a de Bruijn graph to combine reads that map on to the same nodes and edges into ‘super-reads’, reducing the number of reads to be dealt with by Celera by a factor of 50 or more.
The String Graph Assembler (SGA) is the first to make assembly of mammalian-sized genomes practical using the string graph approach [31]. The problem of computing the whole set of overlaps is solved by making use of the Ferragina–Manzini index data structure, which allows overlaps to be quickly calculated while greatly reducing storage requirements for the reads [41]. In principle SGA can assemble a human genome using only one machine, although in practice using a cluster will reduce time requirements. This assembler also implements particularly successful routines to correct single-base errors, mainly by finding bases in reads that are not covered by frequently occuring k-mers.
Following on from the Phusion long-read assembly pipeline [42], the Phusion2 assembler uses a strategy of read clustering and ‘local assembly’ followed by a merging step [43]. In clustering, reads are divided into sets that are expected to be close to each other in the assembly. Using a table of k-mers found in the reads, a relation matrix is built up that records, for each pair of reads, the number of shared k-mers. If reads are considered as nodes, and pairs with more than some minimum threshold of shared k-mers are considered connected by an edge, clusters are connected components in this graph. After obtaining small-read clusters with a controllable size (~100,000 reads), SOAPdenovo and ABySS are run separately on each cluster to obtain a combined assembly. Reads are aligned back to the draft assembly and the Gap5 tool is used to generate the final consensus sequences [44].
Improvements and additions to these tools continue to raise the quality of results for short-read assemblers. For example, ALLPATHS-LG uses shared-memory parallelization and can assemble mammalian genomes [45], and a recent update enables ‘patching’ contigs with long reads (similar to scaffolding with mate pairs) from 454 or PacBio sequencing. Similarly, Velvet1.1 includes mutlithreaded assembly and new algorithms for scaffolding using mate pairs and long reads [46]. Some data on the performance and requirements of each large genome assembler are collated in Table 2.
Table 2
| Assembler | Resources employed for large genome | Test genome | Assembly time | Max. memory usage | Parallel- ised | Data type used and information of PS, RL and MP | Contig N50 (Kb) | Scaffold N50(Kb) | Assembled size and coverage | General data requirements | Summary comments | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ABySS | 168-core cluster, 2.66-GHz CPU | Human | 87 h | ~250 Gb (<16 Gb/ node) | Yes | llumina PE PS 210 bp; RL 2×35-46 bp of 45× | 2.4 | N/A | 2.2 Gb 80.6% | Any short-read libs | Low RAM requirement; good local contig accuracy, but sometimes produces duplicated contigs | [113] |
| ALLPATHS- LG | 48 processors with 512-Gb RAM | Human | 25 days | <512 Gb | Yes | llumina PE PS 180 bp; RL 2×100 bp of 45×; MP 3, 6 and 40 kp of 51× | 24 | 11,543 | 2.55 Gb 91.1% | Requires one overlapping PE and one MP, can now add long-read libs | Very good on contig continuity and accuracy; restrictive read-library requirements | [114] |
| CABOG | Computer grid and 16 processors with 256-Gb RAM | T. Devil | Not given | <256 Gb | Yes | llumina PE PS 300 bp; RL 2×75 bp of 49× Roche 454 single and MP 6–15 Kb of 8× | 11 | 146.8 | 2.93 Gb >95% | Accepts both long (Sanger, 454) and short (lllumina, SOLiD) reads | Versatile: accepts multiple data types required computing resources are relatively big | [115] |
| Phusion2 | 32 processors with 512 Gb RAM | T. Devil | 70 h | <512 Gb | Yes | llumina PE PS 450 bp; RL 2×100 bp of 85×; MP 3–10 Kb of 10× | 28.9 | 2244.5 | 2.93 Gb >95% | Any short-read libs | Flexible, good contig continuity but read clustering is sensitive to the evenness of read coverage | [116] |
| SGA | ~100 cores | Human | 1417 CPU hours or 6 days | 53 Gb | Optional | lllumina PE PS 380 bp; RL 2×100 bp of 48× | 9.9 | 25.1 | 2.69 Gb 95.4% | Any short-read libs | Low RAM requirement and excellent contig accuracy Contig continuity needs improvement | [117] |
| SOAPdenovo | 32 processors with 512-Gb RAM | Panda | 40 h | <256 Gb | Yes | llumina SE, PE 150 bp, 500 bp; RL 2×45–67 bp of 50×; MP 2.5, 5 and 10 Kb of 23× | 40 | 1220 | 2.3 Gb >95% | Any short-read libs | User-friendly, efficient assembler, good long-range continuity Contig accuracy needs improvements | [118] |
Requirements for, and performance of, large genome assemblers. Figures are given for assemblies reported in publications on each assembler cited in the text (figures and notes on ABySS also use information from private communication with Jared Simpson). Caution is necessary when comparing the figures on assembly quality: it is important to note that the input data in each case is far from equivalent, and details of the calculations of statistics may also vary. Each assembler can be expected to improve over time and so relative performance of older assemblers may be understated (e.g., the ABySS assembly does not show its ability to scaffold). SGA was run with only trivial parallelization and could be run serially. No maximum memory requirement was reported for ALLPATHS-LG and so the total memory of the cluster used is given here [31,36-39,45]. Contig: Contiguous sequence; CPU: Computer processing unit; libs: Libraries; MP: Mate pair; N/A: Not applicable; PE: Paired end; PS: Pair size; RAM: Random access memory; RL: Read length; SGA: String Graph Assembler; T. Devil: Tasmanian Devil.
Of these assemblers, almost all will accept any read-pair libraries, although results will be much improved by supplying a range of different insert sizes from overlapping pairs to inserts of several tens of Kb. One study indicates that, at least for particular bacterial-size genomes, ABySS and SGA gain most from the use of multiple short-insert libraries rather than the inclusion of 3 Kb-insert size data (although this could be due to the quality of the 3 Kb library used), whereas most other assemblers benefit more from the latter [47]. ALLPATHS-LG differs, in that it will not run without at least one overlapping read pair library and at least one longer insert library. Because of the current prevalence of Illumina short-read data, most assemblers are optimised for this data type. Use of other short-read platforms is not ruled out by this, but most short-read assemblers exclude the use of 454 reads because there is no support for their larger rate of indel errors. Euler-SR will accept 454 data and CABOG was designed to accept Applied Biosystems, 454 and Sanger reads. As noted above, ALLPATHS-LG will now also accept PacBio reads [102], while SGA is still at an experimental stage and is likely to be developed to handle third-generation long reads [105].
Major NGS-oriented assemblers generally include their own routines for error correction, scaffolding and gap closure that are designed and tuned to work well with the other parts of the pipeline. There also exist a number of standalone software packages for these tasks. Error correction tools include Quake [48,106] and HiTEC [49,107]. For scaffolding with NGS data, there are SSPACE [50], SOPRA [51], Bambus [52] and the recently released MIP scaffolder [53]. There is little in the literature at present to suggest that any scaffolder greatly excels over all others in terms of scaffold length or accuracy.
Assessing performance
It is important to ask to what extent NGS technology trades off costs with assembly quality. While many projects have set out to sequence large numbers of species and individuals for studies of evolution and disease, some researchers have suggested that inherent limitations in using short reads preclude assemblies of the quality necessary for these ends, and have recommended a combination of NGS with other techniques. Alternatively, it may be that improvements in read pair data and its use in assembly turn out to be more useful than combining NGS data with expensive long reads.
It is also interesting to establish how results vary among the various available tools. Judgements here depend on the uses to which the assembly will be put. When structural variation detection is the aim, one would prefer an assembler with high local accuracy, particularly in coding regions of the genome, whereas for de novo projects seeking a draft assembly of a new species this is less of a consideration than finding long contigs and scaffolds. Some important questions are: which NGS assembly tools perform best on different parameters; what read pair libraries to prepare and what settings to use for the best assembly; what results can be expected in terms of contig and scaffold size, errors and coverage of the genome.
To assess these, various metrics are used. Of course, errors can only be assessed to the extent that there exists a reference, meaning that the target genome, or some part of it is known. Without this only length of contigs and scaffolds can be assessed. The N50, defined previously, provides a summary statistic for contig and scaffold lengths; if the genome length has been estimated, this can be substituted for the total length of all contigs in the definition to calculate the ‘NG50’ (and similarly for scaffolds). Contigs or scaffolds can be broken at locations where the match to a reference changes or fails, and the N50 of the resulting blocks gives a more telling estimation of assembly quality. The correct contiguity (CC)50 gives a measure of the long-range continuity of the assembly that is tolerant of small errors. Leaving aside some details, the CC50 is the median separation between pairs of bases that can be considered aligned to approximately the correct relative locations in the reference [54].
A number of recent studies have set out to address these questions. Alkan et al. compared NGS assemblies of two human genomes to the human reference genome and assemblies using older technologies [55]. As might be expected with short reads, the study found major problems with repeats. It is estimated that 99.4% of all true pairwise segmental duplications are absent, resulting in the loss of 16% of the genome (compared with around 8% when using Sanger-type sequencing) and significantly affecting the coding regions of the genome. Segmental duplications are also relevant to studies of disease and evolution, creating problems for the end use of assemblies. The authors conclude that using purely NGS data to sequence large genomes may not be viable. Other studies also emphasize the creation of false segmental duplications in assemblies, which sometimes occur when heterozygous sequences from two haplotypes are assembled into separate contigs and are scaffolded adjacent to each other rather than being merged [56]. However, new results from ALLPATHS-LG show that 40% of true segmental duplications are covered by their short-read assemblies of the mouse and human genomes [45]. This approaches what is possible with Sanger reads, and other assembly performance statistics are even closer to ideal levels. There are also indications from Velvet’s new scaffolding tools that, in some cases at least, good use of mate-pair reads may be more useful than adding a small number of long reads to NGS data [46].
Other studies have compared different assembly techniques. Some previous studies, while focusing on assemblers capable of assembling only short sequences, do provide some interesting results. For example, measuring assemblies using simulated reads from a number of sequences including two human chromosomes, they indicate that SOAPdenovo achieved a better N50 than ABySS whereas ABySS excelled on accuracy. The study also points out that assembly quality is sensitive to the number of base-call errors only when the coverage is low (i.e., before the coverage is so high that increasing it further does not significantly increase assembly quality) [57,58].
The Assemblathon takes the form of a competition in which organizers and outside groups attempt to assemble a given set of reads [54]. To allow a better comparison to the ‘reference’, a simulated genome and read set were used, produced by subjecting a sequence of human DNA to simulated evolution. Significantly, the resulting genome contained only around half the number of 100-base repeats as the original human DNA. Because of this, the competition does little to answer the questions raised above on duplications. The total length was also chosen to be fairly small, at 112.5 Mb. Contigs and scaffolds were then aligned back to the reference and various metrics were used to find assembly quality. For length, ‘paths’ from contigs and scaffolds representing a correct assembly (including combinations of sequence from the two haplotypes, which were exactly known here) were considered, and the ‘contig path N50’, representing the N50 after breaking at points of misassembly, was used as a summary statistic (and similarly for scaffolds).
The three most successful assemblies overall were deemed to be those produced by the Broad Institute (MA, USA; using ALPATHS-LG), Beijing Genomics Institute (BGI; using SOAPdenovo), and Wellcome Trust Sanger Institute (using SGA). None of these dominated on all measures. While BGI’s SOAPdenovo produced the largest contig path N50 of 8.25 × 104, closely followed by the Broad Institute’s ALLPATHS-LG assembly, the scaffold path N50 of the Welcome Turust Sanger Institute (WTSI) SGA assembly was more than double that achieved in the other two at 4.95 × 105, and similarly the Broad Institute’s CC50 was more than double that of the nearest competitor of these three at 2.66 × 106.
While several other assemblies were comparable with the top three on some metrics, these achieved the best size and lack of errors overall, with some tradeoff apparent between major structural errors (such as joins between sequences mapping to distant locations on the genome) and contig size. It is interesting to note that the proportion of base substitution errors to genome size varied hugely. The WTSI SGA assembly achieved a result of 1.3 × 10−7, while the Broad Institute and BGI assemblies contained many more such errors, by a factor of approximately 22 and 92, respectively. The SGA assembly contained only one structural error while others varied from 3 to 20.
Overall results were good, and there was no one assembly that was clearly far ahead of the rest, apart from on substitution errors. However, conclusions based on the Assemblathon must be limited as they may not apply to larger and more realistic datasets (which will in any case vary amongst themselves), especially those with a more challenging repeat structure.
The recent Genome Assembly Gold-standard Evaluations (GAGE) project differed in that it used real data and only assemblies constructed by the organisers using openly available protocols [47]. Real data could give results that are more comparable to typical assembly, but on the other hand parameter choices for assemblers can make a large difference to performance; they may well be less well optimized in the GAGE methodology than in the Assemblathon, where each assembler is run by teams that are highly familiar with it (e.g., in the GAGE project, Velvet was run with a k-mer length of 31, which would normally be much lower than optimal with reads of length 100).
Two bacteria with good finished reference genomes of size 2–5 Mb, as well as a human chromosome 14 and a bee genome of size 250 Mb (which had no available reference) were used. ALLPATHS-LG, SOAPdenovo, ABySS, Velvet and SGA were tested alongside overlap assemblers like CABOG. For both bacteria, ALLPATHS-LG again was more successful than its nearest competitors in terms of N50 of contigs and scaffolds broken at misassembly points, its nearest competitors here being Bambus2 and MSR-CA, while the other assemblers designed for large genomes lagged behind. For the human data, CABOG was marginally more successful on (error-broken) contigs than ALLPATHS-LG, but again ALLPATHS-LG was superior on scaffold length, and the best of the large-genome assemblers overall. Results on the bee genome were best for SOAPdenovo; however, in this case the overlapping read pairs required by ALLPATHS-LG were absent, and errors were not accounted for. It may be the case that the stricter and less-detailed contig-breaking used in the GAGE project is a disadvantage for less locally accurate assemblers. While SOAPdenovo managed a much larger N50 than others, after contig breaking the advantage disappeared; however, results from the Assemblathon showed that this assembler performed well on the small error-tolerant CC50 metric. The weakness here seems to be that SOAPdenovo produces some short ‘indels’ (erroneously inserted or deleted sequences) during gap closure. This tool may still be a better choice for draft de novo assembly.
While these results are useful, most of the recent technical advances have been in handling large eukaryotic genomes. Assembly pipelines should be expected to perform very differently when running on the type of data for which they were primarily designed, while some other assemblers will not be able to run at all on large genomes. Because of this, more wide-ranging and comprehensive comparative studies focused on large genomes will have to be carried out to reach more solid conclusions about the pros and cons of each assembler when they are applied to such datasets. Fortunately, further comparison studies are planned that should substantially improve matters. The dnGasp project is another collaborative effort based on a large, simulated genome [108], while the Assemblathon 2 project will use real, large genomes, from species of snake, bird and fish [109]. Both studies are now closed to new entries, and results will soon be available to compare to those above, providing significant tests of current tools against large and repeat-rich genomes.
Conclusion
WGS genome assembly remains an active area of innovation, which has been greatly affected by the introduction of NGS sequencing, even if its fundamental problems remain largely the same. Assemblies built from NGS reads alone are far from perfect, exhibiting, in particular, many errors involving counts of segmental duplications. Early on, it was suggested that such short-read technology may not be viable for de novo assembly of large genomes without some help from more expensive sequencing methods. However, with the rapid development of assembly techniques, the quality of NGS assemblies is beginning to approach that which is possible by other means. Some assemblers can achieve much better results on local errors than others (and without apparent costs elsewhere for some types of errors), showing that improvements are possible. New assembly analysis studies are set to show how much of a gap still exists between the quality of NGS assemblies and finished sequence.
We have also seen a number of apparent trade-offs. When choosing how to create reads, longer read length often implies more errors, especially when using the new PacBio technology. Most interestingly when designing a study, assemblers that excel on long-range continuity in contigs perform badly on suppressing local errors such as indels (such as SOAPdenovo) or vice versa (such as SGA). The choice made here in different genomic studies will vary depending on the intended use of the assembly.
Future perspective
This observation on the current tradeoff between accuracy and continuity suggests avenues for future improvements in assembly. The best results might be achieved by using accurate assemblies to correct errors in long-scaffold assemblies, or by developing special tools built on similar principles to correct errors after the scaffolding and gap closure stages. There is room for other improvements at the scaffolding stage, where, as has happened at the assembly stage, we are seeing a move from naive, greedy algorithms to more subtle graph-based techniques.
Another developing area is the explicit construction of haplotypes from reads, to the extent that this is possible. At present, producing one representative haplotype is normally taken as the aim of assembly, and alternative alleles are merged when identified. As we have seen, differences between alleles are normally treated in the same way as errors, and they are often scaffolded sequentially when not identified, causing misassemblies. Routines to explicitly identify alternatives during contig construction would help to reduce such errors as well as providing extra genetic information to end users. Pipelines such as CABOG do attempt to identify alleles after the main assembly steps [40]; however, exploiting variance information optimally for error avoidance during assembly is still an open problem.
In the future, as well as improvements in assemblers themselves, there are likely to be improvements in sequencing. Of particular importance are improvements in the production of mate-pair libraries, in terms of accuracy of insert-size estimation and suppression of errors. All this suggests that the way forward may lie with exploiting NGS technologies with improved mate-pair libraries to guide long-range assembly accuracy.
Second- and third-generation technology may soon dramatically improve. This would change assembly methods and greatly improve results. With the release of Illumina’s HiSeq 2500, a platform that aims for a ‘genome in a day’, users can expect 2×150bp reads in late 2012 [110]. Life Technologies’ Ion Torrent plans to launch 2 × 200 bp paired-end reads and 400-base single ends. With two Ion Proton™ chips on the way, the Proton I chip will be targeting exome sequencing, while the Proton II chip, to be released in early 2013, is intended for whole-genome applications. For the latter, it claims the ability to sequence a human genome at about 20× to 30× coverage for US$1000 in total, including sample preparation, chip and reagent costs [111]. The biggest potential player, Oxford Nanopore® (Oxford, UK), could enter the market with two low-cost DNA strand sequencing instruments: a higher-throughput version, named GridIon and a disposable MinIon system. The latter instrument is in the size of a USB memory stick and costs less than US$900. The GridIon system, scalable like a computer cluster, takes disposable reagent cartridges that contain the nanopores. Running 20 GridIon instruments each with 8000 pores in parallel will enable users to sequence a human genome at 15-fold coverage in 15 min for less than US$10 per Gb [112]. Both released systems are expected to produce read length of up to 100 Kb with a reasonable level of base accuracy. A 48-Kb phage λ genome has been sequenced as a single contiguous read and the error rate was reported to be approximately 4.0%. These advances will undoubtedly change the landscape of genomics and its applications, pharmacogenomics included. The genomics community could find itself in a situation in which data is produced in matter of hours, but it takes days or even weeks to assemble a human genome using the fastest assembler. For de novo sequencing on new species or assembly-assisted variation detection, the efficiency of assembly process may be the bottleneck, and new algorithms are ultimately needed to speed up the process as well as cope with the new data.
Finally, the suppression of local errors in assemblies, well as improving the assembly for end-use purposes, has other effects. Segmental duplications will differ in a few of their bases, and we see, unsurprisingly, that more copy-number errors are reported for duplications with a lower proportion of differing bases. With fewer local assembly errors it may be possible to increase sensitivity to differences here, going some way to solving a major outstanding problem with current (and past) assembly techniques.
Acknowledgements
The authors would like to thank J Simpson for information on the SGA and ABySS assemblers.
This work is supported by the Wellcome Trust.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
■ of interest
■■ of considerable interest
Websites
Full text links
Read article at publisher's site: https://doi.org/10.2217/pgs.12.72
Subscription required at www.futuremedicine.com
http://www.futuremedicine.com/doi/pdf/10.2217/pgs.12.72
Free to read at www.futuremedicine.com
http://www.futuremedicine.com/doi/abs/10.2217/pgs.12.72
Citations & impact
Impact metrics
Article citations
Complementary approaches to dissect late leaf rust resistance in an interspecific raspberry population.
G3 (Bethesda), 14(10):jkae202, 01 Oct 2024
Cited by: 0 articles | PMID: 39172650 | PMCID: PMC11457092
Ameliorating the effects of multiple stresses on agronomic traits in crops: modern biotechnological and omics approaches.
Mol Biol Rep, 51(1):41, 29 Dec 2023
Cited by: 1 article | PMID: 38158512
Review
Genome assembly of the pioneer species Plantago major L. (Plantaginaceae) provides insight into its global distribution and adaptation to metal-contaminated soil.
DNA Res, 30(4):dsad013, 01 Aug 2023
Cited by: 2 articles | PMID: 37228100 | PMCID: PMC10254747
Next-Generation Sequencing Technology: Current Trends and Advancements.
Biology (Basel), 12(7):997, 13 Jul 2023
Cited by: 109 articles | PMID: 37508427 | PMCID: PMC10376292
Review Free full text in Europe PMC
Identification and Characterization of a QTL for Growth of Fusarium circinatum on Pine-Based Medium.
J Fungi (Basel), 8(11):1214, 16 Nov 2022
Cited by: 0 articles | PMID: 36422035 | PMCID: PMC9696871
Go to all (53) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The present and future of de novo whole-genome assembly.
Brief Bioinform, 19(1):23-40, 01 Jan 2018
Cited by: 102 articles | PMID: 27742661
Review
Comparing memory-efficient genome assemblers on stand-alone and cloud infrastructures.
PLoS One, 8(9):e75505, 27 Sep 2013
Cited by: 10 articles | PMID: 24086547 | PMCID: PMC3785575
Evaluation of nine popular de novo assemblers in microbial genome assembly.
J Microbiol Methods, 143:32-37, 19 Sep 2017
Cited by: 4 articles | PMID: 28939423
De novo assembly of Dekkera bruxellensis: a multi technology approach using short and long-read sequencing and optical mapping.
Gigascience, 4:56, 26 Nov 2015
Cited by: 13 articles | PMID: 26617983 | PMCID: PMC4661999
Funding
Funders who supported this work.
Wellcome Trust (1)
Wellcome Trust Sanger Institute - generic account for deposition of all core- funded research papers
Prof Sir Michael Stratton, Wellcome Trust Sanger Institute
Grant ID: 098051




